Abstract
Microarray probes and reads from massively parallel sequencing technologies are two most widely used genomic tags for a transcriptome study. Names and underlying technologies might differ, but expression technologies share a common objective—to obtain mRNA abundance values at the gene level, with high sensitivity and specificity. However, the initial tag annotation becomes obsolete as more insight is gained into biological references (genome, transcriptome, SNP, etc.). While novel alignment algorithms for short reads are being released every month, solutions for rapid annotation of tags are rare. We have developed a generic matching algorithm that uses genomic positions for rapid custom-annotation of tags with a time complexity O(nlogn). We demonstrate our algorithm on the custom annotation of Illumina massively parallel sequencing reads and Affymetrix microarray probes and identification of alternatively spliced regions.
Introduction
The measurement of gene expression is one of the most fundamental problems in understanding how genetic information is transformed into active processes in organism. Beyond sequencing of several genomes and the identification of many genes, transcription regulation and expression analysis has become one of the most popular research topics in molecular biology. Many different methods are available for measuring gene expression on a genome-wide scale. 3′-IVT microarrays are the most widely used platform, replacing previous technologies such as SAGE and MPSS, followed by exon microarrays and massively parallel sequencing (MPS) technology platforms. Although underlying biochemistry and what is being measured might differ, above technologies are based on the capacity to use specific tags – we define a tag as a string of DNA sequences: ‘probe’, ‘read’ and ‘tag’ are used interchangeably throughout this study. Regardless of the technology being used, investigators eventually want to get the mRNA abundance measurement (e.g. fold-change or absolute signal intensity) at the gene-level. However, at least for microarrays, it has been shown that re-examining the genomic location of probes may improve the interpretation of the biological results1. The investigation and the selection of a “good” probe that does not cross hybridize to multiple genes, withstands alternative splicing (AS) and does not contain Single Nucleotide Polymorphism (SNP) have become important since they affect on the magnitude of gene expression2, 3. Such probe selection is based on the task of matching probes against a biological reference database. Even with an up-to-date high quality annotation of probes, different studies might not have used the common gene or transcript identifier, which makes comparison of experiments from different platforms or technologies difficult. In this situation, probes have to be annotated again on the common reference. Comparable and disambiguous probe annotation is important since it affects the downstream analysis of microarrays such as a two-sample test between the treated and the control samples, sample classification, and Gene Ontology or pathway analysis. Annotation issues of microarray probes directly generalize to other technologies such as MPS, since they all utilize tags.
There have been many efforts to re-annotate microarray probes, but they are inefficient because a user can query only a handful list of probes on the web-browser2 or a user needs to download custom-made chip description file (cdf) that might not contain user’s preferred reference1. This problem gets worse for MPS reads where each sample run produces a new set of sequences. While one annotation result was shared for all the samples using a particular platform in microarray, each sample should be treated as a new platform in MPS.
We have developed a generic algorithm that efficiently matches genomic positions of the source against the reference. We applied this algorithm to the problem of the custom annotation of the tags in gene expression data. An important goal of our method is to empower researchers to create their own annotations by giving them appropriate tools, as opposed to just helping them once by providing them with a yet another annotation. There are as many ways to match tag sequences to the genome as there are gene databases. For this reason, we have decoupled these problems from our algorithm by enabling to replace the reference transcript database to get a customized mapping.
Methods
Generic Matching Algorithm
Our generic matching algorithm efficiently determines by which reference object’s interval, defined as two genomic positions a start and an end, the source object’s interval is covered. In this section, we restricted genomic positions to reside within a single chromosome and a strand to make the explanation simple. An object can be any biologically meaningful feature with genomic positions. Examples are a microarray probe, an MPS read, a transcript, a SNP or a CpG island. In most cases the interval of the source is shorter than that of the reference. For probe-to-gene annotation, a probe is the source and a gene is the reference. A match for a tag is defined to have occurred against a gene if the probe’s interval is completely contained within the gene’s interval. In search of a SNP-free probe, however, a SNP becomes the source and a probe is the reference since the probe either covers the SNP or not. Instead of comparing every interval with every other interval, the intervals are split into start and end positions, sorted and iterated linearly, keeping track of which interval is active at a certain time-point. Intersection Algorithm was first described by Marzullo4 where the minimum of start positions and the maximum of end positions was defined as an overlapping interval of two input intervals. Time complexity of this algorithm is O(n2). In our proposed algorithm, this computationally intensive task of all pairwise comparison is avoided by “genomic walking” reducing its time complexity down to O(nlogn + n).
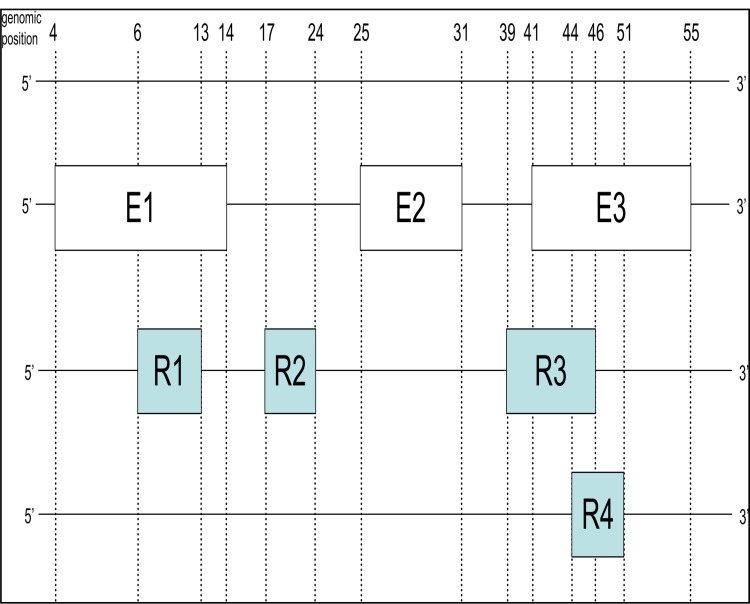
A schematic illustration of the proposed algorithm is shown in Figure 1 and Table 1. The exons and the reads from the MPS technology are positioned along the genome in Figure 1. The start and end positions of the objects are displayed in the top horizontal line. A complete run of the algorithm is depicted in Table 1: First, every tuple which is a set of (object, start-position, end-position) is split into two tuples (object, ‘start’, start-position) and (object, ‘end’, end-position). This list is sorted according to position and terminal type (‘start’ or ‘end’) which can be done in (n log n) time. Then, the list is processed linearly. We keep an extra set of identifiers, the active set.
Figure 1.
Overlapping exons and reads along the genome. The top horizontal line is a genome (5′ -> 3′). The exons (white squares) with the prefix ‘E’ and aligned in the second line. And the reads (blue squares) with prefix ‘R’ are located in the bottom line.
Table 1.
Illustrative run of PositionMatcher using the source and the reference in Figure 1.
| Pos | Object | Terminal | Active Set | Action | Yield |
|---|---|---|---|---|---|
| 4 | E1 | Start | () | E1 in | |
| 6 | R1 | Start | (E1) | R1 in | |
| 13 | R1 | End | (E1,R1) | R1 out | (E1,R1) |
| 14 | E1 | End | (E1) | E1 out | |
| 17 | R2 | Start | () | R2 in | |
| 24 | R2 | End | (R2) | R2 out | |
| 25 | E2 | Start | () | E2 in | |
| 31 | E2 | End | (E2) | E2 out | |
| 39 | R3 | Start | () | R3 in | |
| 41 | E3 | Start | (R3) | E3 in | |
| 44 | R4 | Start | (R3,E3) | R4 in | (E3,R4) |
| 46 | R3 | End | (R3,E3,R4) | R3 out | |
| 51 | R4 | End | (E3,R4) | R4 out | |
| 55 | E3 | End | (E3) | E3 out |
This is the set of objects that currently occur if we imagine “walking along” the genome. Whenever the active set is non-empty and another object is added, it designates an overlap, as can be seen the column “Yield” of the Table 1.
Input format of the source and the reference data is a set of the space-separated text file per chromosome and strand pair. Each file has three columns; the starting position of in the genome, the ending position and the object identifier (Table 2). The genomic positions of the biological objects like gene and SNP can be obtained from the biological database directly. But the position of artificial features like NGS reads or microarray probes are acquired by running the sequence alignment program.
Table 2.
Example of .BED file used in PositionMatcher input. The name of this sub-file is ‘SRR002323_chr1_forward.bed’.It is a tab-delimited and three-column text file. First two columns are genomic position and third column is identifier. ‘SRR002323’ above is NCBI SRA identifier for MPS read.
| start | end | id |
|---|---|---|
| 14452297 | 14452332 | SRR002323.4813773 |
| 14452321 | 14452356 | SRR002323.4158755 |
| 14478182 | 14478217 | SRR002323.5179826 |
Implementation
The proposed algorithm is implemented in Java so that it can run under many different operating systems. The informatics challenge to deal with MPS technology was the size of the data. For example, the biggest MPS file size we used was 11G with 109,712,542 reads. File loading is a Input/Output intensive operation but does not require CPU load for the computer5. We exploit this known fact and break the task into many sub tasks to execute them simultaneously. Before any processing we split both source and reference into sub-files of forward and reverse strands for each chromosome. Each chromosome-strand subset is matched and processed per thread simultaneously, and final results are aggregated. This divide-and-conquer technique not only exploits CPU and processing capabilities to improve algorithm runtime, but also helps solve the problem of dealing large dataset reads. Multithreaded functionality is transparent to our algorithm’s primary function—to match source tuples to reference tuples. The source code is available at http://dbmi.ucsd.edu/confluence/display/PositionMatcher.
Custom Annotation
We performed a custom-annotation of MPS reads and microarray probes to demonstrate the performance of our algorithm. The goal was to match the tags to their target genes. We downloaded the raw Illumina sequencing data from the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) with the submission id SRA000299. This contains six runs; three kidney tissue samples and another three liver samples, all from human. We also downloaded the probe sequences of three human Affymetrix 3′-IVT microarray platforms from NetAffx Analysis Center (http://www.affymetrix.com/analysis/index.affx). The sequences of the probe and the read were fed into an alignment program to obtain genomic positions. With a unprecedentedly large size of the reads data, the new alignment tools based on hash-table or indexing are widely used replacing BLAST6 or BLAT7. We used Bowtie8 for our alignment tool since it is faster and uses less computational resources than competing programs like Maq9 or SOAP10. Then the alignment result was split into 46 pairs of (chromosome, strand).
We used AceView (2007 build) as our transcriptome reference. AceView is a comprehensive compilation from several sources of sequences that can be seen as a complete transcript database11. AceView contains the genomic positions of the exons that constitute the transcripts per gene. We kept only genes containing an official name, HUGO symbols and transcripts showing the canonical and two additional splice sites (GT-AG, GC-AG and AT-AC). Filtering out genes without an official name reduced the total number of genes from 57,810 to 22,349, which is closer to the expected number of human genes12. The splice site filter marks off about 0.3% percent of all introns and therefore reduces the number of accepted transcripts only slightly. The last step is preparing a list of regions of known SNPs. For this purpose we downloaded the genome mapping of dbSNP (build 129) available in the UCSC Genome Browser. Both AceView and SNP were also split into 48 pairs for each chromosome and strand. In a Unix console, tag-to-gene annotation was done by
| $PositionMatcher | Affy-HG-U133A | AceViewGene |
| $PositionMatcher | IlluminaSeq | AceViewGene |
The command had three words, the name of our tool followed by two arguments; the source and the reference. ‘IlluminaSeq’ is the source folder having 48 genomic positions of millions of the reads. Then the presence or the absence of SNP in the tag was checked by
| $PositionMatcher | dbSNP | Affy-HG-U133A |
| $PositionMatcher | dbSNP | IlluminaSeq |
This time the tags are the reference since the tag sequences are longer than SNP. Lastly, we identified the alternatively spliced (AS) regions by running PositionMatcher with AceView against itself. The matcher performed a comparison between all transcripts of a gene and identified differences in their exonic and intronic composition. Previously, in the basic matching algorithm, we only recorded the source object completely covered by the reference object. But in finding the AS region, we modified the algorithm to identify the overlapping regions between exons and introns. And additional pre- and post-processing were performed as followings:
Mark terminal exons.
Locate exons that overlap with introns.
- Declare the overlapping regions in 2 as the alternatively spliced region with the next cases as the exceptions
- it is the first exon of a transcript and it starts inside the intron
- it is the last exon and ends inside an intron
Results
We aligned and matched six Illumina MPS runs from human kidney and liver samples13. Firstly, we downloaded raw files (.fastq format) from the public repository, NCBI SRA. Then we applied Bowtie, the alignment program, to produce six output files all mapped onto human genome (March 2006 build). 11 ∼ 13% of reads were aligned to genome (Table 3). The alignment yield was smaller than that (40%) of Marioni et al.’s13. This is because they allowed up to two mismatches for alignment while we required the perfect match to deal with the effect of SNP addressed in Introduction. The median execution time of alignment for six runs was 510 seconds on a laptop computer (Mac OS 2.6 GHz, with 2 GB RAM). Then we split each aligned result into 46 sub-files per chromosome and strand pair. Next we used our PositionMatcher to match already aligned reads as a source to the AceView transcriptome database as a reference. Although the number of survived reads was smaller than the number of the initial reads (Table 3), gene-coverage stayed between 64% ∼ 75% which is similar to that (72%) of Marioni et al. The median execution time for matching six runs was 63.5 seconds. With our matching tool together with an existing alignment program, we were able to achieve read-to-gene annotation within 12 minutes per run.
Table 3.
Annotation results of Illumina sequencing reads of six samples from human kidney and liver tissue13. The units of the second and the third columns are in Millions(M). The reference gene database was AceView that had a total 22,349 genes.
| Run ID | Init. | Aligned | Matched | Exec. Time (sec.) | |
|---|---|---|---|---|---|
| read (M) | read (M) | read (M) | gene | ||
| SRR002320 | 79 | 10 | 2.4 | 16,580 | 96 |
| SRR002321 | 110 | 13 | 2.8 | 16,081 | 118 |
| SRR002322 | 37 | 6 | 1.3 | 15,162 | 57 |
| SRR002323 | 30 | 3 | 0.7 | 14,242 | 34 |
| SRR002324 | 35 | 6 | 1.5 | 16,082 | 63 |
| SRR002325 | 55 | 7 | 1.7 | 16,170 | 64 |
We compared the accuracy and the execution time of PositionMatcher to a naïve algorithm that performs all pairwise comparison of intervals between source and reference. We downloaded microarray probe sequences three 3′-IVT microarray platforms from Affymetrix and aligned them onto genome with Bowtie. Each platform took 2 ∼ 4 minutes for alignment. For comparison, we restricted to the shortest human chromosome, chr21, and forward strand without the loss of generalizability. The matched results of PositionMatcher had 100% agreement with those of naïve method. Reduction in the execution time was observed in all platforms and reads (Table 4). The difference of gain in execution time increased with the increase of the number of tags.
Table 4.
Comparison of PositionMatcher (PM) and naïve method in chromosome 21 and forward strand. Microarray platforms were all from Affymetrix human 3′IVT array. MPS reads are identified by NCBI SRA id’s. Time is measured in milliseconds (1/1000 seconds).
| Technology | Platform/Read | Number of tags (probes/reads) | Execution Time (millisecond) | |
|---|---|---|---|---|
| PM | naïve method | |||
| Microarray | HGFOCUS | 622 | 350 | 694 |
| Microarray | HGU133A | 1,326 | 360 | 1,612 |
| Microarray | HGUPLUS2 | 3,406 | 430 | 3,869 |
| MPS | SRR002323 | 11,499 | 513 | 9,599 |
| MPS | SRR002325 | 22,988 | 572 | 168,115 |
| MPS | SRR002320 | 30,096 | 660 | 280,332 |
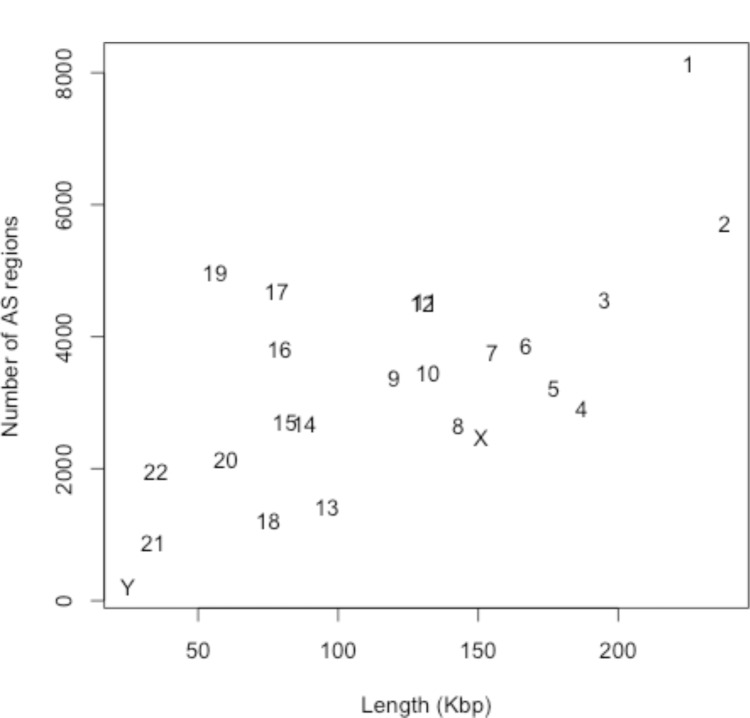
We also constructed an alternatively spliced (AS) region data by matching AceView against itself,. We found 79,970 such regions. Overall, the longer chromosome had the more AS regions (Figure 2). We found AS regions in 70% of human genes. This proportion is similar to the reported estimate of the AS undergoing genes (73%)14. We matched the MPS reads against AS region data and found out that 34∼44% of aligned reads fell in AS regions. From 15 different Affymetrix microarray platforms we found that only 3.4% of 1,602,926 different probes map to multiple genes, 11.1% map to alternatively spliced regions, 2.5% match to regions with known SNPs and 0.4% map both to AS regions and SNPs.
Figure 2.
A plot of the number of AS regions along the chromosome length (Kbp: 1,000 base-pairs).
Conclusion
We have developed an efficient tool that allows the custom annotation of MPS read, microarray probes or any tag-based technologies to measure mRNA abundance. Once the genomic positions are obtained through external alignment program, PositionMatcher will provide tags with rich annotation information by matching against biological references such as genome, transcript, SNP, CpG island, microsatellite and more. Since our algorithm is generic and fast, users can easily create a custom annotation of MPS or any tag type gene expression data against the most recent versions of references. We have shown that our tool is already fast in a usual desktop computer but it can be much faster with a simple modification to run in distributed computing environment by utilizing multi-threading.
Acknowledgments
This project was funded by grant FAS0703850 from the Komen Foundation and grant D43TW007015 from the Fogarty International Center, NIH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33(20):e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferrari F, Bortoluzzi S, Coppe A, Sirota A, Safran M, Shmoish M, et al. Novel definition files for human GeneChips based on GeneAnnot. BMC Bioinformatics. 2007;8:446. doi: 10.1186/1471-2105-8-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benovoy D, Kwan T, Majewski J. Effect of polymorphisms within probe-target sequences on olignonucleotide microarray experiments. Nucleic Acids Res. 2008 Aug;36(13):4417–23. doi: 10.1093/nar/gkn409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marzullo K. Maintaining the Time in a Distributed Systesm: An Example of a Loosely-Coupled Distributed Service. 1984. PhD disseration, Stanford University, Department of Electrical Engineering.
- 5.Hitchens R, Java NIO. Sebastopol, CA: O’Reilly Media, Inc; 2002. [Google Scholar]
- 6.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 7.Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002 Apr;12(4):656–64. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008 Nov;18(11):1851–8. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008 Mar 1;24(5):713–4. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 11.Thierry-Mieg D, Thierry-Mieg J. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7(Suppl 1):S12, 1–4. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Consortium IHGS. Finishing the euchromatic sequence of the human genome. Nature. 2004 Oct 21;431(7011):931–45. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 13.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008 Sep;18(9):1509–17. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark TA, Schweitzer AC, Chen TX, Staples MK, Lu G, Wang H, et al. Discovery of tissue-specific exons using comprehensive human exon microarrays. Genome Biol. 2007;8(4):R64. doi: 10.1186/gb-2007-8-4-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]