Abstract
Translational research requires large patient populations. A single research institute is not able to build up such a population in a short period of time. The String of Pearls Initiative (in Dutch “Parelsnoer Initiatief”, PSI) is a joint effort by the eight academic medical centers in the Netherlands to built an infrastructure for joint biobanking as to meet this challenge of establishing large collections of data and samples in relevant medical domains.
Introduction
With the emergence of the notions of translational research and personalized health care there is an increasing need for large study populations. Recent studies on genomewide association studies used large amounts of cases which cannot be collected in one single centre. For example, the study to identify risk alleles for multiple sclerosis included nearly 8000 cases1. The collaboration among the eight academic medical centers (UMCs) in the Netherlands to build a biobank for Inflammatory Bowel Disease (IBD) has served as a model for an initiative to develop a national infrastructure to support sharing of data and samples in specific medical domains. This contribution describes the objectives of this initiative, known as PSI (shorthand for ParelSnoer Initiatief = String of Pearls Initiative§). We will outline the general principles that will govern the development of various biobanks that are part of this four years project.
Objectives of PSI
The main objective of PSI is to build an infrastructure that will facilitate the collection of clinical data and biological samples for multi-center studies among the eight UMCs in the Netherlands. This infrastructure allows for sharing data that are collected both in the clinical process and specifically for the biobank at hand as well as information about samples that are available from the included cases.
As to demonstrate the feasibility of the approach, eight medical domains have been identified for which prospective biobanks will be developed. These eight domains are IBD, diabetes, cerebral vascular accidents, leukemia, neuro-degenerative diseases, arthritis/arthrosis, hereditary/familial colorectal cancer and kidney failure.
By doing so, we are building eight biobanks that cover a large part of the Dutch patient population with the selected problems. This will allow us on the one hand to quicker address research questions that arise in those domains and on the other hand get more powerful results due to the larger collection of cases available for analysis.
We use a broad definition of biobank. In PSI, a bio-bank is a collection of a) clinical information – observations, lab tests, image data etc – of patients with a specific condition and b) a description of the samples obtained from processing body material – tissue, blood – from those patients and c) the samples.
Development issues in PSI
The PSI project has 4 domains for development.
a). The clinical domains
Each clinical domain is defining the content of their biobank. The clinical representatives of the UMCs are also responsible for the implementation of the mechanisms to collect the agreed upon data and material for the biobank. The exploitation of the biobanks is not part of the PSI project.
b). Central Infrastructure
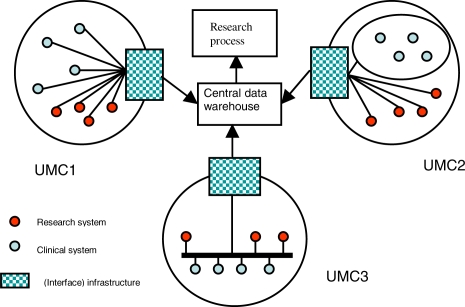
The general architecture is based on the notion that each UMC will publish its consolidated data in a shared datawarehouse (see figure 1). This dataware-house will have an export function as to make the data available for research purposes. For the time being the central infrastructure will only deal with the data and sample domains. The central infrastructure provides access to clinical data and to descriptions of the samples stored in the various UMCs. For a specific research project, these samples first need to be further analyzed before genomic data will be available for bioinformatics and statistical analysis. Hence we will not provide – at least for the time being –bioinformatics tools through the central infrastructure. We will explore whether data that is generated by genomics analysis of the samples should be made available in the central database for future use. Alternatively, a reference to another data store where these data are available could be included.
Figure 1:
Global architecture of the IT infrastructure
c). Local infrastructure and implementation
A main effort in PSI is the implementation of the workflows and IT infrastructure at the UMC level to facilitate data and sample collection. As is evident from figure 1, each UMC can and will have its own infrastructure that has to deliver the data to the central infrastructure. In our concept the interface messages to deliver the data to the central infrastructure will be defined. It is up to each UMC to decide how these messages will be implemented. Developing a local infrastructure for systematic collection of high quality data and samples in the specified domains goes beyond the implementation of a data capture tool. It has to do with structuring clinical processes and in some circumstances with a redesign of the patient flow as to accommodate the additional activities necessary for obtaining samples and data to be included in the biobank. It has been argued that secondary use of data from an electronic patient record has a great potential for clinical research2. However, clinicians with research experience have expressed their concern that a gap exists between the quality requirements for data for clinical care and those for data used for research purposes. As to bridge this quality gap a careful implementation of data acquisition processes for the PSI biobanks is necessary. The best approach also depends on the type of cases to be included and the local working environment.
d). Frame of Reference
A special working group has developed a frame of reference that defines largely the rules for the development and implementation of the biobank infrastructure as well as for the collaboration in the clinical domains. It covers the business architecture (how to do biobanking), the information architecture (the choice of standards and rules for how to develop and maintain the information models for the biobanks) as well as the technical architecture of the infrastructure. The purpose of the frame of reference is to support a coherent development of the infrastructure as well as of the biobanks for the eight clincal domains. It defines general principles that hold for each UMC and/or each clinical domain. It also provides the basic principles on which the central infrastructure is going to be developed.
In the business domain the following aspects have been elaborated: Legal aspects, Ethical aspects, quality aspects and information security. Since it is foreseen that the biobanks could be used by commercial partners, there has to be a legal framework that defines clearly the privacy aspects of the data and samples provided by the patients, the rights the patients have with respect to the use of their data and samples. On the other hand the participating UMCs have an interest in protecting and valorization of their intellectual property. Within the legal domain clear guidelines are provided how to specify the rules for ownership of the biobank. It also identifies issues to be included in the procedures on how to release data and samples from a biobank to interested parties. Ethical aspects dealt with include the nature of the informed consent and the approval by Institutional Review Boards for secondary use of clinical data, collection of additional data and obtaining body materials for biobanking purposes without yet a clear research question and study design. Guidelines exist in The Netherlands for the use of clinical data en body materials, left over from diagnostic procedures and surgery3,4. These guidelines only partly apply to the PSI activities. In most selected clinical domains additional body materials (in particular blood) will be collected. A few UMCs do already have broad informed consent forms to collect body materials for future research. Some haven’t specified the scope of the research, others have restricted the scope to the clinical condition (more or less broadly defined) of the patient that has provided the material. Given these differences it has not been possible to develop one general informed consent for PSI. There is a reference informed consent that can be adapted locally to the particular situation in the UMC.
It is recognized that data and samples have to be of the highest quality to be competitive with others that offer access to patient data and samples. Guidelines for quality management within the eight selected domains have been defined. The main focus is on the collection and processing of the material to be included in the biobanks. Approaches for quality control of the collected data have been proposed as well. It is recognized that the highest possible quality is desirable, but the UMCs should be able to implement these guidelines in their organization. A balance has to be found between what is theoretically desirable and practically feasible.
It is imperative that the privacy of the donors of the material in the biobanks is protected. The highest level of protection would be the storage of anonymous data. However, such an approach would make the inclusion of follow-up data difficult. We have chosen a pseudonymisation approach. Some identifying data will be used to create a code number that is not directly translatable in identifying information. The code creation is standardized among the UMCs. It is expected that this year a law will pass that will require the use of our Citizen Service Number (BSN) in health care to support sharing clinical data among health care organizations. This number will form the basis for the pseudonymisation process. By using the BSN, patients that move from one part of the Netherlands to another part still can contribute their clinical (follow-up) data and material to the biobank. As to further protect the privacy of the patients the data export implemented in the central infrastructure will have a second pseudonymisation which is export dependent. This guarantees that data in two different extracts can not be linked on the basis of the identifiers. The central infrastructure should be able to trace back from the extract pseudo code to the central pseudo code as to be able to link the data in the extract with the samples in the biobanks at the various UMCs. The UMCs are responsible for being able to trace back from the central pseudo code to the BSN and hence to the patient and its data.
In the information architecture domain the main topic addressed is the PSI Information Model, which forms the basis for the specification of each of the eight biobanks (both for the clinical data as for the description of the samples, possibly including also the storage of results of -omics analysis on the samples in the biobank). We have chosen to base this PSI Information model on the base classes of the HL7v3 RIM (Act, ActRelationship, Entity, Role, Participation, RoleLink). Concept models will be developed for information that will be collected in the different clinical domains. Examples are Smoking status, Donor/patient, previous diagnoses, biobank sample. In contrast with the clinical environment, there is less need for process information. Hence the models can be simpler than those developed for the clinical applications. For these developments we also consider other standardization activities like the CDISC efforts to describe their models in HL7v3 format and the work in progress on the representation of –omics data in the HL7v3. Standardization in this respect may also make local implementation a more tractable task. A further concern is the use of coding systems for the systematic recording of clinical observations and laboratory results. SNOMED CT, LOINC and other relevant coding systems are being considered. Another issue that has to be addressed is the traceability of the data. Study sponsors may require that it can be proven that no data tampering has taken place. This means that each data item has to be traceable to its source. This also means that at the central infrastructure version management may be required. This issue needs further study and determines also to a certain extent the data quality that can be guaranteed upfront.
The technical architecture domain mainly deals with the central infrastructure. The latter can be largely considered as an integration architecture that should be able to integrate the data from the UMCs that participate in a clinical domain into one coherent data set. Rather than directly storing the collected data in the central data base, it is foreseen that the UMCs will upload data to the central infrastructure at regular time intervals. The pseudonymisation code allows linkage of data of the same donor over time as to allow follow-up data collection schedules (e.g. during yearly check-ups).
Timeline of the PSI project
The PSI project formally started at 1-1-2007. The first 11 months of the project have been used to develop the frame of reference and the detailed definitions of the clinical domains in terms of the type of patients to be included, the clinical and laboratory data and the body material that will be collected and how the material will be processed. Implementation of all eight clinical domains at the same time is risky hence one domain has been selected to be implemented early in the project in all eight medical centres (IBD) while the others will follow later. Each UMCs will define their own development and implementation plans for the years 2008–2010. This includes the order and timing of the implementation of the eight clinical domains. Figure 2 shows the various phases of PSI.
Figure 2.
Phases of the PSI project
The development of the central infrastructure will take place in 2008, including a complete test of the functionality for data integration.
Each of the UMCs will start to implement IBD domain early in 2008. The timeframe of the implementation of the other domains will depend on the local situation.
In the course of 2010 all biobanks should become operational in all UMCs. Furthermore, it is foreseen that in 2010 the data of at least one biobank will be used to answer specific clinical research questions.
The PSI from a local project manager perspective
Implementation of the processes and procedures to collect data and material for each of the eight domains in the local environment of an UMC is a big challenge. At this level the following aspects are to be dealt with: a) data and material collection and processing procedures, b) the IT infrastructure for capturing all relevant data and c) the physical realization of the (centralized) freezer capacity of the biobank. There is no general approach to these issues since what has to be done depends on the local situation. In the following we describe the early experiences with the local implementation at the Maastricht University Medical Center (MUMC+).
The clinicians from the various clinical domains do have a high level of ambition. PSI is not only seen as a means to advance clinical research, but also as a good reason to review how clinical research can best be integrated with clinical care. Due to the variety of diseases covered, various situations occur. There are large differences in how the clinical data can be collected. On the one hand, one has the leukemia patients who get their treatment in the hospital. Many of these patients already participate in clinical trials. Building up a biobank for this kind of patients should be rather straightforward. Still, there are challenges to redesign the data collection process for the clinical trials in such a way that it can also be used for the PSI biobank. At the other end of the spectrum there is the biobank on kidney failure that tries to capture patients in the early phase of the disease before they develop end-stage kidney failure requiring dialysis or kidney transplantation. To address the problem of early detection of kidney failure a special clinic will be developed. This clinic would be an excellent entry point for patients to be included in the study. Since such a clinic will be largely run by nurse practitioners who are supervised by nephrologists, proper record keeping is a prerequisite. Here it will be possible to integrate data collection for the biobank with clinical work processes. There are, however, logistic issues tp be dealt with since the special clinic will be located at a distance from the biobank laboratory.
There are other diseases where patients – after they have been stabilized – are handed over from specialized care to primary care. For such situations a provision has to be developed that will approach patients that participate in a biobank for a follow-up visit. Such activities take place outside the regular care provision. Hence a kind of outpatient facility for research has to be developed that will take care of these situations.
Given these different contexts in which the data and material has to be collected, the implementation for each clinical domain is approached as a project on its own. These projects will be coordinated through the local PSI project management as to identify common issues that may require a common solution.
The IT infrastructure at each of the UMCs is quite different. This means that also at the local level, different solutions will be developed to address the challenges of PSI. The academic hospital of MUMC+ is replacing their IT infrastructure, including ERP and EPR functionalities. This offers us the opportunity to develop a research IT infrastructure that is integrated with the clinical IT. Research oriented electronic data capture forms will be an integral part of the EPR system. These forms should be accessible for both clinicians and research nurses. Other aspects of support of clinical research like registration of informed consent, order management of samples and tests for research purposes, will be integrated in the Hospital Information System as well. The IT infrastructure to support PSI is being developed in such a way that also local clinical research projects can be facilitated as well.
MUMC+ has already a centralized biobank facility. Material can be processed according to agreed upon Standard Operating Procedures. and stored in freezers at different temperatures. A Biobank Information System (BIS) keeps track of all samples in the biobank. It documents the processing of the samples at the biobank and provides information on the study, donor, type of material, processing steps and storage location of the subsamples. Integration of the BIS with the new IT infrastructure is one of the local objectives in the PSI project.
Discussion
PSI is based on experiences in The Netherlands with earlier registrations of data of HIV/AIDS patients. These registrations have been the basis for several research projects in this domain. The availability of a comprehensive set of data on these patients has attracted funding for such research projects. The business model for PSI is that the availability of bio-banks is attractive for parties with particular research questions. Since the data collection has already taken place to a large extent. The biobank can be used as a pool from which the most appropriate cases can be retrieved. In addition, the participation of the eight UMCs in PSI will create a much larger biobank than any of the UMCs could have created on its own.
There are other initiatives to facilitate (translational) research. In The Netherlands, PALGA provides since 1971 a central archive of all reports created by the pathologists in the 70 pathology departments in Dutch hospitals5. The PALGA data base is being used both for clinical care and for research purposes. It allows researchers to find cases, based on the diagnoses in the pathology reports and to retrieve the samples from the participating centers for additional analysis. PALGA is limited in the sense that it covers only cases from which tissue has been obtained during the care process. Hence it covers not all diseases. Also clinical observations are not included and have to be retrieved later on.
Similar to PALGA there is the Shared Pathology Informatics Network (SPIN) in the USA6. Rather than having a centralized data base of the pathology reports, SPIN provides access to data at the participating sites.
There are other initiatives that aim at linking various data and information sources for translational research. Examples are caGRID (building the infrastructure) and caBIG aiming at the development of the bioinformatics tools that will bring together data, tools, organizations and scientists in a federated environment7. The networks focus on cancer research. The scope of PSI is much boader as it will support also non-cancerous diseases.
Conclusion
PSI is a challenging project that will enable the UMCs in The Netherlands to build biobanks that extend beyond what is possible in any of the UMCs alone or by domain specific collaborations. Due to the collaborative developments that go beyond a single domain, it is possible to define general principles that should be applicable for any new clinical domain that would like to use the infrastructure. Reuse of information models, standard operating procedures and the organizational infrastructure will make it easier to set-up future collaborations. The focus on standardization of procedures, data management and data validation in the UMCs combined with central quality assurance will make these biobanks attractive for others.
Acknowledgments
The project is funded by the Dutch Ministry for Education, Culture and Science: 35 M (46 M$) and matched by the UMCs by a total of 32 M.
We would like to thank the numerous partners in PSI that are contributing to shaping the PSI project and the development of the frame of reference.
Footnotes
In our metaphor, the infrastructure is the string that will link the eight UMCs to enable the creation of the biobanks (the pearls).
References
- 1.The International Multiple Sclerosis Genetics Consortium Risk alleles for multiple sclerosis identified by a genomewide study. NEJM. 2007;357:851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- 2.Powell John, Buchan Iain. Electronic health records should support clinical research. J Med Internet Res. 2005;7(1):e4. doi: 10.2196/jmir.7.1.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grobbee DE, et al. Code for Proper Secondary Use of Human Tissue in the NetherlandsFederation of Medical Scientific Societies, 2002, Available from http://www.federa.org/ (last visited 30 jan 2008)
- 4.Anonymous: Code of conduct: “Use of Data in Health Research”. Federation of Medical Scientific Societies, 2002, Available from http://www.federa.org/ (last visited 30 jan 2008)
- 5.www.palga.nl (English version present, last visited 30 jan 2008).
- 6.http://www.cancerdiagnosis.nci.nih.gov/spin/ (last visited 30 jan 2008)
- 7.https://cabig.nci.nih.gov/workspaces/Architecture/caGrid/ (last visited 30 jan 2008) [Google Scholar]