Abstract
Metabolic network provides a unified platform to integrate all the biological information on genes, proteins, metabolites, drugs and drug targets for a comprehensive system level study of the relationship between metabolism and disease. In recent times, drug-target identification by in silico methods has emerged causing a phenomenal achievement in the field of drug discovery. This paper focuses on describing how microbial drug target identification can be carried out using bioinformatic tools. Specifically, it highlights the use of metabolic ‘choke point’ and ‘load point’ analyses to understand the local and global properties of metabolic networks in Pseudomonas aeruginosa and allow us to identify potential drug targets. We also list out top 10 choke point enzymes based on the load point values and the number of shortest paths. A non-pathogenic bacterial strain Pseudomonas putida KT2440 and a related pathogenic bacteria P.aeruginosa PA01 was selected for the network anlaysis. A comparative study of the metabolic networks of these two microbes highlights the analogies and differences between their respective pathways. System analysis of metabolic networks will help us in identifying new drug targets which in turn will generate more in-depth understanding of the mechanism of diseases and thus provide better guidance for drug discovery.
1. INTRODUCTION
Metabolic pathways are a central paradigm in biology. The metabolic network is of special interest among the different types of biological networks because it can integrate the experimental data for different types of molecules (transcriptomics for genes, proteomics for enzymes and metabolomics for metabolites). Moreover metabolites are more closely related to the phenotype of an organism and the health and disease states of human can be described more meaningfully by the metabolic state of human cells, tissues, organs and the organism as a whole. Thus network-based pathways are emerging as an important paradigm for analysis of biological systems. Despite the advent of the high-throughput techniques sparked by the genomics revolution, discovery and development of new antibiotics has lagged in recent years due to the serious problem of evolution of antibiotic resistance [1].
Comparative genomics has provided a gradual increase in large-molecule targets. It has actually created the new problem of how to select potential drug targets from a larger pool [2]. Putative targets must be essential for the survival of the pathogen and enzymes that do not have human homologs may also be attractive drug targets [3]. Among the numerous criteria used in the selection of antimicrobial compounds, certain criteria are clearly influenced by the choice of target. These include breadth of spectrum, selectivity of the agent for microbial systems and frequency of resistance.
Anti-microbial targets can now be identified and evaluated by automatically comparing all relevant pathogen genomes and the human host genome. Genes that are conserved across different pathogens represent attractive target candidates for new broad-spectrum antibiotics [4]. Antibiotic resistance has increased over the past two decades and has reduced the usefulness of effective antibiotics.
Pseudomonas aeruginosa is a major life-threatening opportunistic pathogen that commonly infects immunocompromised patients. The organism is inherently resistant to many drug classes and is able to acquire resistance to all effective antimicrobial drugs and has the ability to adapt and thrive in many ecological niches, including humans [5]. Besides P. aeruginosa continues to be a major pathogen among patients with immuno-suppression, cystic fibrosis, malignancy, and trauma [6]. A thorough understanding of the metabolism of P. aeruginosa is thus pivotal for the design of effective intervention strategies. On the other hand, Pseudomonas putida KT2440 is a metabolically versatile and non-pathogenic saprophytic soil bacterium that has been certified as a biosafety host for the cloning of foreign genes. Although there is a high level of genome conservation (85%) with the pathogenic P.aeruginosa, key virulence factors including exotoxin A and type III secretion systems are absent in P.putida. The non-pathogenic nature of P. putida has been used potentially in agriculture, biocatalysis and bioremediation [7].
Prioritizing targets will therefore be an essential process in industrial drug discovery. Targets have become plentiful, yet new antimicrobial agents have been slow to emerge from this effort. A prerequisite for mapping metabolic pathways on genomic data is the annotation of proteins with metabolic information. These metabolic pathways are functional units of metabolic networks and help us in understanding a detailed analysis of network robustness and complex reaction networks. The commonly known Enzyme Commission (EC) numbers have been given to characterized proteins in order to classify the enzymatic chemical reactions of proteins.
Graph theory based pathway analysis will be very useful in analyzing metabolic networks consisting of reactions, metabolites and enzymes [8]. Metabolic networks can be represented as a metabolite graph consisting of nodes (metabolites) and edges (reactions) with large number of connecting links. Such representation of network allows the characterization of the metabolic pathways with respect to degree of metabolite (nodes) connectivity defined as possible number of reactions by a metabolite; and the degree of interconnectivity or average network diameter defined as the average shortest path length [9].
In our previous study, a differential genome analysis of metabolic enzymes in Pseudomonas aeruginosa for drug target identification was performed and identified potential drug targets in unique and common pathways [10]. In the present work, we extend our analysis to the identification of potential drug targets based on the concept of ‘load points’ and ‘choke points’. This approach enables to understand the local and global properties of the metabolic network thereby allowing us to identify potential drug targets in the pathogenic bacterium.
Using the current knowledge about proteins, an accurate prediction of protein druggability can capitalize on the huge investments already made in structural genomics initiatives by identifying highly druggable proteins and thereby leading to target identification and validation. While our current knowledge may be limited the ability to assess protein druggability in a fast and reliable manner is simply one of many tools that can help to streamline and enhance this process, especially when integrated with other computational and experimental approaches to target identification and validation.
2. METHODOLOGY
2.1. Load Point and Choke Point analyses
‘Load point’ of a metabolite in a metabolic network is defined as the ratio of number of k-shortest paths passing through the metabolite (enzyme) and its nearest neighbour links [11]. These load point values give a global view of the metabolic network and help in the anlaysis of the metabolic pathway reactions. Pathways that are highly connected in the metabolism of the cell tend to have high load values. Moreover the lethality of a metabolite/enzyme depends on the number of connections it has in the whole metabolic network [12]. Enzymes with large number of connections are found to be three times more essential than the proteins that interact with only a few other neighbors.
On the other hand, ‘choke point’ enzymes are those taking part in a reaction that consumes unique specific metabolite (substrate) or uniquely produces specific metabolite (product) in the metabolic network [13]. These choke point enzymes are crucial points in the metabolic pathway and inactivation of these important enzymes may lead to the disruption of the metabolic network of the bacterium.
2.2. Identification of Potential Drug Targets
To identify potential drug targets, chokepoint and load point analyses was carried out in the metabolic network of P.aeruginosa PAO1. The complete genome sequence of P.aeruginosa PAO1 and Homo sapiens is available [5, 14]. Metabolic pathway information was obtained from Kyoto Encyclopedia of Genes and Genomes (KEGG) [15]. In our previous study [10], we listed out a total of 361 enzymes as potential drug targets by differential genome analysis between the pathogen P.aeruginosa and the host H.sapiens by subjecting to BLAST [16] search against human protein sequence database at an E-value cutoff of 10−2. Here we extend our study to choke point and load point analysis using the Pathway Hunter Tool (PHT) [17] to identify enzymes that are essential for the bacterial network.
Using the above tool we calculated shortest path distribution, the average path length and average alternate paths in the P.aeruginosa metabolic network. The important aspects of global similarity and local similarity need to be considered while performing choke point analysis since higher the similarities smaller will be the network diameter and the average degree of nodes [17]. In this study we chose the local similarity score as 20% and the global similarity score as 10%. We then identified top 10 choke point enzymes based on the number of shortest paths using the tool. A comparative study was performed between the pathogen choke point enzymes with the human metabolic network to differentiate human choke points with that of the bacterial choke points. Finally for the predicted list of choke points in the pathogen we performed a homology search against the human genome using BLAST.
We also carried out a comparative study of metabolic pathways based on shortest path analysis between the pathogenic bacterium P.aeruginosa and non-pathogenic P.putida to highlight the analogies and differences between their respective pathways. We calculated the shortest path distribution, the average path length and average alternate paths.
3. RESULTS AND DISCUSSION
Network models are crucial for shaping our understanding of complex networks and help to explain the origin of observed network characteristics. Based on the network characteristics, metabolite information can be used to calculate the k-shortest paths between metabolites (substrate and product). Distance in metabolic networks can be measured in terms of path length, which represents the number of links passing between two nodes. As there are many alternative paths between two nodes, the shortest path with the smallest number of links between the selected nodes has a special property. The average path length represents the mean over the shortest paths between all pairs of nodes in the entire network of the bacterium [9].
3.1. Metabolic Network Topology of P.aeruginosa and P.putida
A comparative study of metabolic pathways based on shortest path analysis was carried out between P.aeruginosa and P.putida. Analysis of the metabolic network is based on shortest path length since large numbers of biochemical reactions follow shortest path rather than longer paths. The metabolic network for P.aeruginosa used in this study included 996 reactions and 1063 metabolites with network diameter of 33 and the average degree distribution (Connectivity) of 3.09 whereas metabolic network of P.putida includes 992 reactions and 1087 metbolites with network diameter of 31 and the average degree distribution of 3.0
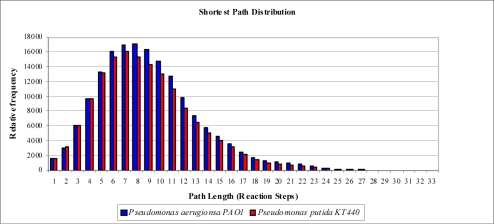
P.aeruginosa and P.Putida have more or less the same network diameter and the same average degree distribuiton of ~3.0 (average connectivity) and approximately the same number of reactions. The average path lengths (shortest path/k-shortest path) for the P.aeruginosa and P.Putida are 8.69/9.06 and 8.43/8.79 respectively. The shortest path distribution for P.aeruginosa and P.Putida is shown below in Figure 1.
Figure 1.
Graph showing the shortest path distribution in P.aeruginosa PA01 and P.Putida KT2440.
Few nodes with a very large number of connectivity links are termed as hubs as they hold many nodes together. However, even the complete removal of nodes that act as hubs that are highly linked in the network can be adjusted through redundancy or alternate pathways that maintain the usual flow of metabolites in the cell. Within a model of a highly interconnected network: if one part of a web is perturbed, other compensatory changes in flow are likely to occur as well, analogous to a ripple effect spreading through the network.
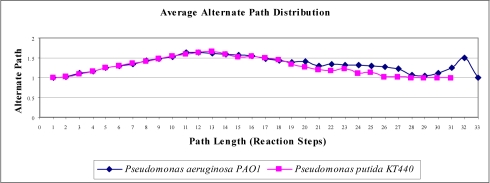
Alternate pathway lengths can thus be used to characterize the large-scale properties of metabolic networks. A contrast in the topological behaviour between the selected model organisms is seen where a shift in the average alternate path in both the organism is observed. P.aeruginosa has a greater number of alternate paths between path lengths 19 and 28 whereas P.Putida has more alternate paths at path lengths between 12 and 18 (Figure 2). This difference in the average alternate path between the two microbes may imply biological significance which can be explored through the network analysis.
Figure 2.
Average alternate path distribution in P.aeruginosa and P.Putida.
It is important to keep track of alternate paths in the metabolic network which is an indicator of the ability of the pathogen to survive under extreme conditions. Most of the enzymes can be replaced by alternative reactions that utilize the same substrate or produce the specific product, thereby allowing a given pathway to operate [18]. Hence blocking a path may not be vital as pathogens can make use of an alternate path performing similar functions. We further carried out ‘load point’ analysis for the two microbes and identified top 10 metabolite load points in them. The loads on the metabolites differ between the two bacterial networks (Table 1 and Table 2).
Table 1.
Top 10 Metabolite load points based on incoming load value in P.aeruginosa (pae) and P. putida (ppu).
| Metabolites | Rank inpae | Rank inppu | Load (in)pae | Load (in)ppu | Links (in)pae | Links (in)ppu | k-shortest path (in)pae | k-shortest path (in)ppu |
|---|---|---|---|---|---|---|---|---|
| 4-Fumarylacetoacetate | 1 | 1 | 2.4963 | 2.59503 | 1 | 1 | 11822 | 11593 |
| 4-Maleylacetoacetate | 2 | 2 | 2.4591 | 2.55418 | 1 | 1 | 11390 | 11129 |
| Homogentisate | 3 | 3 | 2.4422 | 2.53082 | 1 | 1 | 11200 | 10872 |
| GTP | 4 | 5 | 2.3215 | 2.17462 | 1 | 1 | 9926 | 7614 |
| D-Ribulose5-phosphate | 5 | 8 | 2.2981 | 2.01551 | 2 | 3 | 19393 | 19482 |
| 3-Phospho-D-glyceroylphosphate | 6 | 6 | 2.2072 | 2.16432 | 2 | 2 | 17709 | 15072 |
| O-Phospho-L-serine | 7 | 7 | 2.1005 | 2.03306 | 1 | 1 | 7958 | 6609 |
| 2-Phospho-D-glycerate | 8 | 9 | 1.9808 | 1.98454 | 2 | 2 | 14121 | 12592 |
| Dephospho-CoA | 9 | 4 | 1.9044 | 2.21238 | 2 | 2 | 13082 | 15814 |
| CTP | 10 | 15 | 1.8358 | 1.74179 | 2 | 2 | 12215 | 9878 |
| D-Xylulose 5-phosphate | 14 | 10 | 1.7219 | 1.94826 | 4 | 3 | 21800 | 18215 |
Table 2.
Top 10 Metabolite load points based on outgoing load value in P.aeruginosa (pae) and P. putida (ppu).
| Metabolite | Rank inpae | Rank inppu | Load (out) pae | Load (out)ppu | Links (out)pae | Links (out)ppu | k-shortest path (out) pae | k-shortest path (out)ppu |
|---|---|---|---|---|---|---|---|---|
| 4-Maleylacetoacetate | 1 | 1 | 2.49628 | 2.59503 | 1 | 1 | 11822 | 11593 |
| 3-Phospho-D-glyceroylphosphate | 2 | 2 | 2.22454 | 2.1889 | 2 | 2 | 18018 | 15447 |
| gamma-L-Glutamyl-L-cysteine | 3 | 8 | 2.20843 | 1.93855 | 1 | 1 | 8865 | 6013 |
| 2-Phospho-D-glycerate | 4 | 4 | 2.0159 | 2.01989 | 2 | 2 | 14625 | 13045 |
| UDP | 5 | 9 | 1.94658 | 1.93033 | 4 | 4 | 27291 | 23855 |
| D-Ribulose5-phosphate | 6 | 3 | 1.90186 | 2.02557 | 3 | 3 | 19573 | 19679 |
| 4-Fumarylacetoacetate | 7 | 7 | 1.83902 | 1.94113 | 2 | 2 | 12254 | 12057 |
| UDP-N-acetylmuramoyl-L-alanyl-D-gamma-glutamyl-meso-2,6-diaminopimelate | 8 | 5 | 1.83592 | 1.97737 | 1 | 1 | 6108 | 6251 |
| Homogentisate | 9 | 10 | 1.78676 | 1.87938 | 2 | 2 | 11630 | 11335 |
| 3-Phospho-D-glycerate | 10 | 14 | 1.75145 | 1.76915 | 4 | 4 | 22453 | 20304 |
| D-Xylulose5-phosphate | 11 | 6 | 1.73365 | 1.9646 | 4 | 3 | 22057 | 18515 |
Although the degree of metabolites and its load in both the organisms are more or less similar but they differ based on the load points. From the above point of view ‘load points’ provide a more global measure of metabolic activity than the local connectivity analysis. The difference in the load values between the above two organisms suggest the need for deciphering the metabolic network topology in a more significant manner.
3.2. Top 10 choke point enzymes in Pseudomonas aeruginosa
Choke point analysis was carried out for the whole metabolic network using the Pathway Hunter Tool. We had reported a total of 361 enzymes in both unique and common pathways of P.aeruginosa as potential drug targets [10]. Here we compared these 361 enzymes (50 enzymes in unique pathways and 311 enzymes in shared pathways) to that of the choke point enzymes obtained in the above metabolic network tool analysis. This approach resulted in 227 targets matching with the previously reported 361 targets of which 25 targets belong to the unique pathways of the pathogen. The remaining 202 targets matched with that of the enzymes in the shared pathways between the pathogen and human. A total of 63% of proposed drug targets are choke point reactions in the P.aeruginosa genome. We identified top 10 choke point enzymes in P.aeruginosa using the pathway hunter tool. These enzymes were ranked based on the number of shortest paths (Table 3).
Table 3.
Top 10 Choke point enzymes in P. aeruginosa ranked by number of shortest paths.
| Enzyme Id | Enzyme Name | Gene Ids | Load value (in) | Load value (out) | k-Shortest paths (in) | k-Shortest paths (out) | Human choke point | Top BLAST hit (identity) (%) |
|---|---|---|---|---|---|---|---|---|
| 2.4.2.7* | Adenine phosphoribosyl transferase | PA1543 | 1.11 | 1.89 | 74459 | 74459 | No | NoHomologue |
| 2.4.2.8 | Hypoxanthine phosphoribosyl transferase | PA4645 | 0.85 | 1.52 | 60639 | 60639 | Yes | 26% |
| 2.7.4.6 | Nucleoside-diphosphate kinase | PA3807 | 1.47 | 1.17 | 55297 | 55297 | Yes | 48% |
| 2.7.4.14 | Cytidylate kinase | PA3163 | 1.37 | 1.59 | 50169 | 50169 | No | NoHomologue |
| 6.2.1.1* | Acetate-CoAligase | PA0887 PA1997 PA2555 PA4733 |
−0.03 | −0.16 | 48355 | 48355 | No | NoHomologue |
| 2.6.1.1 | Aspartatetransaminase | PA2828 PA3798 PA4722 PA4976 |
0.54 | 0.34 | 40463 | 40463 | Yes | 33% |
| 2.2.1.1 | Transketolase | PA0548 | 1.36 | 1.46 | 40019 | 40019 | Yes | 26% |
| 4.2.1.11 | Phosphopyruvate hydratase | PA3635 | 2.28 | 2.07 | 38260 | 38260 | Yes | 54% |
| 3.6.1.7* | Acylphosphatase | PA0954 | 4.07 | 1.87 | 28547 | 28547 | No | NoHomologue |
| 6.3.5.4* | Asparagine synthase (glutamine-hydrolysing) | PA0051 PA2084 PA3459 |
0.03 | −0.05 | 25142 | 25142 | No | NoHomologue |
Choke point enzymes reported as potential drug targets in the common pathways shared between the pathogen and human.
Our approach identified top 10 choke point enzymes of which 4 of them were already reported as potential drug targets in our previous work. An additional criterion for being a chokepoint enzyme is that it must not have isozymes which would make it more likely to be a potential drug target. Comparing the choke point enzymes with the human genome sequence will help in differentiating pathogen choke points and the human choke point enzymes.
The top 10 choke point enzymes were BLASTp searched against human protein sequence database at an E-value cutoff of 10−2. Five out of ten enzymes were identified as choke points only in pathogen and not in human. The enzymes adenine phosphoribosyl transferase (EC 2.4.2.7), cytidylate kinase (EC 2.7.4.14), acetate-coA ligase (EC 6.2.1.1), Acylphosphatase (EC 3.6.1.7) and asparagine synthase (EC 6.3.5.4) do not share any significant homology with the human genome (Table 3). Therefore targeting these enzymes might cause the lethality of the pathogen. Adenine phosphoribosyltransferases catalyze the Mg2+ dependent reaction that transforms a purine base into its corresponding nucleotide and they are present in a wide variety of organisms. They play major roles in purine salvage among most living organisms. Most of them rely on multiple purine salvage pathways to replenish their purine nucleotides. A simultaneous inhibition of all the major purine salvage enzymes will be necessary for depleting purine nucleotides from these pathogenic organisms.
ATP or GTP formation by substrate level phosphorylation of ADP or GDP at the expense of the free energy of the thioester bond of acyl-CoA is a well known and much studied biochemical process. Enzymes (acetate-coA ligase) catalyzing this process play central roles in energy metabolism. Potential drug targets should adversely affect the pathogen but not the host human and therefore if the drug target has a homologous enzyme in human it should not be essential.
4. CONCLUSION
It must be noted that chokepoints may not be essential if they create unique intermediates to an essential product and exhibit alternate pathway reactions. Alternatively, a model might overestimate the number of redundant reactions or pathways; this can be due to errors in annotation and unaccounted regulation. Equally true, each component of the reaction network may be present in the target organism but not expressed under the conditions to be examined. The above discussed computational approaches herein yield a larger pool of candidates which are not biased, thereby providing a wider range of new potential drug targets.
References
- 1.Schmid MB. Novel approaches to the discovery of antimicrobial agents. Current Opinion in Chemical Biology. 1998;2:529–534. doi: 10.1016/s1367-5931(98)80130-0. [DOI] [PubMed] [Google Scholar]
- 2.Read TD, Gill SR, Tettelin H, Dougherty BA. Finding drug targets in microbial genomes. Drug Discovery Today. 2001;6:887–892. doi: 10.1016/s1359-6446(01)01914-6. [DOI] [PubMed] [Google Scholar]
- 3.Galperin MY, Koonin EV. Searching for drug targets in microbial genomes. Current Opinion in Biotechnology. 1999;10:571–578. doi: 10.1016/s0958-1669(99)00035-x. [DOI] [PubMed] [Google Scholar]
- 4.Galperin MY, Koonin EV. ‘Conserved hypothetical’ proteins: Prioritization of targets for experimental study. Nucleic Acids Research. 2004;32:5452–5463. doi: 10.1093/nar/gkh885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stover CK, Pham XQ, Erwin AL, et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 6.Hutchison ML, Govan JRW. Pathogenicity of microbes associated with cystic fibrosis. Microbes and Infection. 1999;1:1005–1014. doi: 10.1016/s1286-4579(99)80518-8. [DOI] [PubMed] [Google Scholar]
- 7.Nelson KE, Weinel C, Paulsen IT, et al. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environmental Microbiology. 2002;4:799–808. doi: 10.1046/j.1462-2920.2002.00366.x. [DOI] [PubMed] [Google Scholar]
- 8.Schuster S, Fell DA, Dandekar T. A general definition of metabolic pathways useful for systematic organization and analysis of complex metabolic networks. Nature Biotechnology. 2000;18:326–332. doi: 10.1038/73786. [DOI] [PubMed] [Google Scholar]
- 9.Barabasi AL, Oltvai ZN. Network biology: Understanding the cell’s functional organization. Nature Reviews Genetics. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 10.Perumal D, Lim CS, Sakharkar KR, Sakharkar MK. Differential genome analyses of metabolic enzymes in Pseudomonas aeruginosa for drug target identification. In Silico Biology. 2007;7:453–465. [PubMed] [Google Scholar]
- 11.Rahman SA, Schomburg D. Observing local and global properties of metabolic pathways: ‘Load points’ and ‘choke points’ in the metabolic networks. Bioinformatics. 2006;22:1767–1774. doi: 10.1093/bioinformatics/btl181. [DOI] [PubMed] [Google Scholar]
- 12.Jeong H, Tombor B, Albert R, Oltval ZN, Barabasi AL. The large-scale organization of metabolic networks. Nature. 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- 13.Yeh I, Hanekamp T, Tsoka S, Karp PD, Altman RB. Computational analysis of Plasmodium falciparum metabolism: Organizing genomic information to facilitate drug discovery. Genome Research. 2004;14:917–924. doi: 10.1101/gr.2050304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 15.Kanehisa M, Goto S, Kawashima S, Nakaya A. The KEGG databases at GenomeNet. Nucl. Acids Res. 2002;30:42–46. doi: 10.1093/nar/30.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altschul SF, Madden TL, Schaffer AA, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rahman SA, Advani P, Schunk R, Schrader R, Schomburg D. Metabolic pathway analysis web service (Pathway Hunter Tool at CUBIC) Bioinformatics. 2005;21:1189–1193. doi: 10.1093/bioinformatics/bti116. [DOI] [PubMed] [Google Scholar]
- 18.Perumal D, Lim CS, Chow VTK, Sakharkar KR, Sakharkar MK. A combined computational-experimental analyses of selected metabolic enzymes in Pseudomonas species. Int J Biol Sci. 2008;4:309–317. doi: 10.7150/ijbs.4.309. [DOI] [PMC free article] [PubMed] [Google Scholar]