Abstract
The conduct of large-scale translational studies presents significant challenges related to the storage, management and analysis of integrative data sets. Ideally, the application of methodologies such as conceptual knowledge discovery in databases (CKDD) provides a means for moving beyond intuitive hypothesis discovery and testing in such data sets, and towards the high-throughput generation and evaluation of knowledge-anchored relationships between complex bio-molecular and phenotypic variables. However, the induction of such high-throughput hypotheses is non-trivial, and requires correspondingly high-throughput validation methodologies. In this manuscript, we describe an evaluation of the efficacy of a natural language processing-based approach to validating such hypotheses. As part of this evaluation, we will examine a phenomenon that we have labeled as “Conceptual Dissonance” in which conceptual knowledge derived from two or more sources of comparable scope and granularity cannot be readily integrated or compared using conventional methods and automated tools.
Introduction
A defining characteristic of the conduct of translational studies is the collection, integration, storage and analysis of large-scale data sets consisting of both phenotypic and bio-molecular variables. Such integrative data sets are used to enable analyses that target the identification and quantification of significant relationships between such variables, which can be used to inform the diagnosis, staging and planning of treatment for pathophysiologic states1–4. However, the current state of knowledge and practice pertaining to the investigation of such bio-marker-to-phenotype relationships commonly relies on either the naïve discovery of potential linkages between variables using statistical or data mining techniques5, and/or the testing of intuitively derived hypotheses1. At the same time, significant volumes of knowledge exist in the form of conceptual knowledge collections such as ontologies and published literature extracts that could be extremely useful in informing or generating such hypotheses2,3,6. Approaches such as Conceptual Knowledge Discovery in Databases (CKDD) have been proposed in prior reports as a means of leveraging such conceptual knowledge collections in order to generate high-throughput hypotheses relative to specific, integrative data sets6. However, as we have previously reported2,3, the use of such techniques, while extremely promising, still presents a number of challenges, including the ability to employ sufficiently scalable validation methods. In this report, we describe an evaluation of the efficacy of employing a natural language processing (NLP) approach to extract conceptual knowledge from published biomedical literature abstracts in order to validate and augment hypotheses concerning bio-marker-to-phenotype relationships derived from common ontologies, such as SNOMED-CT 7 and the NCI Thesaurus8. This evaluation was conducted in the specific experimental context of the Chronic Lymphocytic Leukemia Research Consortium (cll.ucsd.edu), which is funded by the National Cancer Institute (NCI). As part of our evaluation, we will examine a phenomenon that we have labeled as “Conceptual Dissonance” in which conceptual knowledge derived from two or more sources of comparable scope, and granularity cannot be readily integrated or compared using conventional methods and automated tools.
Background
Based upon the objective described in the preceding introduction, the following section will briefly review contributing work related to the experimental methodology and context of our study.
Conceptual Knowledge Engineering
Knowledge engineering (KE) is a process by which knowledge is collected, represented and subsequently used by computational agents to replicate expert human performance in an application domain9. It incorporates four major steps: 1) knowledge acquisition, 2) computational representation of that knowledge, 3) implementation or refinement of the knowledge-based agent, and 4) verification and validation of the output of the knowledge-based agent9. Conceptual knowledge, one of three primary types of knowledge that can be targeted by KE, can be defined as a combination of atomic units of information and the meaningful relationships among those units9. The knowledge sources used during the knowledge acquisition stage of the KE process can take many forms, including standard terminologies, ontologies, narrative text, databases and domain experts. The work described in this manuscript utilizes a conceptual knowledge acquisition approach known as conceptual knowledge discovery in databases (CKDD)10. At a high level CKDD is concerned with the utilization of automated or semiautomated computational methods to derive knowledge from the contents of databases. The use of domain-specific knowledge collections, such as ontologies, is necessary to inform this knowledge induction process since commonly used database modeling approaches do not always incorporate semantic knowledge corresponding to the database contents10. This overall approach is the basis for a specific CKDD methodology known as constructive induction6. In constructive induction, data elements defined by a database schema are mapped to concepts defined by one or more ontologies. Subsequently, the relationships included in the mapped ontologies are used to induce semantically meaningful relationships between the mapped data elements. The induction process generates “conceptual knowledge constructs (CKCs)” concerning the contents of the database, which are defined in terms of data elements and semantic relationships that link those elements together in a meaningful manner2.
Experimental Context
The specific experimental context for the work presented in this report stems from a collaboration with the Chronic Lymphocytic Leukemia Research Consortium (CLL-RC), an NCI-funded translational research program consisting of eight sites. The CLL-RC coordinates and facilitates basic and clinical research on the genetic, biochemical and immunologic bases of Chronic Lymphocytic Leukemia (CLL), which is the most common adult leukemia in the United States11. The incidence rate of the disease appears to be on the rise, and environmental and genetic factors have been shown to contribute to its development11. The clinical course and phenotypic presentation of CLL is highly heterogeneous, and as such, there are no known curative strategies12. The research portfolio of the CLL-RC focuses primarily on the discovery and evaluation of novel biologic and pharmacologic treatments for CLL, with particular emphasis on the identification of phenotypic ↔ bio-molecular relationships that may improve clinical staging and/or assist in evaluating patient responses to novel therapies. A critical facility supporting the ability of the CLL-RC to engage in such research is the use of a central data repository, associated data collection instruments, and data mining and analysis tools, which are known collectively as the CLL-RC Integrated Information Management System (CIMS)13,14. CIMS facilitates the collection and storage of numerous high-throughput, multidimensional data sources generated by instrumentation and methodological approaches including quantitative and qualitative immunophenotyping, multiple modalities of gene expression analysis, and Fluorescent In Situ Hybridization (FISH) analyses of cytogenetic abnormalities.
Contributing Prior Work
In prior reports, we have demonstrated the efficacy of applying constructive induction using a novel platform known as TOKEn (Translational Ontology-anchored Knowledge-discovery Engine) in order to: 1) discover potential hypotheses linking bio-molecular and phenotypic variables within the CIMS data repository2; and 2) validate and prioritize such hypotheses based upon the results of human-mediated meta-analysis of published literature abstracts3. However, a limitation in our earlier reports has been the scalability of available validation and prioritization techniques, given their reliance on human intervention. This limitation is the primary motivation for the work reported here.
Methods
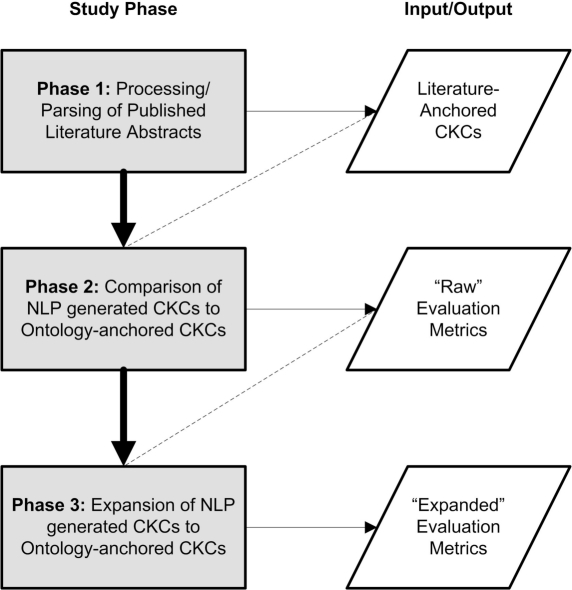
Given the preceding motivation, this three-phase study focuses upon evaluating the efficacy of employing natural language processing (NLP) techniques in order to extract conceptual knowledge from published literature abstracts, and compare that knowledge to the previously generated conceptual knowledge constructs (CKCs). This work focuses on two primary research questions:
Is the conceptual knowledge that can be extracted from published biomedical literature using the SemRep NLP platform syntactically and semantically comparable to that extracted from common ontologies using TOKEn; and
Is the conceptual knowledge encoded from the published biomedical literature significantly different from that found in ontological knowledge sources.
Phase One: In the first phase of our study, a Pubmed query was run in order to retrieve all literature published within the past three years (as of August 2008) that had either been indexed using the MeSH category of “Leukemia, Lymphocytic, Chronic, B-Cell” and/or contained lexical variants of the term “Chronic Lymphocytic Leukemia” in their title or abstract. The UMLS Knowledge Source Server (umlsks.nlm.nih.gov) was utilized to identify lexical variants by determining all of the Metathesaurus concepts with a common Lexical Unique Identifier corresponding to Chronic Lymphocytic Leukemia. These retrieved references were exported using the Medline text output format. This output file was processed in order to remove all references that did not include a textual abstract, and sub-select the PubMed ID (PMID), Title (TI) and Abstract (AB) for each entry. The resulting text was then submitted for parsing using the publicly available SemRep NLP platform maintained by the National Library of Medicine11,12. SemRep was invoked both with and without the SemGen option, which is used to enable the parsing of genomic concepts. The output generated by SemRep from these literature abstracts was post-processed in order to sub-select only unique CKC triplets consisting of CUI-relationship-CUI patterns.
Phase Two: Using a Perl script, CKC triplets that began and terminated with concepts that were manually mapped by subject matter experts (SMEs) to variables in the CIMS repository (during the course of the previously introduced contributing study2,3) were compared with those generated by SemRep in Phase One, in order to identify any direct matches between the two sets of CKCs. The original CIMS-derived CKCs utilize UMLS Metathesaurus relationship types extracted from the MRREL raw text file using a graph theoretic algorithm implemented as a Perl script2, and the SemRep CKCs utilize UMLS Semantic Network relationship types assigned automatically by the SemRep service. Due to these differing relationship types, additional processing was necessary to normalize the two data sets by classifying the CIMS-derived CKC component CUIs according to their semantic type(s), and determining any corresponding Semantic Network relationships between them.
Phase Three: In this fina l phase, the SemRep generated CKCs were iteratively expanded by using a Perl script to traverse the UMLS MRREL file and select descendant concepts relative to the initial CKC concepts, thus generating new, more granular CKCs. This analysis was done to evaluate the effects of such expansion on the degree of overlap between the SemRep derived CKCs and the prior TOKEn generated CKCs at increasing levels of granularity.
Results
In the following section, we will summarize the results associated with each of the preceding study phases:
Phase One: The previously described literature search strategy yielded a total of 1945 abstracts that included full text abstracts. These abstracts yielded a total of 6599 unique triplets using both SemRep and the previously described post-processing approach. Examples of these triplets are included in Table 1.
Table 1:
Examples of literature-derived CKCs.
| Initial Concept | Relationship | Terminal Concept |
|---|---|---|
| Chromosomes, Human, Pair 8 | LOCATION_OF | IGH@ gene cluster |
| IGH@ gene cluster | ASSOCIATED_WITH | Disease Progression |
Phase Two: There were 5800 CKCs, comprised of two to five concepts, generated by the TOKEn algorithm in our prior study. These CKCs were broken down into 1626 distinct transitive triplets that were subsequently classified using the UMLS Semantic Network as described in our methods. The corresponding Semantic Network relationships were assigned to these initial triplets, resulting in an expanded set of 10759 triplets (i.e., each initial triplet could be expanded to include one or more semantic relationships). When comparing these triplets to those resulting from SemRep, there were no exact matches.
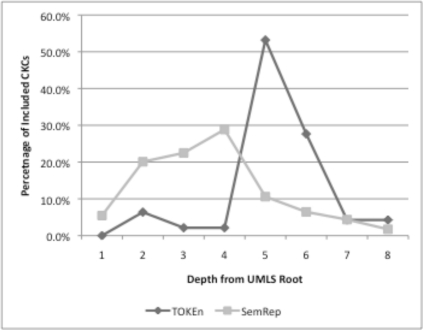
Phase Three: In order to better understand why no exact matches between the TOKEn and SemRep generated CKCs occurred in Phase Two, a heuristic evaluation of the CKCs was performed by two SMEs who had participated in our prior studies. At a high level, the factors assessed by the SMEs included: 1) the existence of common semantic meaning between initial or terminal concepts in the two sets of CKCs; 2) the comparative granularity of the initial or terminal concepts in the two sets of CKCs; and 3) the presence or absence of overlap between positive control CKCs in the two sets (e.g., CKC’s identified by the SMEs as being both valid and pertaining to well established basic science or clinical domain knowledge). This evaluation led to the preliminary conclusion that the concepts included in the SemRep generated CKCs were more general (i.e., less granular) than those included in the TOKEn generated CKCs. This phenomenon was further illustrated by a quantitative analysis of the incidence of concepts included on both types of CKCs at increasing depths from the UMLS root (a surrogate measure for concept granularity that has been used in our prior evaluations of constructive induction2,3), as illustrated in Figure 2.
Figure 2:
Incidence of concepts included in TOKEn and SemRep generated CKCs at increasing granularity levels (e.g., depths from UMLS root).
Building upon these findings, the 6599 unique triplets that were derived from the SemRep output during Phase One were iteratively expanded to include descendants up to 10 steps away from the initial concept, resulting in over 22 million unique triplets. When comparing these triplets to those resulting from TOKEn, there were still no exact matches. As a result, we examined the intersection of unique CUIs comprising the TOKEn and SemRep triplets, ignoring the linking semantic relationships and any descendants. The TOKEn and SemRep triplets were comprised of 47 and 2513 unique CUIs, respectively. Of these, only one CUI corresponding to “Chronic Lymphocytic Leukemia Refractory” was in both data sets. 18/121 (14.9%) and 27/6136 (0.44%) of the unique concept-concept pairs extracted from the TOKEn and SemRep triplets, respectively, contained this concept. Further review of these results by our SMEs yielded a number of qualitative findings, which will be further described in the Discussion section, and that were explanatory as to potential reasons for this continued lack of intersection between the two sets of CKCs.
Discussion
Is the conceptual knowledge that can be extracted from the published biomedical literature using SemRep syntactically and semantically comparable to that extracted from common ontologies using TOKEn?
While it was possible to compare the CKCs extracted from the published literature using SemRep to those previously generated using TOKEn, the difference in relationship types and the need to map between them from Semantic Network relationship types to those found in the NCI Thesaurus and SNOMED-CT made the process significantly resource-intensive. Furthermore, we heuristically observed that the use of Semantic Network relationship types in the CKCs generated using SemRep limits the expressiveness of the linkages between included concepts in comparison to those relationship types available in other knowledge sources, such as those used by TOKEn.
Is the conceptual knowledge encoded from the published biomedical literature significantly different from that found in ontological knowledge sources?
Our findings would initially appear to indicate that the CKCs generated using SemRep were significantly different than those generated using TOKEn. However, heuristic analyses of the CKCs that intersected between the two sets based upon the occurrence of a single common initial or terminal concept led us to conclude that a number of factors contributed to the lack of shared knowledge constructs, namely:
Mapping granularity mismatch: The granularity of mappings between “raw” concepts and ontology-anchored concepts differed greatly between the UMLS Knowledge Source Server (UMLSKS) as used by TOKEn and SemRep, with the UMLSKS employing much more granular or specific mappings. For example, the average distance to the root for CUIs in the TOKEn and SemRep CKCs was 5.2 and 3.8, respectively.
Processing scope mismatch: The scope of mappings and/or knowledge anchored reasoning varied between TOKEn and SemRep, with the TOKEn approach being holistic across all database schema-defined concepts, and the SemRep approach being limited to lexically distinct phrases. For example, though the chromosomal abnormality del(17p13) is mentioned in the same abstract as refractory CLL, concepts that are transitively related in the TOKEn data set, they are never in the same sentence and thus not related by SemRep.
Semantic context mismatch: The assignment of semantic relationships by the two approaches was materially different, which led to limited comparability across the two sets of CKCs. That is, SemRep seems to use a rule-based assignment of Semantic Network relationships based on the syntax and semantics of the extracted concepts, while the TOKEn post-processing used an algorithmic approach that classified concepts related via Metathesaurus relationships and assigned the Semantic Network relationships post-hoc.
We believe that collectively, the three preceding mismatch types, and their quantitative and qualitative manifestations illustrate a scenario that we have labeled as Conceptual Dissonance. In this phenomenon, CKCs derived from two or more conceptual knowledge sources of qualitatively comparable scope, granularity and semantics, using commonly available tools, cannot be readily integrated or compared using automated methods. In response to this challenge, we believe that the development of automated methods for the detection and normalization of Conceptual Dissonance are necessary. One potential approach to addressing this challenge is the use of graph-theoretic methods for semantic normalization, leveraging the isomorphic nature of sub-sets of semantically similar knowledge in the graph-like representations of large-scale ontologies or terminologies. Such an approach has been validated in prior studies concerning semantic search across disparate knowledge sources15, and we intend to apply it to the problem space described in this report as part of our future work.
Conclusion
The ultimate goal of the work described in this manuscript is to further refine a novel approach to employing conceptual knowledge sources in the support of translational hypothesis discovery and testing. Though we initially intended to demonstrate the ability to improve the scalability and reproducibility of such techniques, our findings have instead led to the identification of a phenomenon of interest that we have labeled as Conceptual Dissonance. This presents a unique challenge to the practical application of conceptual knowledge engineering approaches in support of translational research, and warrants further exploration.
Figure 1:
Overview of study phases.
Table 2:
Example of a TOKEn-based triplet.
| TOKEn CKC | Transitive Triplets |
|---|---|
| Gain of Chromosome 6 - [may be cytogenetic abnormality of disease] - stage I childhood liver cancer - [disease may have finding] - Alanine aminotransferase increased | Gain of Chromosome 6 - [may be cytogenetic abnormality of disease] - stage I childhood liver cancer |
| stage I childhood liver cancer - [disease may have finding] - Alanine aminotransferase increased |
Acknowledgments
This work was support by the NCI (2 P01CA081534-07A1 and 1R01CA134232-01).
References
- 1.Mathew JP, Taylor BS, Bader GD, Pyarajan S, Antoniotti M, Chinnaiyan AM, et al. From bytes to bedside: data integration and computational biology for translational cancer research. PLoS computational biology. 2007 Feb 23;3(2):e12. doi: 10.1371/journal.pcbi.0030012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Payne PR, Borlawsky TB, Kwok A, Dhaval R, Greaves AW, editors. AMIA 2008 Translational Bioinformatics Summit. San Francisco: 2008. Ontology-anchored Approaches to Conceptual Knowledge Discovery in a Multi-dimensional Research Data Repository. [PMC free article] [PubMed] [Google Scholar]
- 3.Payne PR, Borlawsky TB, Kwok A, Greaves A. Supporting the Design of Translational Clinical Studies Through the Generation and Verification of Conceptual Knowledge-anchored Hypotheses. AMIA 2008 Annual Symposium; Washington, D.C. 2008. [PMC free article] [PubMed] [Google Scholar]
- 4.Butte AJ. Medicine. The ultimate model organism. Science. 2008 Apr 18;320(5874):325–7. doi: 10.1126/science.1158343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu H, Motoda H. Feature Extraction, Construction and Selection: A Data Mining Perspective. Norwell, MA: Kluwer Academic Publishers; 1998. [Google Scholar]
- 6.Joseph P, Bruce GB. Ontology-guided knowledge discovery in databases. Proceedings of the international conference on Knowledge capture; Victoria, British Columbia, Canada. 2001. [Google Scholar]
- 7.SNOMED-CT (www.snomed.org)
- 8.NCI Thesaurus (ncicb.nci.nih.gov)
- 9.Payne PR, Mendonca EA, Johnson SB, Starren JB. Conceptual knowledge acquisition in biomedicine: A methodological review. J Biomed Inform. 2007 Oct;40(5):582–602. doi: 10.1016/j.jbi.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liou YI. Knowledge acquisition: issues, techniques, and methodology. Orlando, Florida, United States: ACM Press; 1990. [Google Scholar]
- 11.Kipps TJ. Immunobiology of chronic lymphocytic leukemia. Current opinion in hematology. 2003 Jul;10(4):312–8. doi: 10.1097/00062752-200307000-00010. [DOI] [PubMed] [Google Scholar]
- 12.Grever MR, Lucas DM, Dewald GW, Neuberg DS, Reed JC, Kitada S, et al. Comprehensive assessment of genetic and molecular features predicting outcome in patients with chronic lymphocytic leukemia: results from the US Intergroup Phase III Trial E2997. J Clin Oncol. 2007 Mar 1;25(7):799–804. doi: 10.1200/JCO.2006.08.3089. [DOI] [PubMed] [Google Scholar]
- 13.Greaves AW, Payne PRO, Rassenti L, Kipps TJ, editors. CRC Tissue Core Management System (TCMS): Integration of Basic Science and Clinical Data for Translational Research. AMIA 2003 Annual Symposium; Washington, D.C. 2003. [PMC free article] [PubMed] [Google Scholar]
- 14.Payne PRO, Greaves AW, Kipps TJ, editors. CRC Clinical Trials Management System (CTMS): An Integrated Information Management Solution for Collaborative Clinical Research. AMIA 2003 Annual Symposium; Washington, D.C. 2003. [PMC free article] [PubMed] [Google Scholar]
- 15.Zhong J, Zhu H, Li J, Yu Y. Conceptual Structures: Integration and Interfaces. In: Goos G, Hartmanis J, van Leeuwen J, editors. Lecture Notes in Computer Science. Berlin: Springer Berlin / Heidelberg; 2002. pp. 92–106. [Google Scholar]