Abstract
With the growing prevalence of large-scale, team science endeavors in the biomedical and life science domains, the impetus to implement platforms capable of supporting asynchronous interaction among multidisciplinary groups of collaborators has increased commensurately. However, there is a paucity of literature describing systematic approaches to identifying the information needs of targeted end-users for such platforms, and the translation of such requirements into practicable software component design criteria. In previous studies, we have reported upon the efficacy of employing conceptual knowledge engineering (CKE) techniques to systematically address both of the preceding challenges in the context of complex biomedical applications. In this manuscript we evaluate the impact of CKE approaches relative to the design of a clinical and translational science collaboration portal, and report upon the preliminary qualitative users satisfaction as reported for the resulting system.
Introduction
Recently, several national-scale biomedical and life science initiatives have emerged that emphasize the creation of multidisciplinary teams to address systems-level scientific hypotheses. Such programs notably include the National Institutes of Health-funded Clinical and Translational Science Award (CTSA) and National Cancer Institute-funded Cancer Biomedical Informatics Grid (caBIG). This shift toward a team science model has created a significant demand for the biomedical informatics community to design and implement computational collaboration platforms capable of supporting such research efforts. However, despite this growing demand, there exists a paucity of literature describing systematic approaches to gathering end-user requirements from such team science communities, and subsequently designing web-based collaboration platforms capable of satisfying their information needs. This gap in knowledge is illustrated by a literature search of the Medline database conducted in January 2010, using the PubMed browser and a set of heuristically selected MeSH terms including “Biomedical Research”, “Communication”, and “Computing Methodologies”, yielding a corpus of 232 articles published between 2007 and present. Upon review by the authors, 13 (5.6%) of these articles completely or partially addressed the design or evaluation of web-based team science platforms. Despite this lack of formative research concerning contemporary methodological approaches to the design of team science platforms and their efficacy or usability, a limited body of literature describing studies concerned with the use of other collaboration platforms, such as interactive web portals, does exist. These works demonstrate the positive impact of such tools on the efficiency, timeliness and quality of biomedical and life science research programs (1–4). The remainder of this manuscript is motivated by the existing gap in knowledge between reproducible and rigorous approaches to the design of collaborative platforms, and promising reports concerning their positive impact on team-science activities.
Background
In the following section, we will review both the state of knowledge concerning the design and use of collaborative teams-science portals, and describe a set of conceptual knowledge engineering (CKE) approaches that are intended to yield highly usable biomedical applications.
Collaborative Team-Science Portals
As introduced earlier in this manuscript, there exists a small but important body of literature describing the benefits of using Internet or web-based software platforms to support the interaction and collaboration among and within multidisciplinary teams of investigators and research staff in the clinical and translational sciences. Taken as a whole, these reports repeatedly demonstrate that the use of such platforms enable the efficient and timely collaboration of team members who are physically, temporally and contextually distributed (2–4). In addition, it is shown that the use of such platforms can often result in higher quality data and research products by virtue of their ability to reduce redundancy and increase transparency surrounding research methods and protocols (3). A thematic analysis of this body of literature identifies several shared features of such collaboration platforms, including: 1) document sharing and management (e.g., version control); 2) contact management; 3) shared calendars; 4) discussion forums; and 5) end user-centric content publication and curation (e.g., wiki’s and equivalent technologies) (2, 4). Similarly, a common set of technology and knowledge management components, including both content management systems (e.g., Joomla, Drupal, Microsoft Office SharePoint Services) and corresponding instance-specific information taxonomies, underlie such functionalities (1). It is important to note in this context that the majority of the previously introduced features are predicated on the use of dynamic content management systems, which are reliant on information taxonomies created by system designers. These knowledge structures directly define the system’s functionality, and its model, view and controller levels.
Conceptual Knowledge Engineering
Conceptual knowledge engineering (CKE) techniques fall with the broader context of knowledge engineering (KE) methodologies, wherein knowledge is collected, represented, and subsequently used by computational agents to replicate expert human performance in an application domain. The KE process incorporates four major steps: 1) knowledge acquisition (KA), 2) computational representation of that knowledge, 3) implementation or refinement of the knowledge-based agent, and 4) verification and validation of the output of the knowledge-based agent. Conceptual knowledge, one of three primary types of knowledge that can be targeted by KE, can be defined as a combination of atomic units of information and the meaningful relationships among those units. This definition have been derived and validated based upon empirical research that focuses on learning and problem-solving in complex scientific and quantitative domains (5). Conceptual knowledge collections in the biomedical domain span a spectrum that includes ontologies, controlled terminologies, semantic networks and database schemas. The knowledge sources used during the KA stage of the CKE process in order to generate such collections can take many forms, including narrative text, databases and domain experts. We have recently described a taxonomy consisting of three categories of KA techniques that can be employed when targeting the conceptual knowledge found in such sources, including the elicitation of atomic units of information or knowledge, the relationships between those atomic units, and combined methodologies that aim to elicit both such atomic units and the relationships between them (5). Commonly used combined conceptual knowledge acquisition methodologies include ethnographic observations and interviews (6), as well as categorical sorting exercises (7). The application of conceptual knowledge collections to inform the design of software components can take numerous forms. In prior reports we have demonstrated the efficacy of using both computational taxonomies and visual sub-languages derived from such knowledge collections to inform the design and functionality of highly usable biomedical applications, such as radiology reporting and clinical trial management systems (8–10). Those same reports have also demonstrated that improved end-user performance and qualitative satisfaction are consistently associated with the use of CKE methods in comparison to heuristic or intuitive system design processes as are commonly used in prevailing software engineering methodologies (9, 10).
Research Questions
The primary objective for this study was evaluate the impact of CKE design methods on the design of a collaborative team-science portal intended for use by members of the CTSA-funded Center for Clinical and Translational Science (CCTS) at The Ohio State University (OSU). In doing so, we have posed the following research questions:
Can CKE methods be applied to inform the design of a collaborative team-science portal capable of meeting the needs of diverse end-user communities?
What is the subjective satisfaction of users of the resulting team-science portal, across various strata of the OSU CCTS community?
Methods
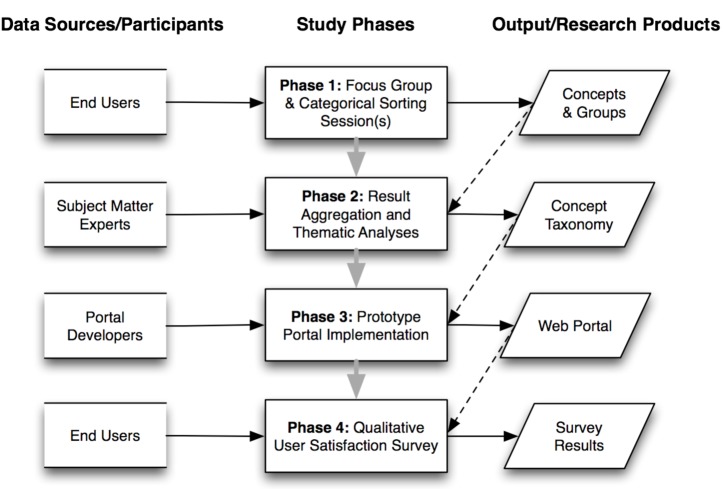
The Center for Clinical and Translational Science (CCTS, ccts.osu.edu) incorporates investigators, research staff, and administrators from all seventeen colleges at OSU, as well as Nationwide Children’s Hospital and the broader central Ohio biomedical research community. As such, it is imperative that the CCTS be able to deploy and maintain a team-science portal capable of supporting the efficient interaction of such geographically and contextually distributed participants. In order to address this need, we set out to design a collaborative team-science portal using an enterprise-wide Microsoft Office SharePoint Service (MOSS) portal platform. Like many other dynamic content management systems, MOSS uses information taxonomies to inform functionality at all levels from underlying information representation and storage, to high-level presentation layer components. As part of the system design process, and in order to address the preceding need for an integrative information needs taxonomy, we conducted a four-phase CKE study (Figure 1), which is described in the following sub-sections.
Figure 1:
Summary of four-phase CKE study used to design and evaluate the impact of an information taxonomy for the CCTS team-science portal.
Phase 1: During the first phase of our study, three focus groups were convened, consisting of the major strata of end-users in the CCTS community. Investigators, research staff, and administrators were invited to engage in these activities via a series of e-mail announcements sent to known CCTS participants. During the 60-minute focus group sessions, in the initial 30 minutes were dedicated to an interactive group brainstorming session during which the participants were prompted by a facilitator to articulate the types of information they would find most useful in the context of a CCTS web portal. For the remainder of the session, the participants conducted a group categorical sorting exercise. The concepts articulated during the first half of the session were transcribed onto note cards, which were then collaboratively sorted into semantically similar groups as defined by the participants (i.e., the group characteristics and size/number were not pre-defined by the investigators). Once the participants were satisfied with the composition of the groups, they provided a name for each group summarizing its common semantic meaning.
Phase 2: During the second phase of the study, two subject matter experts with over ten years of experience each in the clinical and translational sciences (PP, RR) performed an iterative, thematic analysis of the categorical groups created during the prior study phase. The nomenclatures used by the study participants to describe their sorting groups were normalized based upon common semantic meaning. These groups were then aggregated based upon those normalizations. Subsequently, the constituent information concepts in the groups were similarly normalized and duplicate concepts censored from ensuing analyses. Finally, the frequency of pair-wise associations between concepts and group names was noted and used to annotate the groups, in order to identify “high priority” concepts within the overall corpus. Based upon these results, an information needs taxonomy was constructed with inferred hierarchical relationships between group name super classes and subsumed information needs concepts.
Phase 3: During the third study phase, a collaborative web portal was implemented using the Microsoft Office SharePoint Service (MOSS) portal platform present at OSU, and deployed for use by all members of the CCTS. This portal incorporates the functionality types described earlier in the background section of this manuscript, and makes use of the information needs taxonomy designed during Phase 2.
- Phase 4: Sixty days after the collaborative web portal was made widely available to the CCTS community, end-users were invited via e-mail to participate in an anonymous survey concerning their impressions of it. The survey was constructed using the SurveyMonkey web application, and designed as a derivative of the standard qualitative user satisfaction (QUS) survey instrument (11), including the following questions:
- What is your primary role in the CCTS (Investigator, Research Staff, Administrator, Other)?
- Did you participate in a CCTS portal focus group (Yes, No)?
- How often do you use the CCTS web portal (Daily, Weekly, Monthly, Never, Other)?
- Rate the usability of the portal (10-point scale from Easy[0]-Difficult[9])
- Rate the usability of the portal (10-point scale from Frustrating[0]-Satisfying[9])
- Rate the organization of the portal (10-point scale from Confusing[0]-Clear[9])
- Rate the consistency of the portal (10-point scale from Inconsistent[0]-Consistent[9])
- Rate the ability of the portal to meet your information needs (10-point scale from Never[0]-Always[9])
- Rate the understandability of the portal (10-point scale from Easy[0]-Difficult[9])
The results of this survey were analyzed using both descriptive statistics and significance testing, stratifying responses by end-user type and participation in the initial CKE-based design sessions (as indicated by self-reporting of such participation via the survey instrument). The overall objective of this phase was to provide for validity checking of the web portal design generated using the earlier CKE methods.
Results
In the following section, we will summarize the results of our design and evaluation study:
Phase 1: Three focus group sessions were conducted, with each session consisting of a convenience sample of CCTS participants corresponding to the following strata: Investigators (n = 21); Research Staff (n = 9); and Administrators (n = 3). Each focus group completed the process described in the preceding methods sections, creating [6,6,8] groups of information concepts, respectively. When taken collectively, the concept groups created by the participants ranged in size from 2–12 subsumed concepts. Examples of the articulated concepts include: “listing of current clinical trials”; “roadmap for starting a clinical trial”; “single calendar of meetings”; and “shared resource inventory”.
Phase 2: Two subject matter experts reviewed and aggregated the preceding sorter groups into a composite information needs taxonomy, which consists of five high level concepts and allows for multiple-hierarchies (Table 1). These high level concepts subsume an average of 8 additional concepts.
Phase 3: As stated earlier, the information needs taxonomy generated via the preceding two phases was used to inform the implementation of a MOSS-based team science portal, which was then deployed for use by all members of the CCTS.
Phase 4: End-users of the CCTS portal were contacted after they had used the platform for between 60–120 days. They were asked to participate in an anonymous survey consisting of nine questions as described in the preceding methods section.
Table 1.
Summary of the high-level concepts comprising the information needs taxonomy (listed in order of importance as indicated by their frequency annotation).
| High Level Information Needs |
|---|
| 1. Training and Education |
|
2. Networking and Collaboration Example subsumed concepts:
|
| 3. Information Dissemination |
|
4. Research Resources Example subsumed concepts:
|
|
5. Research Planning Example subsumed concepts:
|
21 individuals participated in the survey. The corresponding user types and usage frequencies are summarized in Table 2. Of note, no respondents indicated they use the site on a daily basis. There were 9 respondents who indicated they had only recently begun using the portal, and therefore did not self-identify as daily, weekly, or monthly users. None of the participants indicated they had never used the site. Of the survey respondents, 7 had participated in a focus group (33%), while the remainder had not. The average responses of the participants to the six Likert-scale questions (4–9) included in the survey are summarized in Table 3.
Table 2.
Summary of surveyed user types and how frequently they reported using the web portal.
| User Type/Frequency | N (%) |
|---|---|
|
Investigators -Daily -Weekly -Monthly -Other |
9 (43)
0 (0) 3 (33) 4 (44) 2 (22) |
|
Research Staff -Daily -Weekly -Monthly -Other |
9 (43)
0 (0) 0 (0) 4 (44) 5 (56) |
|
Administrators -Daily -Weekly -Monthly -Other |
3 (14)
0 (0) 0 (0) 1 (33) 2 (67) |
Table 3.
Summary of QUS survey responses.
| Question | Average Response (Range) | Scale Directionality |
|---|---|---|
| 4 | 2.85 (1–7) | 0-best, 9 worst |
| 5 | 5.85 (2–8) | 0-worst, 9-best |
| 6 | 5.69 (2–9) | 0-worst, 9-best |
| 7 | 6 (2–9) | 0-worst, 9-best |
| 8 | 5.77 (2–8) | 0-worst, 9-best |
| 9 | 3.92 (0–8) | 0-best, 9 worst |
Statistical significance testing indicated that there were no significant differences in response to the preceding six questions based upon participation in the portal design focus groups, frequency of portal use, or role in the CCTS. While these results are not derived from a sufficiently large sample size to infer general characteristics of the web portals design, they do provide for sufficient qualitative and quantitative validity checking as needed to ensure that the portal design is responsive to the needs of a broad variety of end users.
Discussion
The preceding results serve to address the initial, research questions that motivated this study, namely:
The use of CKE methods was able to produce a consensus information needs taxonomy that could in turn be used to inform the design of collaborative team-science portal, and
The usability and subjective satisfaction of users of the resulting team-science portal was uniform, regardless of strata (e.g., role, portal usage frequency, and whether individuals had participated in a portal design focus group).
These results lend support to the position that the use of CKE methods, such as those employed in our study, can provide for a systematic approach to the design of team-science platforms. Furthermore, the use of convenience samples from the targeted end-user community to engage in such CKE methods (assuming a relatively equitable distribution of the roles of such members in that community) can yield a product that is broadly usable and accepted by the intended adopters. It should be noted that this study is limited in that: 1) elements of the methodology relied on the subjective and potentially biased input of a small number of subject matter experts; 2) it only reports on a single design cycle and a limited period of system utilization; and 3) it was conducted at a single site. In response to these limitations, we intend to repeat this study locally as well as at one or more additional sites in order to evaluate the reproducibility and durability of our results. Finally, we plan to make the information needs taxonomy we have generated available via the project.bmi.ohio-state.edu gForge web site.
Conclusions
These results demonstrate that an CKE-based approach to the design of collaborative, team-science platforms is tractable, systematic, and yields desirable results in terms of end user satisfaction and initial system usability. Such perceived, face validity of a systems usefulness has been widely recognizes as being central to effective end-user adoption of information technology. Furthermore, we have also demonstrated that such approaches can be rendered practicable by virtue of the ability to employ a small, convenience sample of users to yield wide-spread and consistent usability and system adoption. Given the importance of such team-science platforms in the modern clinical and translational science environment, we anticipate that the usefulness of and need for these types of approaches will only continue to increase.
Acknowledgments
This research was support in by part by NIH/NCRR (1U54RR024384-01A1 [PP]), and NIH/NLM (K22-LM008534 [PE], R01-LM009533 [PE]).
References
- 1.Ruttenberg A, Clark T, Bug W, Samwald M, Bodenreider O, Chen H, et al. Advancing translational research with the Semantic Web. BMC Bioinformatics. 2007;8(Suppl 3):S2. doi: 10.1186/1471-2105-8-S3-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agostini A, Albolino S, Boselli R, De Michelis G, De Paoli F, Dondi R, editors. Stimulating knowledge discovery and sharing GROUP ‘03: Proceedings of the 2003 international ACM SIGGROUP conference on Supporting group work. Sanibel Island, Florida, USA: ACM Press; 2003. [Google Scholar]
- 3.Payne PR, Johnson SB, Starren JB, Tilson HH, Dowdy D. Breaking the translational barriers: the value of integrating biomedical informatics and translational research. J Investig Med. 2005 May;53(4):192–200. doi: 10.2310/6650.2005.00402. [DOI] [PubMed] [Google Scholar]
- 4.Patrick TB, Craven CK, Folk LC. The need for a multidisciplinary team approach to life science workflows. J Med Libr Assoc. 2007 Jul;95(3):274–8. doi: 10.3163/1536-5050.95.3.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Payne PR, Mendonca EA, Johnson SB, Starren JB. Conceptual knowledge acquisition in biomedicine: A methodological review. J Biomed Inform. 2007 Oct;40(5):582–602. doi: 10.1016/j.jbi.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rahat I, Richard G, Anne J. A general approach to ethnographic analysis for systems design Coventry. United Kingdom: ACM Press; 2005. [Google Scholar]
- 7.Rugg G, McGeorge P. The sorting techniques: a tutorial paper on card sorts, picture sorts and item sorts. Expert Systems. 1997;14(2):80–93. [Google Scholar]
- 8.Payne PR, Borlawsky T, Kwok A, Greaves A. Supporting the Design of Translational Clinical Studies Through the Generation and Verification of Conceptual Knowledge-anchored Hypotheses. AMIA Annu Symp Proc. 2008:566–70. [PMC free article] [PubMed] [Google Scholar]
- 9.Payne PR, Mendonca EA, Starren JB. Modeling participant-related clinical research events using conceptual knowledge acquisition techniques. AMIA Annu Symp Proc. 2007:593–7. [PMC free article] [PubMed] [Google Scholar]
- 10.Payne PR, Starren JB. Quantifying visual similarity in clinical iconic graphics. J Am Med Inform Assoc. 2005 May-Jun;12(3):338–45. doi: 10.1197/jamia.M1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shneiderman B. Designing the user interface : strategies for effective human-computer-interaction. 3rd ed. Reading, Mass: Addison Wesley Longman; 1998. [Google Scholar]