Abstract
PubAtlas (www.pubatlas.org) is a web service and standalone program providing literature maps for the biomedical research literature. It accepts user-defined sets of terms (PubMed queries) as input, and permits ‘BLASTing’ of one set against another: for all terms x and y in these sets, deriving the results of the pairwise intersections x AND y. This all vs. all capability extends PubMed with a literature analysis interface. Correspondingly, the basic form of literature map that PubAtlas provides for exploring associations among sets of terms is an interactive tabular display, in heatmap/microarray format.
PubAtlas supports development of specialized lexica -- hierarchies of controlled terminology that can represent sets of related concepts or a ‘user-defined query language’. PubAtlas also provides historical perspectives on the literature, with temporal query features that highlight historical patterns. Generally, it is a framework for extending the PubMed interface, and an extensible platform for producing interactive literature maps.
Finding Associations in PubMed
PubMed2 has become an essential resource in many fields of the biomedical research. It provides a web browser interface for searching the millions of documents in the MEDLINE database, providing a variety of different indexes and features. Because of the rapidly increasing volume of publications, ability to search and summarize the scientific literature has become increasingly important.
A consequence of this information explosion is an increasing interest in cross-disciplinary efforts, linking fields that have been poorly connected. These efforts will require new ways of gaining perspective on relationships between different literatures, and seeing how they can be unified in new ways.
The largest ‘association base’ we have is the literature, and it is our richest medium for connecting information. PubMed/MEDLINE is already used to link information from a multitude of sources. Still, there are many extensions.
Literature Mapping with PubAtlas
PubAtlas is an extension of PubMed that focuses on exploration of information associations. Given one or more sets of user-specified terms (PubMed queries), it generates a literature map -- an active visual interface that provides statistics about the documents retrieved by PubMed for each term. The most basic form of literature map is a cross-tabular layout, a heatmap measuring associations between terms. The table includes co-occurrence measures, reflecting the number of documents in which two terms occur.
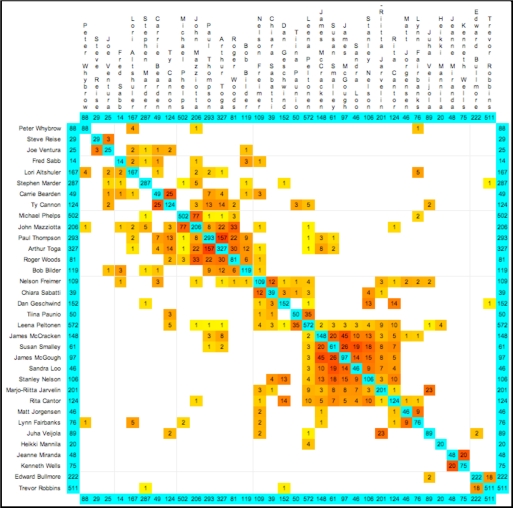
Figure 1 shows sample PubAtlas output, highlighting stronger associations in the literature.
Figure 1.
PubAtlas output showing literature co-occurrence patterns among investigators in the Consortium for Neuropsychiatric Phenomics (CNP) at UCLA. This summary is the result of BLASTing the lexicon of investigators against itself -- answering queries x AND y for all pairs of investigators x and y. Notice that neuroscience investigators cluster in the upper left, while geneticists cluster in the lower right. Robert Bilder spans the neuroscience spectrum, while Nelson Freimer spans the full spectrum of interests.
By their nature, PubMed queries are imprecise specifications for concepts, so these associations are also imprecise. However, PubAtlas is useful for gaining an overview of the literature, and of broad patterns in the coverage of the literature. PubAtlas also offers other views, and permits interactive access to literature in PubMed.
BLASTing queries have promise where the lexica are from different disciplines. We have found it useful in phenomics -- the systematic study of phenotypes on a genome-wide scale -- in identifying associations across multiple scales of science. Phenotypes are measurable traits or characteristics, and important ones acquire stylized terminology, and often this terminology can be used as a very precise ‘query language’, permitting useful associations to be obtained from PubMed. Carefully chosen terminology can span translational boundaries, and link related concepts in different organisms. Furthermore, PubAtlas may also aid with knowledge management or social connections in science, such as linking researchers with complementary interests, or studying collaboration histories. It can provide maps of interactions not only between concepts, but also between researchers. These capabilities all are useful in interdisciplinary research fields like phenomics.
PubAtlas obeys access restrictions on PubMed, including restrictions on frequency of queries. It uses cacheing and optimization to avoid unnecessary queries, and uses algorithmically sophisticated representations of hit lists and streaming execution to provide reasonable performance. When given two term sets of length m and n, it submits m+n queries to PubMed. Thus, although not fast for term sets with hundreds or thousands of queries, PubAtlas can support term sets of this magnitude.
PubAtlas is not just a visual front-end for PubMed. One obvious difference is that it also provides temporal information, an important dimension of scientific literature. Also however PubAtlas provides bulk query features, including management of user-defined concept lexica and query expansion7,8. It can be extended to include other user-defined query features. A goal of PubAtlas is that literature maps be useful as machine-generated ‘interactive review papers’. Although not of the quality or depth achievable by a human expert, they can be tailor-made on demand and reflect all indexed literature.
Existing Extensions of PubMed
Many extensions have been developed for the PubMed interface, although its current interface is very popular. It is most effective in specific modes of use, since documents responding to a query are provided in modest screenfuls, with more recent publications first. Many proposals emphasize improvements for exploratory searches or specialized kinds of query processing.
This situation has spawned ‘Pub....’ interfaces that provide some extension of the interface for PubMed/MEDLINE search. The number of extensions is large, and includes for example: AliBaba (PubMed as a graph), Anne O’Tate (search results summarized by key features), Arrowsmith (finding disconnected literatures), BioIE (extracting informative sentences), ClusterMed (clustering of search results), ConceptLink (spatial mapping of search results by concept), GoPubMed (PubMed and GO linking), HubMed (a ‘new interface’, with novel features and RSS), PubFocus (ranking of results and incorporating controlled vocabularies), PubGene (gene-related query extensions), PubMed Assistant (improving interaction via better interface design), PubMed Interact and SLIM (improving interaction via web interface constructs), PubMed On Tap (improved PDA access), PubReMiner (summarizing distributions of journals, authors, and keywords), Relemed (relevance scoring), and XplorMed (summarizing the subjects of search results).
Three systems that influenced our ideas initially were VisualNet1 (an early ‘thematic map’ interface with a geographic flavor), PubMatrix5 (a heatmap interface reflecting co-occurrence strengths for general terms), and PubNet4 (an interactive graph interface reflecting co-occurrence strengths among terms specifying authors or title/abstract keywords).
PubAtlas
PubAtlas draws on many of the systems above. It tries to go beyond them in scale (permitting more detailed exploration of larger tables than PubMatrix, say), new features (temporal analysis, query expansion), methodology (predefined lexica, optimization, etc.). It also draws on our earlier systems PubBrain (www.pubbrain.org) and PubGraph (www.pubgraph.org).
Each PubAtlas lexicon is a set of term definitions of the form t : q, where t is a ‘term’ and q is a corresponding PubMed query. A term t is a name of a specific concept of interest, and q is a query that identifies relevant publications. Ideally, to represent its concept exactly, q would be chosen so as to have perfect precision and recall. That is, q should retrieve all relevant publications for the concept denoted by t, and only these. Although achieving this ideal is not usually achievable, imprecise queries often do very well in identifying patterns. Furthermore their precision can be improved by using powerful PubMed query features, such as MeSH terms3. Because of the challenges in attaining high precision and recall with PubMed queries, though, it can require considerable time and experience to develop high-quality lexica.
PubAtlas permits use of specific MeSH subhierarchies as lexica. If one wants to explore the literature for neurotransmitter receptors, for example, one can use the MeSH Receptor, Neurotransmitter subhierarchy (D12.776.543.750.720). The result of finding out which CNP investigators have published work on any of 177 MeSH neurotransmitter-related terms is shown in Figure 3.
Figure 3.
PubAtlas output showing the result of BLASTing a lexicon of MeSH neurotransmitter-related terms against CNP investigators. PubAtlas can address larger-scale queries, involving hundreds of terms. Zooming is possible with the controls on the left, and hovering over over a particular entry produces a display summarizing history.
Currently about 50 lexica have been defined for use within CNP, and about half of these are MeSH hierarchies. PubAtlas also allows definition of a global context — a background query, relative to which all user queries are framed. In other words, given a context query c, every lexicon query q is rewritten to (q AND c), filtering its results. As is suggested by Figure 4, the context can be limited to year ranges, publications types, and so forth, but it also can involve very advanced PubMed queries.
Figure 4.

PubAtlas allows active display of histories of associations. These images show collaboration among CNP investigators in even years from 1998 to 2008. Growth patterns give insight; collaboration has jumped with the arrival of each investigator. Until 2002 neuroscience (upper left quadrant) and genetics (lower right) had few collaborations, but since then cross-disciplinary effort has expanded rapidly. Informatics-based collaborations (upper-left corner) have also expanded significantly.
The table resulting from a BLASTing operation can be viewed in different ways: as a distribution, graph, movie, etc. Each such view is a literature map, rendering information as an information geometry that can highlight patterns and facilitate certain kinds of exploration. Since temporal information is an important part of the literature, many different kinds of literature maps are possible. Currently PubAtlas summarizes associations in a tabular format, but will include views like those in PubBrain and PubGraph.
Both BLASTing and heatmap/microarray maps have proven their worth as exploratory tools in bioinformatics, and have accumulated a vast set of related tools and techniques. PubAtlas suggests new adaptations to literature mining.
PubAtlas permits Concept Query, and Phenomics
The central idea in the development of PubAtlas is one of allowing scientists to define their own concepts (controlled vocabulary in the lexica), to use these concepts routinely in querying, and to be able to visualize patterns of association among them. Like MeSH terms3, these hierarchical lexica can define concept spaces in which BLASTing is a very natural process for finding associations.
If well-constructed, PubAtlas lexica can function as user-defined query languages. BLASTing them (particularly with the use of contexts) is then a way of exploring their combination. The ability to develop new concepts and explore their connections is a primary goal of Phenomics -- the systematic study of phenotypes on a genome-wide scale.
Phenotypes are detectable/measurable characteristics of an organism -- outward manifestations of the interaction of its genotype with its environment -- including diseases and disorders. For example, neuropsychiatric disorders are among the most costly and disabling illness phenotypes. They pose enormous challenges to biomedical discovery, due to their complexity and the broad gaps between laboratory science models and the clinical phenomena they seek to explain. Bridging these gaps requires interdisciplinary work, and particularly: a vocabularly of concepts shared by collaborators, and vehicles for exploring and mapping associations among these.
Neuroscientific phenotypes by definition involve measurements that can be difficult to summarize with traditional informatics tools. For example, neuropsychiatric research connects both highly mathematical concepts about geometry with very empirical psychiatric concepts. A goal of phenomics projects is often to associate phenotype concepts across many scales with concepts at the level of the genome. A set of phenotypes defines a language; scientists collaborating in this area often want to explore the literature for results about these phenotypes, and communicate using this language.
Lexica also give a way to provide query expansion6,7,8. For example, the term ‘N-back test’ can expand to the query (“nback” OR “n back” OR “wisconsin card sorting” OR “Sternberg” OR “Stroop” OR “choice reaction time” OR “paced auditory serial addition” OR “digit span” OR “delayed match to sample”). Although this query expansion mechanism is similar to PubMed’s Automated Term Mapping facility, it can be domain-specific or even user-specific. Similarly a hierarchical lexicon can be used to relax queries, and produce more hits. They can also be expanded to cover dfferent but related concepts,6,7,8.
Conclusion
If we view the scientific literature as an ‘association base’, it is clear that there are many ways we can extend it, and improve our ability to find scientific connections. Although PubMed is already used to link information from a multitude of sources, a number of previous efforts have shown ways to improve upon the PubMed interface.
There are different ways to look at PubAtlas: a visual front-end for PubMed, a means for visualizing many query results (à la BLAST), and a historical or temporal query scheme (providing animations that show change over history). Moreover PubAtlas is a system for analyzing lexica -- hierarchical sets of controlled terminology or ‘user-defined query languages’, and a system for producing machinegenerated interactive review papers. Generally, PubAtlas is a framework for extending PubMed -- adding support for indexing, new query expansion methods, and interactive literature maps.
The implementation of PubAtlas includes handling of hierarchical lexica, user-specified contexts for queries, cacheing, query optimization, and aspects of analyzing associations. These capabilities, and the underlying architecture, overlap with those of database management systems, and the BLASTing discussed here is an important kind of query. An extensible database platform with support for new indexing schemes, query expansion methods, and modes of display is a natural architecture for continued growth.
Our development of PubAtlas has been motivated by needs in the emerging field of phenomics -- the systematic study of phenotypes on a genome-wide scale. Phenotypes are naturally modeled as literature queries, and phenomics is naturally interdisciplinary, touching on many fields and literatures. PubAtlas was conceived as a way to span scientific disciplines, formalize shared vocabulary, and support analysis of associations. PubAtlas is currently being developed as research infrastructure for this purpose at the UCLA Center for Computational Biology and Consortium for Neuropsychiatric Phenomics.
Figure 2.
Two small lexica and their PubAtlas result, when run in the context “PubMed”[TIAB]. The first is a list of systems that extend the PubMed interface, and the second is a list of some characteristic features. The table shown is the PubAtlas output summarizing results of all pairs of queries x AND y, where x is a system and y is a feature, omitting rows and columns having no intersection. For this is simple pair of lexica, with no pretense of completeness, the resulting machine-generated table can serve as an interactive review paper - an overview of publications about these systems. The table is also a web-based interface that spawns interactions with PubMed.
Footnotes
This research supported by NIH grants RL1LM009833, UL1DE019580 (UL1RR024911), P20RR020750, P20MH065166, RO1MH082795, and the UCLA Center for Computational Biology (CCB).
References
- 1.Boulos MNK, VisualNet The use of interactive graphical maps for browsing medical health Internet information resources. International Journal of Health Geographics. 2003;2(1) doi: 10.1186/1476-072X-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.PubMed:www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=pubmed
- 3.MeSH (Medical Subject Headings) http://www.nlm.nih.gov/mesh/
- 4.Douglas SM, Montelione GT, Gerstein M. PubNet: a flexible system for visualizing literature-derived networks. Genome Biology. 2005;6:R80. doi: 10.1186/gb-2005-6-9-r80. (formerly www.pubnet.org) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker KG, Hosack DA, Dennis G, Jr, Lempicki RA, Bright TJ, Cheadle C, Engel J. PubMatrix: a tool for multiplex literature mining. BMC Bioinformatics. 2003 Dec 10;4:61. doi: 10.1186/1471-2105-4-61. (formerly www.pubmatrix.org) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qiu Y, Frei HP. ‘Concept-based query expansion’. Proc ACM SIGIR. 1993 [Google Scholar]
- 7.Chu Wesley W, Zhenyu Liu, Wenlei Mao Q. Zou ‘KMeX: A Knowledge-Based Digital Library for Retrieving Scenario-Specific Medical Text Documents’. In: Feng D, editor. Biomedical Information Technology. Elsevier; 2007. [Google Scholar]
- 8.Zhenyu Liu, Chu Wesley W. ‘Knowledge-based query expansion to support scenario-specific retrieval of medical free text. Info Retrieval. 2007 [Google Scholar]