Abstract
The need for easy, non-technical interfaces to clinical databases for research preceded translational research activities but is made more important because of them. The utility of such interfaces can be improved by the presence of a persistent, reusable and modifiable structure that holds the decisions made in extraction of data from one or more datasources for a study, including the filtering of records, selection of the fields within those records, renaming of fields, and classification of data. This paper demonstrates use of the Web Ontology Language (OWL) as a data representation of these decisions which define a study schema.
Introduction
The need to query multiple clinical databases for research data without intricate knowledge of either query languages or database structure is well recognized and is increasingly being addressed in clinical research literature. “Translational research” refers not only to the interaction between pre-clinical and clinical research (“T1”) but also refers to the cycle of research results informing clinical practice while clinical practice data informs clinical research (“T2”). In the T2 translational framework, then, the ability to access that clinical practice data accurately and easily is crucial, and it is in that light that the following work is presented.
Various graphical interfaces for querying data have been suggested and are coordinated with common approaches for storage of data including use of a single schema across federated databases, schema integration of disparate databases, and transformation with query of EAV or XML data structures. One aspect of this query process that is seldom addressed, however, is the representation of a study schema, expressed either on a single database or across multiple disparate databases, such that the study schema is both persistent and reusable. Such a persistent and reusable structure would allow a data analyst to repeat studies or to modify previous studies without recreating them, and encapsulates important knowledge for study interpretation.
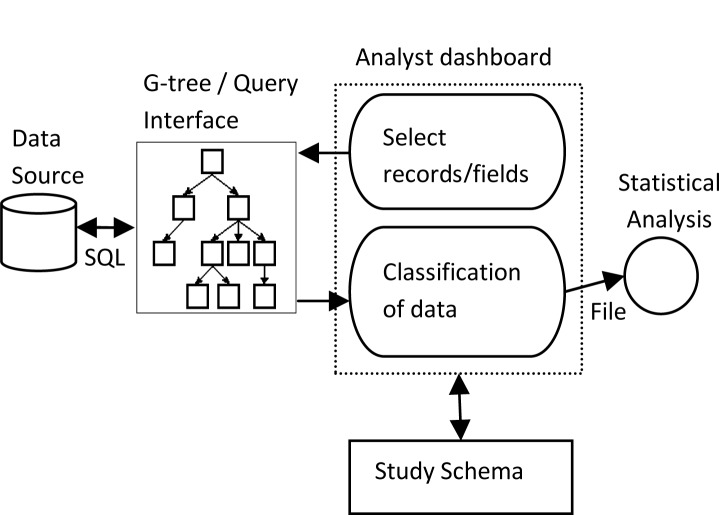
Our current work on study schema representation builds on prior research by this team on concepts we call GUI-As-View (GUAVA) and MultiClass1–4 as shown in Figure 1. It is our contention that the metadata required for accurate selection of data for a clinical study, when that data was collected using a forms-based user interface, is contained in the user interface. This includes both common types of metadata such as data type, and on-screen selection options, but also uncommonly captured metadata such as on-screen prompts or legends, the relationship of one piece of data to another and one screen to another, and whether or not the data item was required from users. We have demonstrated the ability to create a forms-based user interface using a common development platform which is self-documenting, allowing automatic creation of a query interface containing this extensive metadata. The in-memory structure that contains the interface knowledge we call a G-tree. From the G-tree, a query interface which represents the entire spectrum of metadata for the analyst can be built automatically and has been demonstrated with both tree structure and forms representations of the query interface.
Figure 1.
Overview of study progression. The G-tree is an in-memory representation of the user interface from which a query interface can be automatically created and through which the datasource can be queried. We are concerned in this work with a persistent representation for the study schema.
With such a query interface, a data analyst, who may be a domain expert but not a database expert, may retrieve data from clinical databases without writing statements in a query language nor knowing the schema of the database holding that data. The “analyst dashboard” proposed here would not support analysis of that data (i.e. aggregation or statistical manipulation) which is best done with statistical analysis tools, but rather the retrieval of datasets for use in that analysis. Functions required for this process include selection of records, limitation of the data fields retrieved for each record, renaming of fields from the stored names to analytically meaningful names, and data classification. The MultiClass principles that we have previously described directly address classification of data retrieved from multiple disparate datasources.
As an example of classification, consider a data element “bowel prep results” as included in gastrointestinal endoscopy procedure notes. Suppose the study requires that “bowel prep results” be classified as either “adequate” or “inadequate”. In one documentation system, this data might be collected in a structured element with 4 choices, “Excellent,” “Good,” “Fair,” and “Poor”. In another documentation system, this data might be collected using the Boston Bowel Preparation Scale (BBPS) which outputs an integer from 0 to 9. Since there is no one right way to classify this data, the analyst might choose, for purposes of the current study, to classify “Excellent”, “Good”, and “Fair” from the first information system and 4–9 from the second information system as “adequate” and all other choices as “inadequate.” Dynamic classification is superior to static classification (i.e. integration) of data like this since, for purposes of other studies, different classifications to “adequate” and “inadequate”, or different expressive scales, might be more appropriate.
The persistent and reusable study schema format must, then, allow for selection of records, choosing needed fields, renaming of fields, and dynamic classification of data obtained from multiple datasources. We will next briefly review related literature and then present our suggested study schema format.
Related work
The collection and retrieval of heterogeneous data is fundamental for both biomedical research and clinical data analysis. Facilitating query creation and expression becomes essential in order to get relevant results. A foremost concern is to reduce the complexity of the query interface such that nontechnical analysts can create simple, accurate and relevant queries.
Query by Example (QBE)5 was probably the first graphical user interface created for this purpose. In this interface, the user specifies the conditions on which data is to be filtered by entering representative data in desired data fields. QBE requires the users, however, to specify the join conditions on the database, and therefore requires the user to have knowledge of the data storage schema. In our analyst dashboard, joins would not need to be specified since the required knowledge is captured in the G-tree on which the query interface is derived and is based on the launch relationship of forms in the user interface. QBE also does not offer a persistent query/study schema storage form.
One of the older graphical interfaces, QUICK6 was built as a query interface to CPL (Collection Programming Language) – Kleisli specifically for querying multi-database systems. An essential part of this system for naïve users is a thesaurus which provides a mapping between user terms and database terms. Persistence and reusability of queries is not a part of QUICK and the system still requires the user to write SQL-type statements. Thesaurus-like functionality (where fields are related to their label or the user interface) in our system is contained in the G-tree (one per datasource) and can be implemented in the analyst dashboard.
XGI7 is an example of a graphical interface developed to help users query XML datasources. The XGI interface allows inexperienced researchers to create queries expressed in XQuery. The expressivity of XGI was tested and found to have moderate limitations, because some required query constructs had not been implemented. Our work in GUAVA is similar to XGI provided the XML element names reflect the labels from the user interface and the additional contextual information is available by some means. Regardless of the language used to express queries, the system must faithfully manage the mapping from the analyst queries to the physical database(s), often relational, and store the queries in an easily analyzable form. Both systems allow for operators such as renaming, but neither allows for more complex structures such as arithmetic computations. It is not the intention of our work to include such complex constructs since our goal is retrieval of a dataset that will subsequently be analyzed using a statistical analysis tool.
The Qure Data Management platform8 has several components for supporting biomedical research including study data collection. All data is stored in databases with an entity-attribute-value (EAV) schema. One component of this system is a query engine which is designed, as is ours, to output a dataset for analysis in special data analysis tools. Because of the hierarchical data structure of the underlying EAV data, joins do not need to be specified in querying; similarly, joins are not required in our analyst dashboard because of the hierarchical structure inherent in forms-based interfaces captured in the G-tree. Despite similarities, this work differs from ours in that the physical database schema in our work is arbitrary. GUAVA components handle the conversion of all data and queries from a “natural” schema which is evident from the G-tree and inferred in the query interface, into any database schema, therefore allowing the use of data from multiple sources without requiring the export or reformatting of data from those sources. The query parameters in the Qure system can be saved, but the format in which that knowledge is saved and the method of reusability is not specified.
These and other published works clearly describe common goals for querying multiple datasources and provide examples of design and functionality in query interfaces. What is either missing or little emphasized in all, however, is a plan for the storage, modification and reusability of the knowledge embodied in a specific study schema. We have explored the Web Ontology Language (OWL)9 which was not developed for this purpose, but we will demonstrate in the following section how it can be used as a persistent and reusable representation of study decisions.
Persistent representation of clinical study decisions
We first use OWL to represent the studies created by data analysts. OWL consists of a family of related languages that can be used to represent ontologies. OWL-Lite, the simplest language of the family, is sufficient for our purposes. OWL is based on a description logic; this allows us to easily describe the classification decisions made by the analysts as they prepare a study using the analyst dashboard. In this section, we describe how we represent the various aspects of the study schema in OWL. The study schema file is stored in OWL-XML which allows our software to programmatically parse and recreate a stored study. Protégé 4.0 was used for these examples.10
In OWL every class is a subclass of the class “Thing” which encompasses the universe in which our ontology (i.e., our representation of study schemas) resides. Thus we define each study as a subclass of the class “Thing”. Once we have defined our class we can create data and object properties that represent the selection, filtering and classification of records from our datasource.
The first step in defining a study schema is the selection of the fields of interest from a given datasource. This information is stored in the data properties of the study ontology. Each data property is named starting with the datasource, followed by the path to the attribute, followed by the attribute itself. In OWL each data property can be given a range which represents the type of the data property (e.g. Integer, Boolean, String). For the input data properties the range corresponds to the data type of the field in the source data. This information is stored in the source Query Interface which can be retrieved by the software without the need for the analyst to know this information.
Figure 2 demonstrates the input fields necessary for a sample study against an application for documenting GI colonoscopy procedures (CORI4) built using GUAVA components. Because we are using a query interface based on the G-tree, the attributes from the CORI4 application are named using the path of forms, controls, and data controls for the attribute, as it appears in the user interface. For example, the CORI4.Indications.TherapeuticIntervention.Dilation OfStricture represents the Dilation of Stricture field on the Therapeutic Intervention form, a form that is launched from the Indications portion of the Colonoscopy form.
Figure 2.
Representation of input schema using OWL
Once the input schema has been defined, the next step is for the analyst to describe how procedures will be selected from the datasource. We store information about the filters analysts place on the records retrieved from the datasource using an anonymous superclass of our study. These filters are value restrictions placed on data properties from the source. Figure 3 shows a common style of filter used to select procedures for a study where the procedures must fall into a given date range and be of a certain type (Colonoscopy, in this example). The “and” syntax shown in Figure 3 results in the selection of the intersection of all records where all three data restrictions hold. The analyst can easily express more complex selections, e.g., that require a particular indication or finding or other constraint on the input data.
Figure 3.
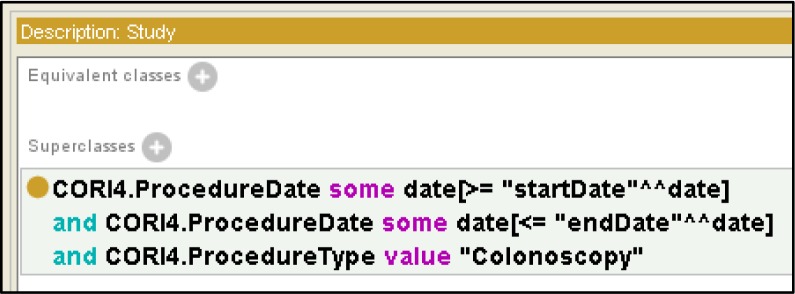
Filtering retrieved data
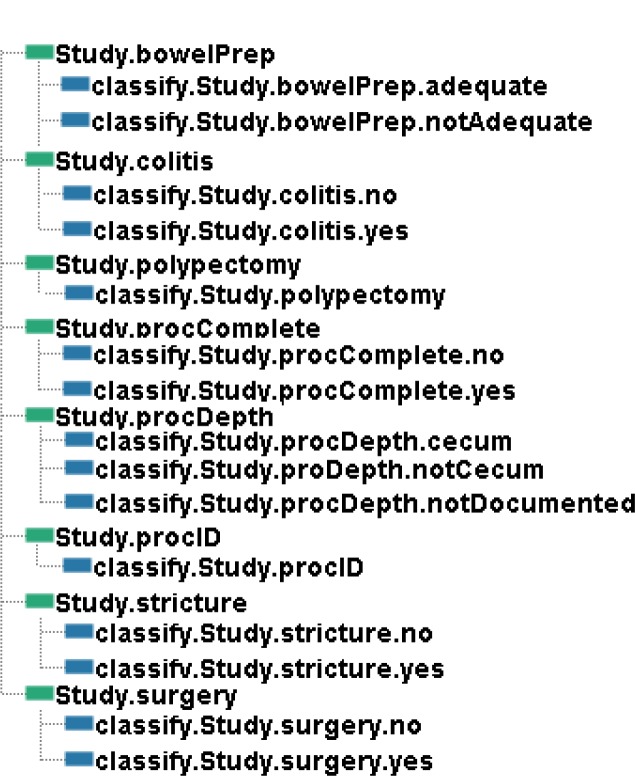
The analyst must then specify and name the fields that will appear in the output file from the study which can then be passed to a statistical analysis program. In addition, the analyst must describe the details of how the values in the output file are to be established by writing classifiers for each output field. Figure 4 shows the fields in the output along with the names of the classifiers that have been defined for each field for this study. We see, for example, that there is one classifier that describes when the Study.BowelPrep field will be set to adequate and another classifier that describes when the Study.BowelPrep field will be set to notAdequate. The analyst can define as many classifiers for a field as the study requires; the Study.ProcDepth in Figure 4, for example, has three classifiers. The data type of output fields can be determined by the software based upon the classifications created by the analyst. The analyst also has the ability to choose the name for the output fields as he or she writes the classifiers.
Figure 4.
Output schema and classifications
OWL’s object properties are used to define classifications on fields in the study. Figure 5 depicts the classification for Study.BowelPrep.adequate. In this case we map all reports with an input field from CORI4 of “QualityOfBowelPreparation” with values of “Excellent”, ”Good”, and “Fair” to the output field “BowelPrep” with a value of “adequate”. Using a similar naming scheme to the input and output data properties we name a classification with the term “classify” followed by the output field we are classifying, followed by the classifier. We use data restrictions on both the domain of the property and the range of the object property to represent the classifier. In the case that an analyst desires to rename a field from the source without modifying any of the source data, we use a simple (i.e., trivial or identity mapping) classification that takes as its domain all values from the source and has the target field as its range. As an example, Figure 6 shows a classifier that renames the source field “PolypectomyOfKnownPolyps” to the target field of “polypectomy” where the values are taken without modification.
Figure 5.
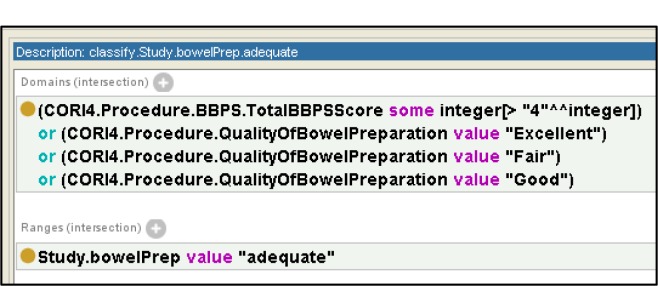
Classification of bowel preparation using OWL
Figure 6.
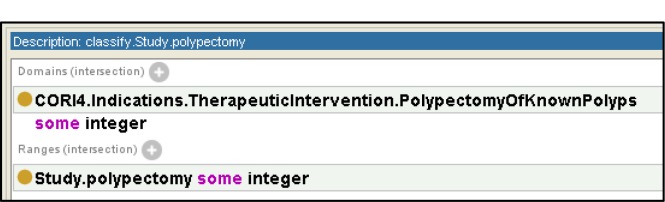
Identity classification to perform rename
Our work demonstrates that we can successfully describe the various decisions and specifications involved in defining a study schema using OWL. OWL is particularly well-suited to reason over study schemas, once specified, to determine for example that the records selected for one study are a subset of or disjoint from those in another study. Similarly, an OWL reasoner can easily compare classifiers to see how they relate.
Given the study schema in OWL, we ran a simple experiment where the CORI4 source data was also represented in OWL and we used the Protégé tool to select, project, and classify the data. The experiment worked correctly but was unable to scale to a reasonably-sized input file. This is not surprising, given that the Protégé tool was not designed to be a large-scale data processing engine. We envision a system where the OWL specifications are automatically translated into appropriate queries to extract source data and to transform data as needed into the output file. Thus the OWL specification will drive the automatic processing but will not be used directly to do the data processing.
The study representation also allows for the addition of multiple heterogeneous data sources. We can add data sources by defining new data properties for those sources and then adding restrictions on those new data properties in the classification object properties of the existing study ontology.
Further Work
We also investigated using the Mapping Specification Language (MSL),11 a language designed to perform schema mapping on XML databases, as a storage format for our study decision. We found MSL able to express the same information as the OWL representation but MSL required some additional overhead, for example input and output schemas, and the study decisions required three separate files for storage. We have favored the use of OWL for the persistent storage and reuse of study decisions to avoid this overhead as well as take advantage of the fact that OWL is based on a description logic which will allow us to perform extra computation over our studies.
From these structures, we plan to build a suite of tools that use an OWL representation of a study to create queries against datasources and then convert the resulting dataset into a form acceptable by standard statistical packages. While developed as a component of a GUAVA-enabled user and query interface, the principles presented here could be used equally well for any number of non-GUAVA data sources. In our GUAVA example we pull the input schema from the G-tree of the application. In the case of an SQL or XML database we can use DDL statements or an XML Schema to import the source schema into our application. Once the source schema has been imported there is no difference in the creation of filters or classifiers. However, once non-GUAVA sources are included the domain analyst must be aware of the underlying physical schema and associated business logic of each source in order to create filters and classifiers.
Acknowledgments
This work was supported in part by grant 1R21LM009550 from the NIH/National Library of Medicine, Judith R. Logan, PI and NSF Grant 053462, Lois Delcambre, PI. The authors also wish to thank James F. Terwilliger, PhD, whose dissertation provided the foundation for this ongoing work.
References
- 1.Terwilliger JF, Delcambre LML, Logan JR. Context-Sensitive Clinical Data Integration. In: Grust T, editor. Proc EDBT 2006 Workshop on Information Integration in Healthcare Applications (IIHA); Munich, Germany. March 26, 2006; pp. 20–31. [Google Scholar]
- 2.Terwilliger JF, Delcambre LML, Logan J. The User Interface is the Conceptual Model. Proc 25th Intl. Conference on Conceptual Modeling (ER2006); Tucson, AZ. November 6–9, 2006; pp. 424–436. [Google Scholar]
- 3.Logan JR, Terwilliger JF, Delcambre LML. Exploiting the User Interface for Tomorrow’s Clinical Data Analysis. Journal on Information Technology in Healthcare. 2008 Apr;6(2):138–149. Reprinted from Today’s Information for Tomorrow’s Improvements 2007, (ITCH 2007). [Google Scholar]
- 4.Terwilliger JF, Delcambre LML, Logan JR. Querying Through a User Interface. Data and Knowledge Engineering. 2007;63:748–768. [Google Scholar]
- 5.Zloof MM. QBE/QBE: A language for office and business automation. IEEE. 1981 May;14(5):13–22. [Google Scholar]
- 6.Tan WC, Wang K, Wong L. Quick: Graphical User Interface to Multiple databases. DEXA Proceedings. 1996 Sep;9/10:404–409. [Google Scholar]
- 7.Li X, Gennari JH, Brinkley JF. XGI: A Graphical Interface for XQuery Creation. AMIA Annual Symp Proc. 2007:453–457. [PMC free article] [PubMed] [Google Scholar]
- 8.Jager M, Kamm L, Krushevskaja D, et al. Flexible Database Platform for Biomedical Research with Multiple User Interfaces and a Universal Query Engine. International Baltic Conference; 2008. [Google Scholar]
- 9.W3C OWL Web Ontology Language Current Status. http://www.w3.org/standards/techs/owl accessed 11/5/2009.
- 10. Protégé. http://protege.stanford.edu/ accessed 11/5/2009.
- 11.Roth M, Hernandez MA, Coulthard P, et al. XML mapping technology: Making connections in an XML-centric world. IBM Systems Journal. 2006;45.2:389–409. [Google Scholar]