Abstract
We describe a corpus-based approach to creating a semantic lexicon using UMLS knowledge sources. We extracted 10,000 sentences from the eligibility criteria sections of clinical trial summaries contained in ClinicalTrials.gov. The UMLS Metathesaurus and SPECIALIST Lexical Tools were used to extract and normalize UMLS recognizable terms. When annotated with Semantic Network types, the corpus had a lexical ambiguity of 1.57 (=total types for unique lexemes / total unique lexemes) and a word occurrence ambiguity of 1.96 (=total type occurrences / total word occurrences). A set of semantic preference rules was developed and applied to completely eliminate ambiguity in semantic type assignment. The lexicon covered 95.95% UMLS-recognizable terms in our corpus. A total of 20 UMLS semantic types, representing about 17% of all the distinct semantic types assigned to corpus lexemes, covered about 80% of the vocabulary of our corpus.
INTRODUCTION
Clinical research eligibility criteria specify who is eligible for a clinical research study and, later, to whom clinical study results can be applied. There is an increasing need to efficiently transform free-text clinical research eligibility criteria into computable formats to provide decision support for clinical phenotype extraction, clinical research participants screening, and evidence-based medicine. Sim et. al have developed an annotation tool [1] to encode eligibility criteria with standard terminologies via The Unified Medical Language Systems (UMLS) [2]. However, this method did not resolve the inherent ambiguities in the UMLS semantic network, where a term can be mapped to multiple concepts and semantic types. A lexicon is central to all forms of medical language processing. At present, there is no semantic lexicon for standardizing the encoding of clinical research eligibility criteria. Many approaches to developing medical lexicons have benefited from the UMLS knowledge sources [3,4,5]. Our goal was to extend Johnson’s approach [3] to reduce the ambiguity in UMLS semantic type assignment during the development of a semantic lexicon for clinical research eligibility criteria automatically from UMLS resources. In the rest of this paper, we describe a pipeline architecture and corpus-based approach to creating a semantic lexicon for clinical research eligibility criteria using the UMLS knowledge sources.
METHODOLOGY
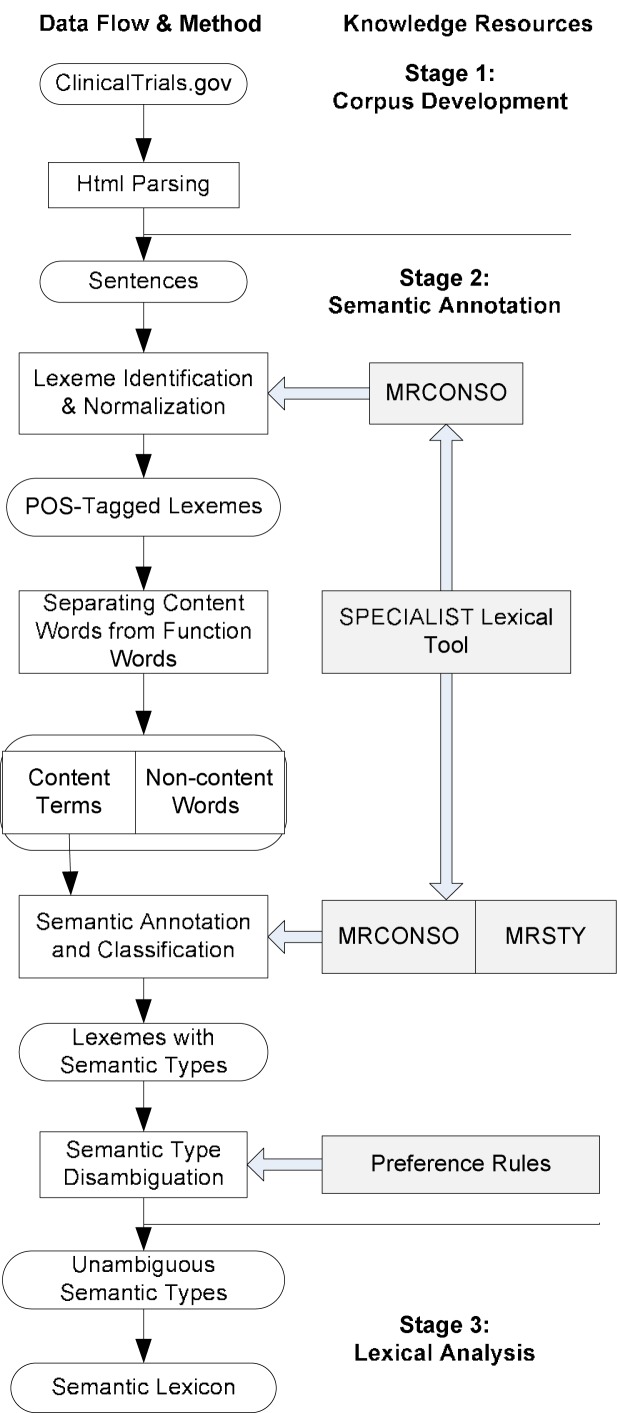
Figure 1 illustrates the steps and knowledge sources used at each step.
Figure 1:
System modules and data flow of the pipeline architecture for creating a semantic lexicon for clinical research eligibility criteria from UMLS
Stage 1: “Corpus Development”. We built a lexical database for free-text clinical research eligibility criteria extracted from the public clinical trial registry maintained by the National Library of Medicine, Clinicaltrials.gov, (http://www.clinicaltrials.gov) [6]. This web site has the most comprehensive information for 80444 clinical trials as of October 30, 2009. We developed a web-crawler application to select random samples of text from the Eligibility Criteria sections of clinical trial entries, parse the HTML web pages, and extract eligibility criteria text in order. A MySql database containing 10,000 eligibility criteria sentences was established for further corpus analysis.
Stage 2: “Semantic Annotation”. We first processed the corpus to identify UMLS-recognizable semantic units, which we refer to as lexemes, a single-word or multiple-word string that matches those occurring in the MRCONSO table of the Metathesaurus. From MRCONSO, we retrieved a Metathesaurus concept unique identifier (CUI) for each word string. Then we used the Stanford tagger [7] and the Penn Treebank tag set [8] for part-of-speech (POS) tagging. All words tagged as nouns, verbs, adjectives or adverbs were considered content words, which potentially had semantic types in the UMLS Semantic Network. In contrast, function words such as “the”, “or”, etc., numerals and operators (e.g., > or <) do not have UMLS semantic types.
MRCONSO contains a small range of lexical variants for lexemes. Matching against these variants is managed through the use of Specialist Lexicon Tools [9], which can be used to reduce lexical variation for general English text, providing normalized strings for a wider range of variants, including tense, number, and part-of-speech variants.
To take advantage of potentially expanded coverage through use of the Specialist Lexicon, we built a version of MRCONSO in which UMLS CUIs were mapped to these normalized strings. Input lexemes were normalized before checking against this version of MRCONSO. Strings in the base version of MRCONSO that did not have normalized terms in the Specialist Lexicon were used in their original form. Although the word vocabularies contained in the UMLS Metathesaurus and Specialist Lexicon were not identical, we identified and assigned semantic types for over 90% of all lexemes in our corpus.
The MRSTY table of the UMLS contains semantic types defined for each CUI in MRCONSO. We mapped input lexemes to these CUI’s as above and looked up their semantic types in MRSTY. We call lexemes successfully annotated with semantic types as semantic terms, or simply terms. The majority of terms were associated with a single semantic type. However, many had multiple types, resulting in ambiguity (See table 1).
Table 1:
Example of semantic assignment before applying semantic preference rules
| Terms | Semantic Types |
|---|---|
| One-to-One Mapping | |
| immunodeficiency | Disease or Syndrome |
| recent | Temporal Concept |
| One-to-Many Mapping | |
| patient |
|
| therapy |
|
| No Mapping | |
| while | |
| bulky | |
When using the UMLS, one general source of ambiguity stems from the fact that the Semantic Network [10] is an ontology intended to cover medicine as a whole, including both medical science and clinical medicine. For example, the word “prednisone” has at least two senses, one describing a steroid chemical with a certain structure, and one describing a pharmaceutical medicine. The Semantic Network provides two types associated with these two senses, namely Chemical Viewed Structurally and Chemical Viewed Functionally. The sense of pharmaceutical medicine might be expected to be more appropriate in clinical text. Using a corpus of hospital discharge summaries, [3], this was verified. Many such cases were examined and, through a manual process of textual analysis, a set of hand-crafted preference rules was developed.
Preference rules have the form: if TYPE-A (or any of its descendants in the UMLS Semantic Network) and TYPE-B (or any of its descendants) are specified for a given lexeme, then retain TYPE-B (or any descendant) and discard TYPE-A (or any descendant). For example, the lexeme “beta Hydroxyphenethylamine” is assigned the types Pharmacologic Substance and Organic Chemical. Pharmacologic Substance is a descendant of Chemical Viewed Functionally. Organic Chemical is a descendant of Chemical Viewed Structurally. Given preference for Chemical Viewed Functionally (the clinical sense) over Chemical Viewed Structurally (the biological sense), the type Pharmacologic Substance would be retained, and the type Organic Chemical would be discarded, for this lexeme. Preference rules can be formulated at any desired level of generality allowed by the Semantic Network. Table 2 shows 5 examples of frequently applied semantic preference rules.
Table 2:
Frequently applied preference rules
| Discarded Type | Preferred Type | Example Lexeme |
|---|---|---|
| Health Care Activity | Diagnostic Procedure | liver biopsy, lumbar puncture |
| Intellectual Product | Health Care Related Organization | intensive care unit, hospital |
| Quantitative Concept | Temporal Concept | minutes, second |
| Spatial Concept | Body Location or Region | mediastinal, pericardial |
| Idea or Concept | Organism Function | recovery, birth, death |
Stage 3: “Lexical Analysis”. We investigated the coverage of the sample corpus provided by our annotation procedure, using the Metathesaurus, Semantic Network, and preference rules. Results are described in the next section.
RESULTS
1. Coverage
The corpus contained a total of 74,188 text tokens, including all content words and other text tokens. The average sentence length was 7.41 (text tokens). There were 47,129 content words (See Table 3). Of these, 15.56% were multiple-word lexemes and 84.43% were single-word lexemes. 95.95% of content words were assigned at least one semantic type. 4.05% were not assigned any type; all of these were single-word lexemes. In the corpus, there were 6,921 unique content words, 90.02% of which were assigned at least one semantic type and 9.08% of which were not assigned any type.
Table 3:
Coverage of the corpus by UMLS types
| Content Words | Occurrences Total Count: 47,129 | Unique Occurrences Total Count: 6,921 | ||
|---|---|---|---|---|
| Count | Percent | Count | Percent | |
| No Type | 1908 | 4.05% | 691 | 9.98% |
| Has Type | 45221 | 95.95% | 6230 | 90.02% |
| Multiple Words | 7334 | 15.56% | 2283 | 32.99% |
| Single Word | 39795 | 84.43% | 4638 | 67.01% |
We also examined coverage of the corpus vocabulary by individual semantic types. Table 4 lists the top 20 types in terms of percent of occurrences of corpus lexemes assigned the individual type.
Table 4:
Coverage of the 20 semantic types
| Semantic Type | % of corpus |
|---|---|
| Temporal Concept | 11.07% |
| Qualitative Concept | 10.60% |
| Functional Concept | 6.19% |
| Laboratory Procedure | 5.16% |
| Therapeutic or Preventive Procedure | 4.62% |
| Disease or Syndrome | 4.58% |
| Intellectual Product | 4.01% |
| Idea or Concept | 4.00% |
| Pharmacologic Substance | 3.70% |
| Organism Attribute | 3.39% |
| Spatial Concept | 2.99% |
| Health Care Activity | 2.92% |
| Finding | 2.87% |
| Organism Function | 2.67% |
| Population Group | 2.62% |
| Professional or Occupational Group | 2.27% |
| Quantitative Concept | 2.11% |
| Neoplastic Process | 1.72% |
| Patient or Disabled Group | 1.71% |
| Body Part, Organ, or Organ Component | 1.40% |
| Total: | 80.60% |
This set of the top 20 types represents 17.9% of the 117 unique types applied to the corpus, but covers 80.6% of the corpus vocabulary. These types can therefore be considered as the primary semantic classification of our randomly selected sample of eligibility criteria text, when analyzed through use of the UMLS ontology.
2. Disambiguation
Using the preference rules, 117 out of 134 semantic types were applicable to the corpus. 22,878 input content words had multiple semantic types and were processed by the preference rules. 24,251 content words were not processed by any preference rule, these being either singly-typed or non-typed content words in the text. The semantic types that were most frequently excluded by our preference rule were listed in Table 5. The table reflects the fact that the more concrete types are preferred to UMLS conceptual types wherever possible.
Table 5:
Ambiguity reduction in the top 5 semantic types after applying the semantic preference rules
| Semantic Types | Occurrence | |
|---|---|---|
| Before | After | |
| Idea or Concept | 10017 | 2965 |
| Qualitative Concept | 8408 | 4489 |
| Intellectual Product | 6134 | 1941 |
| Conceptual Entity | 3135 | 218 |
| Manufactured Object | 2470 | 121 |
Before applying preference rules, 88,594 semantic types were assigned to content words. After preference rules were applied, only 45,221 semantic types were assigned. Before applying preference rules, 2324 (33.57%) of unique content words had two or more types. After applying the preference rules, all these terms only had one UMLS semantic type each.
3. Non-content tokens in the corpus
All input tokens having POS tags classifying them as nouns, verbs, adjectives and adverbs were considered content words potentially having semantic types. The rest of the input text consists of what are traditionally referred to as function words, such as articles, prepositions, and others, as well as numbers, symbols, abbreviations, or units (See Table 6).
Table 6:
Functional words and their examples
| Non-content tokens | Examples |
|---|---|
| Function words | The, of, can, if, while… |
| Number Strings | 18, 60, 1979, 2005 |
| Symbols Strings | -, #, >=, @, +, ?, * |
| Abbreviations | GLD, HCV, NICHD |
| Units | mm3, ph, mmhg, l, kg |
Such input tokens still contain valuable semantics for interpreting eligibility criteria text. We will demonstrate the usage of both content words and non-content words below.
EXAMPLES OF ANNOTATION
With the semantic lexicon, we can automatically annotate eligibility criteria sentences with unambiguous semantic types. We compared the performance of our annotation tool to MetaMap Transfer (MMTx 2.4C version) [11] as illustrated by the following two examples.
Example 1:
Sentence: Estimated creatinine clearance > 50 mL/min.
Our Annotation:
{Estimated creatinine clearance| Laboratory Procedure} {>|SYMBOL} {50|NUMERAL} {mL|UNIT} {min|Temporal Concept} {.|.|}
MMTx 2.4C:
{Estimated creatinine clearance > 50 mL|Laboratory Procedure} {/min.|Temporal Concept}
Example 2:
Sentence: Patients with complications such as serious cardiac, renal and hepatic disorders.
Our Annotation:
{Patients|Patient or Disabled Group} {with|} {complications Pathologic Function} {such|} {as|} {serious|Qualitative Concept} {cardiac|Body Part, Organ, or Organ Component} {renal|Body Part, Organ, or Organ Component} {and|} {hepatic|Body Location or Region} {disorders|Disease or Syndrome} {.|.|}
MMTx 2.4C:
{Patients|Patient or Disabled Group} {with complications|Pathologic Function} {such as serious cardiac, renal|Idea or Concept} {and|} {hepatic disorders.|Disease or Syndrome}
The examples showed that our method produced finer-grained results than MMTx 2.4C. MMTx returned “such as serious cardia, renal” as a single constituent, which was questionable. In contrast, our annotation tool effectively decomposed the phrase into more granular semantic units: “such”, “as”, “serious”, “cardiac”, and “renal”.
DISCUSSION
Previously developed lexicons had a coverage of 79% [3] for discharge summaries, and 77% [4] for non-clinical biological text. Our lexicon has 95% coverage of the vocabulary of our corpus of eligibility criteria. Approximately 80% of the corpus vocabulary was covered by only a small set of 20 types (17% of distinct occurring types). By contrast, Verspoor found a much smaller set of distinct types (3% of distinct occurring types) providing 77% coverage of his particular corpus.
It can be seen that medical text varies considerably in the breadth of its vocabulary (affecting coverage by resources like the UMLS Metathesaurus), and in the specificity of its semantics (after the vocabulary is reduced to its conceptual content).
We compared our annotations with MMTx mainly because MMTx is a widely used, general-purpose tool for UMLS-based semantic annotation, providing many conveniences and options. It would be possible to obtain more precise results from MMTx by passing it more detailed word strings (i.e. lexemes, as we have defined them) rather that full sentences. But this would require prior implementation of the lexeme-identification methods employed in this paper. Use of MMTx would then be redundant, because lexemes and their types can be directly looked up in MRCONSO and MRSTY.
CONCLUSION AND FUTURE WORK
We developed an annotation procedure which provides a UMLS-based, unambiguous semantic lexicon with 95% coverage for a random sample of eligibility criteria text (10,000 sentences). We also identified 20 semantic types defined by UMLS that can serve as a preliminary classification of terms in eligibility criteria text. These observed restrictions on type occurrence suggest that there are specific semantic constraints operating in the language of eligibility criteria text that can be studied further.
As part of our future work, a sublanguage of clinical research eligibility criteria will be explored, wherein only certain restricted sentence types and predications can be expected to occur. This would further aid the development of procedures for extraction and standardization of eligibility criteria.
We will also study whether the UMLS type classification that we have observed is optimal for developing a standard ontology for eligibility criteria. The ultimate goal is construction of an automated extraction procedure mapping raw text to a standards-based formal structure for eligibility criteria.
Other research has been done in the area of eligibility criteria modeling [12]. Semantic classes highly specific to eligibility criteria have been defined, such as Assessments (of a patient), Interventions (performed on a patient) and Behavior (of a patient). These are entered into templates summarizing the criteria for a research study [13]. It may therefore be necessary to better align UMLS classes to those of optimal models. For example, the UMLS types Laboratory Procedure, Organism Attribute, Health Care Activity and Organism Function may all map to the class Assessments. This may provide a more concise representation. Mapping of UMLS types to models is also important because the UMLS Metathesaurus remains a crucial resource for text-based extraction procedures.
Acknowledgments
This research was funded under NLM grant 1R01 LM009886-01A1 and CTSA award UL1 RR024156. Its contents are solely the responsibility of the authors and do not necessary represent the official of NIH.
REFERENCES
- 1.Tu S, Peleg M, Carini S, Bobak M, Rubin D, Sim I. A Practical Method for Transforming Free-Text Eligibility Criteria into Computable Criteria. Journal of American Medical Informatics Association, JAMIA. 2009. Under Review in 2009. [DOI] [PMC free article] [PubMed]
- 2.McCray AT, Bodenreider O, Malley JD, Browne AC. Evaluating UMLS Strings for Natural Language Processing. Proceedings AMIA Annual Symposium. 2001:448–452. [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson SB. A Semantic Lexicon for Medical Language Processing. Journal of the American Medical Informatics Association. 1999;6:205–218. doi: 10.1136/jamia.1999.0060205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verspoor K. Comparative and Functional Genomics. Wiley InterScience; 2005. Towards a semantic lexicon for biological language processing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedman C, Liu H, Shagina L, Johnson S, Hripcsak G. Evaluating the UMLS as a source of lexical knowledge for medical language processing. Proceedings AMIA Annual Symposium; Washington, D.C.. 2001. pp. 189–193. [PMC free article] [PubMed] [Google Scholar]
- 6.McCray AT. Better Access to Information about Clinical Trials. Annals of Internal Medicine. 2000;133:609–614. doi: 10.7326/0003-4819-133-8-200010170-00013. [DOI] [PubMed] [Google Scholar]
- 7.Toutanova K, Klein D, Manning C, Singer Y. Feature-Rich Part-of-Speech Tagging with a Cyclic Dependency Network. Proceedings of HLT-NAACL. 20032003:252–259. [Google Scholar]
- 8.Marcus MP, Santorini B, Marcinkiewicz MA. Building a Large Annotated Corpus of English: The Penn Treebank. Computational Linguistics. 1993;19:313–330. [Google Scholar]
- 9.Browne A, Divita G. The SPECIALIST Lexicon and NLP Tools. WorldVistA Conference Presentation. 2009.
- 10.McCray AT. The UMLS semantic network. Annual Symposium on Computer Applications in Medical Care; Washington; DC. 1989. pp. 503–507. [Google Scholar]
- 11.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program.. Proceedings AMIA Annual Symposium; 2001. pp. 17–21. [PMC free article] [PubMed] [Google Scholar]
- 12.Tu S, Peleg M, Carini S, Rubin D, Sim I. ERGO: A Template Based Expression Language for Encoding Eligibility Criteria 2008.
- 13.Tu SW, Campbell JR, Glasgow J, Nyman MA, McClure R, McClay J, Parker C, Hrabak KM, Berg D, Weida T, Mansfield JG, Musen MA, Abarbanel RM. The SAGE Guideline Model: Achievements and Over-view. Journal of the American Medical Informatics Association. 2007;14:589–598. doi: 10.1197/jamia.M2399. [DOI] [PMC free article] [PubMed] [Google Scholar]