Abstract
An integrated data repository (IDR) containing aggregations of clinical, biomedical, economic, administrative, and public health data is a key component of an overall translational research infrastructure. But most available data repositories are designed using standard data warehouse architecture that employs arbitrary data encoding standards, making queries across disparate repositories difficult. In response to these shortcomings we have designed a Health Ontology Mapper (HOM) that translates terminologies into formal data encoding standards without altering the underlying source data. We believe the HOM system promotes inter-institutional data sharing and research collaboration, and will ultimately lower the barrier to developing and using an IDR.
Introduction
An integrated data repository (IDR) containing aggregations of clinical, biomedical, economic, administrative, and public health data is a key component of an overall translational research infrastructure. Such a repository can provide a rich platform for a wide variety of biomedical research initiatives. Examples might include correlative studies seeking to link clinical observations with molecular data, data mining to discover unexpected relationships, and support for clinical trial development through hypothesis testing, cohort scanning and recruitment. Significant challenges exist to the successful construction of a repository, and they include the ability to gain regular access to source clinical systems and the preservation of semantics across systems during the aggregation process.
Most available data repositories are designed using standard data warehouse architecture that employs arbitrary, legacy data encoding standards. The traditional approach to data warehouse construction is to heavily reorganize and frequently to modify source data in an attempt to represent that information within a single database schema. This approach to data warehouse design is not well suited for the construction of data warehouses to support translational biomedical science because researchers require access to the true and unmodified source of information and simultaneously they need to view that same data with an information model appropriate for each researcher’s specific field of inquiry. In this paper we describe the development and functioning of the Health Ontology Mapper (HOM), which facilitates the creation of an IDR by directly addressing the need for terminology and ontology mapping in biomedical and translational sciences and by presenting a discovery interface for the biomedical researcher to effectively understand and access the information residing within the IDR. HOM can facilitate distributed data queries by normalizing local representations of data into formal encoding standards.
Background
There are several challenges posed by IDR projects geared toward biomedical research: 1) integrity of source data - a clear requirement in the construction of an IDR is that neither source data nor their interpretation may ever be altered. Records may be updated, but strict version control is required to enable reconstruction of the data that was available at a given point in time. Regulatory requirements and researchers demand clear visibility to the source data in its native format to verify that it has not been altered; 2) high variability in source schema designs – an IDR imports data from many unique software environments, from multiple institutions, each with their own unique encoding schema; 3) limited resources for the data governance of standardization - widespread agreement on the interpretation, mapping and standardization of source data that has been encoded using many different terminologies over a long period of time may be infeasible. In some cases the owners of the data may not even be available to work on data standardization projects, particularly in the case of historical data; 4) limited availability of software engineering staff with specialized skill sets - interpretation of source data during the data import process requires a large and highly skilled technical staff with domain expertise, and talent often not available or available only at considerable expense; and 5) multiple interpretations of data - there are valid, yet sometimes contradictory interpretations of the clinical meaning of source data depending on the researcher’s domain of discourse. For example, two organizations may use the same diagnosis code differently and clinical and research databases often encode race and ethnicity in differing ways. We have developed an alternative approach to provide researchers with data models based on their own preferences, including the ability to select a preferred coding/terminology standard if so desired. We believe that such an approach will be more consistent with typical research use cases, and that it will allow investigators to handle the raw data of the repository with the degrees of freedom to which they are accustomed.
An ontology-mapping component is essential for providing successful and cost effective data integration for two main reasons:
to streamline data acquisition and the identification process by a) mapping in a just-in-time fashion, instead of requiring that all data be loaded into the IDR in a single common format, and b) not requiring that all data be stored within a single centralized database schema.
to develop a standards-based technical infrastructure by a) allowing the researcher to view and extract data using the standards-based data encoding appropriate to that researcher’s domain of expertise b) providing a knowledge management system that allows less technical users to apply existing maps to fulfill information needs, and c) facilitating inter-institutional data sharing and distributed query despite different data encoding standards at each participating site.
Consider the following two use cases. In the first instance, an investigator wishes to identify all patients who have received antibiotics known to treat anaerobic organisms. In general, IDRs contain drug dictionaries that are hierarchical and based on structural classes such as penicillins, cephalosporins, macrolides, quinolones, etc. Medications that treat anaerobic organisms are scattered throughout the existing drug dictionary. Currently, an investigator can manually select all medications across all drug classes that are used to treat anaerobic organisms and run a query. However, once the task is complete, this new set of medications grouped by anaerobic effectiveness would not be available to the next research project that may want to leverage the same set of medications. Invariably, this leads to redundant work and inconsistent querying since the new query may not incorporate the same set of anaerobic-covering medications as the first query. HOM will not automatically create the new mapping to anaerobic antibiotics. However, HOM provides the infrastructure to create that new mapping, and once that map is created, it is incorporated into a library that fosters reusability. In the second use case, an investigator wishes to query across IDRs from distinct health systems, one of which uses ICD9 to encode diagnoses while the other uses SNOMED. Since an ICD9-SNOMED mapping already exists, HOM would enable seamless queries for patients with related diagnoses from both institutions without the end user having to be concerned with the different coding schema in use at each institution.
Methods
HOM is an ontology mapping software service that runs inside of an IDR. This service provides the capability to map data encoded with different terminologies into a format appropriate for a single area of specialty, without preempting further mapping of that same data for other purposes. This approach represents a fundamental shift in both the representation of data within the IDR and a shift in how resources are allocated for servicing translational biomedical informatics environments.
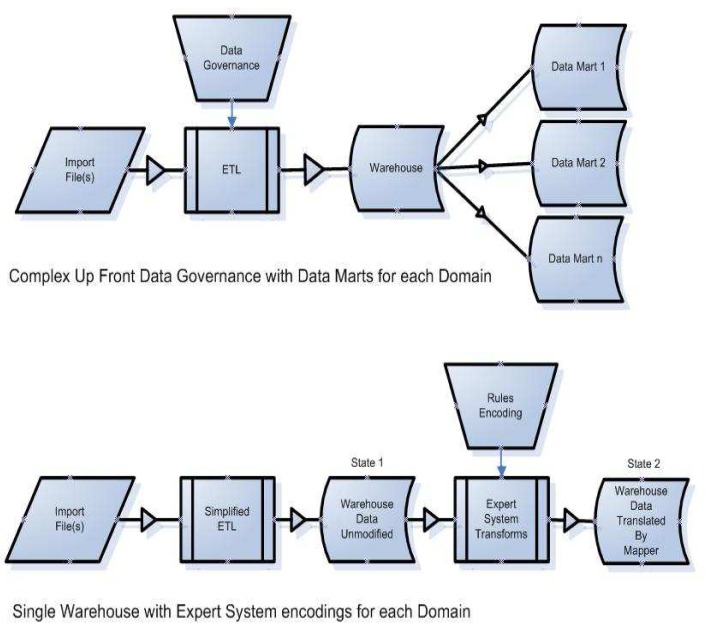
Instead of relying on an inflexible, pre-specified data governance and data model, HOM shifts resources to handling user requests for data access via dynamically constructed views of data (Fig.1). Therefore, data interpretation happens as a result of an investigator’s specific request and only as required.
Figure 1.
Complex data governance (top) can be exchanged for rules encoding (bottom)
User interactions with an IDR that implements the Health Ontology Mapper differ from those with a traditional data warehouse in two important respects: 1) Data Discovery - in models where up-front data governance has been applied, the data governance and standardization process generates a large amount of documentation that is required to describe the source data, raising a barrier to researcher utilization. In the Health Ontology Mapper, the knowledge required of the researcher has been significantly reduced, and the researcher only needs enough information about the data available to formulate specific criteria for query. 2) Translation - the translation of data from its source terminology into the ontology required by the researcher is no longer completed during the extract, transform and load (ETL) phase. The ontology mapping is completed after the source data has already been imported into the IDR. As a result of that alternate data translation workflow, the HOM enhanced IDR contains both the source system data and the formally encoded mapped results simultaneously and both the raw source data and its derivative representations can be made available to the researcher.
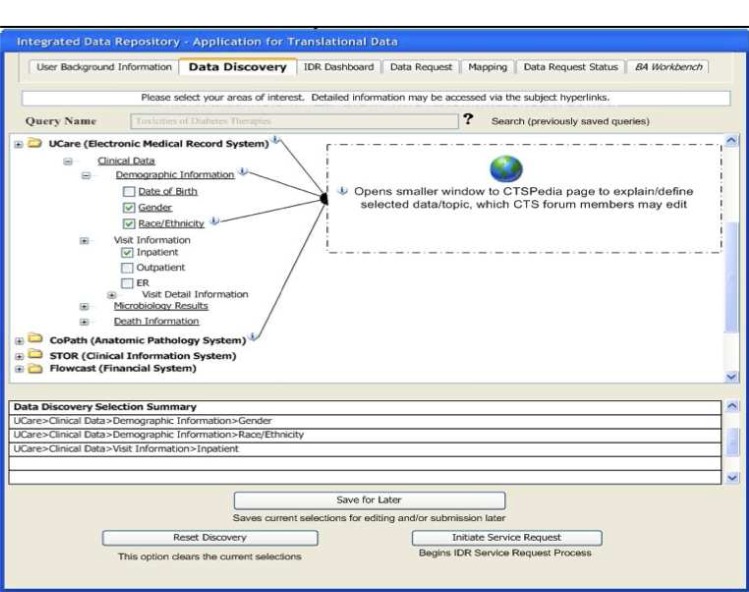
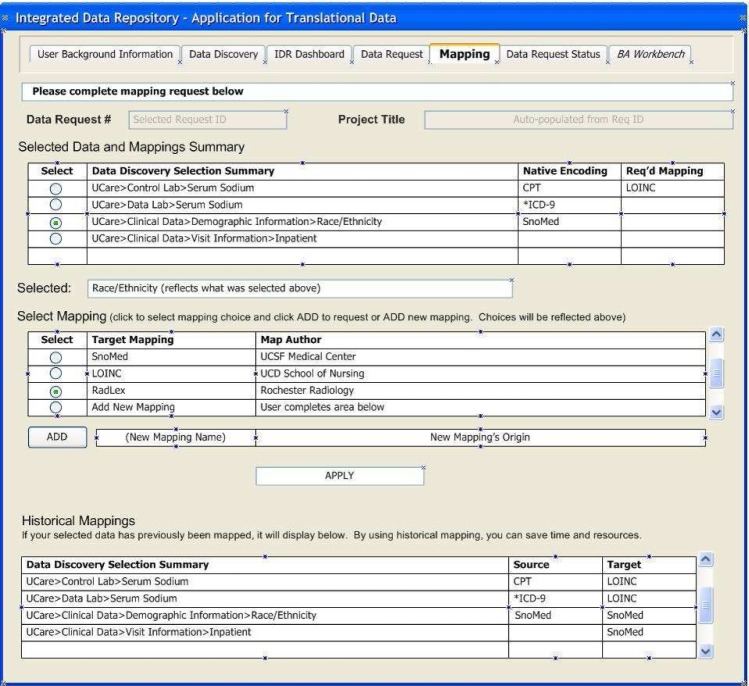
To support these distinctions, we have developed two technologies that make this approach practical: 1) A Rule Based Ontology Mapper – the source data is translated into the ontology that the biomedical researcher requires for a particular domain of expertise. The IDR uses an XML rule-based system to perform this mapping of source data format to the researcher’s ontology of choice. 2) A Discovery Interface – because all source data will not be analyzed in detail at the time of the initial ETL process that brings data into the warehouse, a mechanism is required to conceptualize the IDR contents. We have developed a web browser-based interface for data discovery and concept mapping so that the researcher can learn what types of data are available prior to requesting institutional review board (IRB) approval for access. These self-service user interfaces (UIs) are illustrated below (Figs. 2–3).
Figure 2.
Data Discovery UI showing IDR contents
Figure 3.
Mapping UI to request alternate encodings
An IDR that utilizes the HOM approach will need a web browser based interface for requesting access to the distributed data. Figure 2 shows how we have implemented that idea as the Discovery Interface for HOM. Researchers are granted access to the Discovery Interface (but not to any source data) prior to IRB approval. The Discovery Interface provides the following specific features: a) a full conceptual view of the data contained within the IDR that describes what the data is and the relationships among data; b) a description of the specific ontology into which source datum is translated; c) help text providing a written description of each particular conceptual element; d) access to the name of the source data environment from which the conceptual element was imported; e) access to researcher annotations regarding each specific conceptual element using a web based annotation interface, and; f) if pertinent and available, a link to the source data owner’s website.
With access to a complete catalog of the raw data available within the IDR investigators can then collaborate with biostatistics professionals to explore how data from different source data systems can be combined in novel ways.
HOM XML map rules are built on a logical data model, which includes work developed by the caBIG community for terminology metadata as well as modeling derived from work by Noy1 et al., Brinkley2 et al., Gennari3 et al., and Advani5 et al. At the center of the logical data model are structures for Metadata, Provenance, and System tables that address high-level administrative and data ownership information requirements. These include: 1) metadata for provenance and institutional affiliation; 2) locally and globally unique and human-readable object identifiers for all objects and actors, including those who are responsible for the mapping (e.g. creator); 3) individuals contributing or performing the activity (e.g. contributors) and; 4) those with primary responsibility such as oversight or review (e.g. curators). Each mapping intrinsically has a source and a target instance and every instance requires a robust set of attributes to uniquely identify the map both locally and globally. These logical model elements also provide information regarding map derivation and details about the nature of the transformation activity. The user requests specific data transformations by interacting with the Mapping Interface (Fig. 3).
The maps, relationships, and data transform structures are represented by each XML ontology map rule. Relationships or associations (including collections) have their own set of metadata such as unambiguous descriptions, directionality, cardinality, etc. Maps have associated identifiers not only about themselves, but also about their relationship to a target table (Fig. 6) where the mapped results are stored. Map rules are textual data that contain an XML encoded mapping rule.
The logical data model and the XML specification for HOM have been adopted into the new HL7 CTSII13 specification on the transmission of mapping rules and that specification has passed functional requirements balloting.
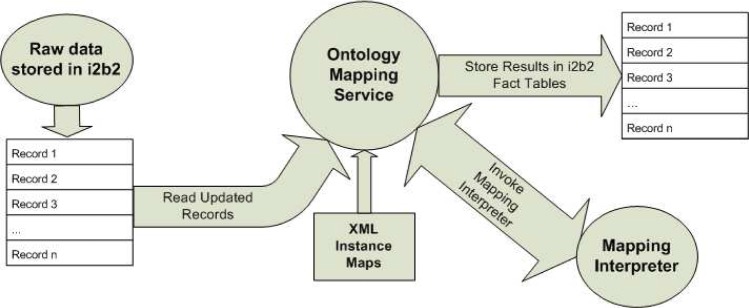
HOM consists of only two runtime components, an Ontology Mapper Discovery Interface (Fig 2) that accepts and tracks user requests and an Ontology Mapping Service and its associated Mapping Interpreter. Ontology Mapping Service runs as a background task and processes data according to a preconfigured schedule.
Project status
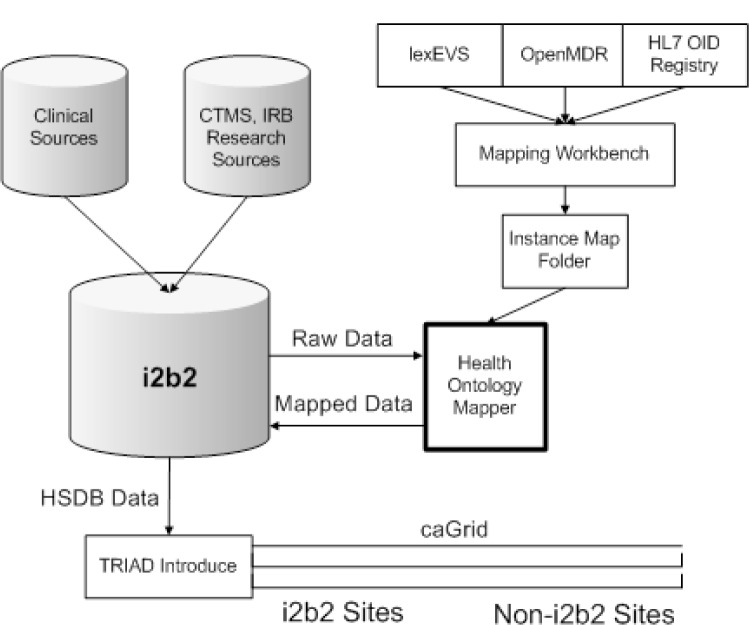
The Health Ontology Mapper project was initiated at the Mayo Clinic CTSA symposium in 2007. Its focus has been on providing syntactic and semantic interoperability for grid computing environments on the i2b2.org6 integrated data repository platform. By supplying syntactic interoperability and by leveraging the semantic interoperability of components developed for caBIG the HOM system has successfully connected i2b2 to caGrid for the HSDB4 (Human Studies Database) project. HOM specifically leverages the caDSR11 (Data Standards Repository) system for providing standard common data element definitions and the lexEVS8 system for terminology services. HOM also has been specifically integrated with caGrid by using the TRIAD12 Introduce10 and OpenMDR7 environments to provide the advanced data standards integration, grid query and terminology services.
The Human Studies Database Project (HSDB) is defining and implementing the informatics infrastructure for institutions to share the design of their human studies. The HSDB team has developed the Ontology of Clinical Research (OCRe) that models study features such as study design type, study interventions and exposures, and study outcomes to support scientific query and analysis. In support of the HSDB project the HOM system and approach was recently successfully applied during the initial development of the HSDB prototype.
First, the TrialBank8 system (which stores study outcomes) was selected as the initial source database. Data from the TrialBank system was imported into i2b2 in its native TrialBank data-encoding format;
Common Data Element (CDE) definitions were selected from the caDSR (Data Standards Repository) that best match the data encoding needs of the HSDB OCRe ontology;
The OCRe ontology is encoded in OWL (Web Ontology Language) and the OpenMDR interface that is used to access caDSR requires that each data standard be encoded in ISO 111–79 (Unified Modeling Language) format. The OCRe ontology was translated from its native OWL format into UML (ISO 111–17 format);
Those elements of the ISO 111–79 formatted model that contain a data payload were annotated with CDE numbers;
A set of HOM instance map files were manually encoded in XML format by a terminologist to describe the translation of TrialBank data to OCRe;
The HOM was run on the TrialBank data stored in i2b2 to produce a syntactically interoperable data set;
The resulting OCRe standard format data was then semantically annotated by HOM in the i2b2 encoding tables; and
The TRIAD Introduce tool was used to expose the HSDB TrialBank data over caGrid.
Our initial queries of that HSDB data were successfully executed using the cQL query language. The components used were standard caGrid and TRIAD software tools, which have been enhanced with the addition of HOM, to provide semantic and syntactic interoperability between caGrid and the i2b2.org platform. The initial HSDB distributed query environment can now be augmented to include many additional source data environments by leveraging that same set of re-usable software components.
Conclusion
The Health Ontology Mapper aims to greatly facilitate biomedical research by minimizing the initial investment that is typically required to resolve syntactic incongruities that arise when merging data from disparate sources. We believe that the use of the HOM rule-based system will make the translation of data into views for a specific researcher more easily and quickly than a traditional data warehouse design while supporting both data standards and data sharing. Our further work will now focus on the development of an Ontology Mapper Mapping Workbench to facilitate XML map authorship and we will seek to use HOM to provide semantic and syntactic interoperability for the Harvard SHRINE grid on the CICTR (Cross-institutional Clinical Translational Research) grant. We also plan to support the launch of the DBRD (Distributed BioBank for Rare Disease), and the HOMERUN (Hospital Reengineering Network) data grids.
Figure 4.
Ontology maps and association with targets
Figure 5.
System Architecture of HOM
References
- 1.Noy NF, Musen MA. The PROMPT Suite: Interactive Tools For Ontology Merging And Mapping. International Journal of Human-Computer Studies. 2003;59(6):983–1024. [Google Scholar]
- 2.Brinkley JF, Suciu D, Detwiler LT, Gennari JH, Rosse C. A framework for using reference ontologies as a foundation for the semantic web. Proc AMIA Symp. 2006:96–100. [PMC free article] [PubMed] [Google Scholar]
- 3.Gennari JH, Musen MA, Fergerson RW, Grosso WE, Crubézy M, Eriksson H, Noy NF, Tu SW. The evolution of Protégé: an environment for knowledge-based systems development. International Journal of Human Computer Studies. 2003;58(1):89–123. [Google Scholar]
- 4. Overview – TrialBank – [cited 2009 October 25] Available at http://rctbank.ucsf.edu/home/hsdb.html.
- 5.Advani A, Tu S, O’Connor M, Coleman R, Goldstein MK, Musen M. Integrating a modern knowledge-based system architecture with a Legacy VA database: The ATHENA and EON projects at Stanford. Proc AMIA Symp. 1999:653–7. [PMC free article] [PubMed] [Google Scholar]
- 6. Overview – i2b2 [cited 2009 October 31] – Available at http://www.i2b2.org.
- 7. Overview -- Metadata Repository -- caGrid.org. [cited 2009 October 20]; Available from: http://cagrid.org/display/MDR/Overview.
- 8.Sim I, Olasov B, Carini S. An ontology of randomized controlled trials for evidence-based practice: content specification and evaluation using the competency decomposition method. J Biomed Inform. 2004 Apr;37(2):108–19. doi: 10.1016/j.jbi.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 9.LexEVS (Version 5.0). [cited 2009 October 20]; Available from: https://cabig-kc.nci.nih.gov/Vocab/KC/index.php/LexEVS_Version_5.0.
- 10.Hastings S, Oster S, Langella S, Ervin D, Kurc T, Saltz J. Introduce: An Open Source Toolkit for Rapid Development of Strongly Typed Grid Services. J of Grid Computing. 2007;5(4):407–427. [Google Scholar]
- 11.NCICB: Cancer Data Standards Registry and Repository (caDSR) [cited 2009 October 31]; Available from: http://ncicb.nci.nih.gov/infrastructure/cacore_overview/cadsr.
- 12. Overview – TRIAD Architecture – [cited 2009 October 25] Available at: http://ccts.osu.edu/content/biomedical-informatics-0.
- 13. Overview – HL7 CTSII Service Functional Model – [cited 2009 October 25] Available at: http://www.hl7.org/dstucomments/