Abstract
In October 2006, the National Institutes of Health launched a new national consortium, funded through Clinical and Translational Science Awards (CTSA), with the primary objective of improving the conduct and efficiency of the inherently multi-disciplinary field of translational research. To help meet this goal, the Ohio State University Center for Clinical and Translational Science has launched a knowledge management initiative that is focused on facilitating widespread semantic interoperability among administrative, basic science, clinical and research computing systems, both internally and among the translational research community at-large, through the integration of domain-specific standard terminologies and ontologies with local annotations. This manuscript describes an agile framework that builds upon prevailing knowledge engineering and semantic interoperability methods, and will be implemented as part this initiative.
Introduction
The National Institutes of Health (NIH) have defined translational research as the process by which “basic scientists provide clinicians with new tools for use in patients and for assessment of their impact, and clinical researchers make novel observations about the nature and progression of disease that often stimulate basic investigation”1. To promote such inter-disciplinary team science, the NIH launched a new national consortium, funded through Clinical and Translational Science Awards (CTSAs), in which participating sites will work together as a “discovery engine” to improve the conduct and efficiency of translational research2. One of the central foci of the CTSA consortium is concerned with the application of clinical and translational research informatics approaches in order to address issues surrounding usability, workflow and interoperability among information systems, and internal and external collaborators2. As part of its efforts satisfy this objective, the Ohio State University (OSU) Center for Clinical and Translational Science (CCTS) has launched a knowledge management initiative (KMI) that focuses on the integration of domain-specific standard terminologies and ontologies with local annotations to facilitate semantic interoperability among administrative, basic science, clinical and research computing systems. This manuscript describes the development of an agile knowledge engineering framework that will be implemented as part of the CCTS KMI.
Background
In the following section, we will briefly introduce the contributing areas of informatics practice and research, and associated gaps in knowledge that serve to inform the development of our framework.
Knowledge Engineering
Over the last several years, the definition of knowledge engineering (KE) has evolved from a process of transferring expert knowledge into a computational format for use by intelligent agents to a model-based perspective on software engineering. These models can be utilized to structure knowledge such that applications can effectively emulate the capabilities of a domain expert3. Though a complete review of KE methods and tools is beyond the scope of this manuscript, we will briefly describe a representative sample of such knowledge and practice. For further details concerning KE methods and theory, the authors recommend the recent reviews provided by Studer, et al.4, Choi, Song and Han5, and Payne, et al.6.
Two widely known KE approaches are CommonKADS7 and the Unified Problem-solving Method Language (UPML)8. CommonKADS is comprised of a set of methods intended to support the creation of models that capture the distinct features of a knowledge-based system (KBS), including: 1) aspects of the organizational environment in which the KBS will operate; 2) the types of knowledge required to solve a particular task (expertise or knowledge model); and 3) the system architecture and computational mechanisms. UPML is an architectural description language for KBSs that seeks to unify and generalize previously developed KE methodologies, including the CommonKADS expertise model. UPML models are comprised of elements such as ontology-anchored tasks, problem-solving methods and domain models, reasoning processes, and semantic inter-relationships. There are several open-source tools available to meet the operational needs of such KE techniques, including the Protégé Ontology Editor and Knowledge Acquisition System9 and Apelon Distributed Terminology System (DTS)10. Protégé is a standards-based system that implements a rich set of knowledge-modeling structures, including UPML. It also provides tools for the creation, visualization and manipulation of ontologies in various representational formats, and the construction of ontology-anchored domain models and knowledge-based applications. Similarly, the DTS is an integrated set of software components that provides for terminology editing and content management services. This platform is intended to support the interoperability of health information through the curation of rich networks spanning national and international data standards as well as local vocabularies.
Model-driven Semantic Interoperability
In addition to enabling semantic interoperability and harmonization across heterogeneous information systems and data sources throughout the OSU CCTS and translational research community at-large, the KMI also aims to capture locally relevant, domain-specific conceptual knowledge. The goal of this objective is to support hypothesis discovery in large-scale, integrative data sets, which is a common aim across the informatics efforts of many other CTSA programs.
The Object Management Group has created a software development strategy known as Model Driven Architecture (MDA)11, which focuses on the use of platform-independent models to describe the functionality of a given application. The design and use of such reference information models (RIMs) capable of enabling semantic interoperability and harmonization among multi-dimensional data sources has been addressed in numerous prior research and development efforts, including the NCI’s cancer Biomedical Informatics Grid (caBIG)12. We will use the Biomedical Research Integrated Domain Group (BRIDG) project, which is part of this initiative, as an exemplary case of such efforts13. This project has and continues to develop research-specific RIMs that are semantically annotated through the use of the centrally curated NCI Enterprise Vocabulary Service (EVS)14 to define the constituent components and reflect the relationships among them. The incorporation of such formal semantics ensures that nomenclature and meaning can be broadly understood and is reusable throughout the end-user community. In addition, the annotations developed as part of the BRIDG project are harmonized with those of other information models, such as the Clinical Data Interchange Standards Consortium (CDISC) Study Data Tabulation Model (SDTM). The BRIDG model has been designed and is curated during collaborative modeling sessions conducted in both real time and asynchronously via computer mediated methods. The logical model is represented as a Unified Modeling Language (UML) class diagram, which is constructed and annotated using standards-based modeling tools (e.g. Enterprise Architect).
There are two primary challenges associated with such RIM development methodologies that should be noted. First, domain experts with the technical expertise necessary to engage in the modeling process are often not readily available. As such, current best practices rely on having non-domain experts (e.g., knowledge engineers) employ systematic KE methods, such as those introduced earlier, in order to define the required information models. Second, the use of centrally curated terminologies and ontologies can make it difficult to build and subsequently employ locally relevant vocabularies in a timely manner. Methods intended to enable widespread semantic interoperability of both RIMs and related standard terminologies or ontologies while retaining the ability to simultaneously use locally relevant or curated vocabularies remain an open area of research.
Description of Proposed Framework
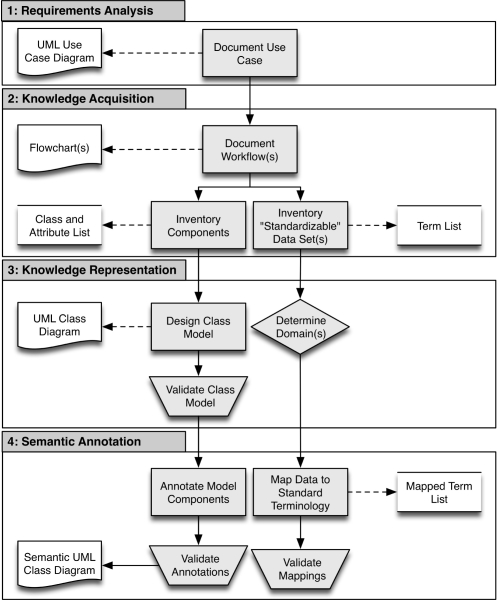
The preceding KE approaches and model-driven methods for ensuring semantic interoperability serve to provide much of the tooling and methods required to support the goals of the CCTS KMI. However, since our focus is on providing for the widespread interoperability of CCTS information systems both internally and throughout the translational research community at-large via the rapid and on-demand creation of RIMs defined by ontology-anchored conceptual knowledge (e.g., standard and locally relevant vocabularies), we have developed an agile knowledge engineering framework (Figure 1) that builds upon the preceding techniques and practices.
Figure 1.
Overview of proposed KE framework.
Given this motivation, our proposed KE framework was iteratively developed during the course of two projects conducted as part of our CCTS program in collaboration with Apelon Inc.15 and the Chronic Lymphocytic Leukemia Research Consortium (CLL-RC)16, respectively, and described later in this manuscript. This framework incorporates several aspects of the CommonKADS, UPML and MDA methodologies, including the development of domain-specific schemas that are modeled using UML class diagrams, and annotation of relevant classes, attributes and associations in terms of standard ontologies and terminologies. The specific methods associated with each phase of our framework are described in the following section.
Phase 1: Requirements Analysis
A knowledge engineer should identify key stakeholders and work with them to establish a clear use case that provides details concerning the initiated project, including necessary resources, and resultant tasks and deliverables. Use case(s) should include a high-level explanation of why the project is being initiated, and establish specific, achievable and demonstrable goals against which project success can be measured. An exemplary generic template for documenting such information is described below:
Overview of motivating use case
Key Stakeholders and their associated roles
Tasks/Activities (e.g., what actions are necessary to achieve the desired outcomes of the use case)
Outcomes/Deliverables associated with the preceding tasks/activities
Resources necessary to perform the activities required to generate the desired deliverables
Assumptions or Limitations associated with the implementation of the use case
Phase 2: Knowledge Acquisition
The knowledge engineer should utilize the motivating use case and its referenced information sources (e.g., database, spreadsheet, paper) to document all end-user workflows. The final decision regarding which modality or combination of data sources will provide the most comprehensive inventory should be the responsibility of the end-user. Based upon these workflows, the knowledge engineer should compile inventories of all object classes, attributes and their associated value domains, associations, and any “standardizable” or coded data sets (e.g., adverse events). Each of these inventories should be cleaned and/or optimized to ensure the development of accurate models and semantic annotations. The following steps, each requiring human review, intervention and final judgment, can be taken to obtain clean data sets:
Remove any objects or terms that are not relevant to the domain of interest.
Identify synonyms. In a coded data set, synonyms should have the same code.
Disambiguate duplicates. True duplicates should be censored.
Correct misspellings. Misspellings in the source system can only be changed by those responsible for its maintenance. However, correct spellings should be used in models and annotations.
Expand abbreviations and acronyms; remove any “jargon “. Maintain a list of all abbreviations and corresponding expanded terms.
Phase 3: Knowledge Representation
The object classes, attributes and their value domains, and associations documented in the previous phase should be utilized to either construct or translate any external data models into an appropriate conceptual model, or RIM. This model should be represented as a UML class diagram. The class model should be iteratively refined until agreed upon by all stakeholders.
The knowledge engineer should assess the domains of any “standardizable” or coded data sets in order to determine which standard or local terminologies and/or ontologies will serve as the mapping target (e.g., for medications, RxNorm is an appropriate terminology, but LOINC is not). The data elements should then be defined in terms of their basic elements (e.g., unique concept name and/or code), attributes (e.g., synonyms), internal relationships and associations with other local or standard terminologies and/or ontologies. If any local data sets do not include explicitly unique concept names and/or codes the knowledge engineer must determine how such properties will be uniquely derived from the available data. The knowledge engineer should import these term lists into their knowledge-editing environment (see the Discussion for examples).
Phase 4: Semantic Annotation
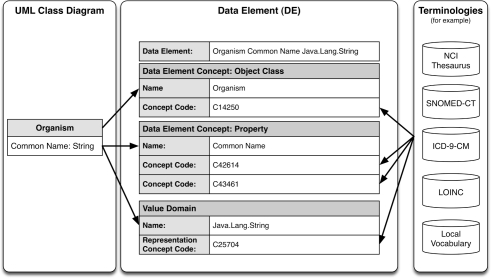
Each component of the class model should be annotated with either an appropriate local or standard concept code, or existing Data Element (DE). New DEs should be curated as necessary to adequately represent local information contained within the domain model (Figure 2).
Figure 2.
Data Element definition (adapted from17).
Each concept in either a “standardizable” or coded data set contained within the underlying data model should be mapped to the determined standard terminology or ontology based upon existing properties (e.g., National Drug Codes in the medication domain) where possible. In cases where no such direct link exists, a terminology mapping solution (see the Discussion section for examples) or other definitional resources (e.g., Micromedex for medications) should be used to map among standard and local terminologies/ontologies. A knowledge engineer must perform an initial review of the resulting object annotations and concept mappings in order to assess their accuracy. Someone, who has not been involved in the original mapping, should conduct a second review. This reviewer should examine all of the annotations assuming that the knowledge engineer was correct, and note any discrepancies. If both reviewers agree to an annotation, it should be accepted as complete and accurate. However, if the two reviewers disagree and are unable to reach a consensus, a subject matter expert should review the annotation and make the final authoritative determination.
Ongoing Knowledge Engineering Projects
The objective for the first KE project, initiated within the Information Warehouse (IW) at the OSU Medical Center (OSUMC) and conducted as part of the OSU CCTS, is to utilize KE methodologies to anchor both coded and un-coded IW data with existing standard terminologies in order to enable the performance of class-based queries (e.g., find all patients prescribed an antibiotic). The IW stores multi-dimensional data from over 70 information systems utilized throughout the enterprise to allow a broad variety of customers, including researchers, clinicians, educators and administrators to view and analyze integrated data sets. Specifically, this pilot project focuses on the domain of medications, and mapping local codes stored in the IW from the enterprise Computerized Physician Order Entry (CPOE) and billing systems to standardized schemas, such as SNOMED-CT. We are currently conducting the final concept mapping validation (KE Phase 4), and developing a custom user interface for the generation of class-based SQL queries that can be run against the IW.
The main goal of the second ongoing KE project is to enable the Chronic Lymphocytic Leukemia Resarrch Consortium (CLL-RC)16 clinical trials management system (CTMS), which is currently being reengineered, to interoperate and exchange key data elements (e.g., patient demographics) with caTissue18 using a Grid-based electronic data interchange infrastructure. The CLL-RC is an NCI-funded multi-institutional program/project, which coordinates and facilitates basic and clinical research on the genetic, biochemical and immunologic bases of CLL. We are currently analyzing the workflows of the existing CTMS (KE Phase 2), and determining how/where they can be optimized, developing corresponding flowchart diagrams, extracting any concepts that will be used as object classes, attributes or associations in the subsequent class diagram, and inventorying the data elements that can be encoded using standard terminologies or ontologies.
Discussion
The projects described above demonstrate that existing KE methodologies and MDA approaches can be adapted and integrated to construct a framework that can provide for the agile and timely execution of knowledge engineering efforts in the context of translational research. Other CTSA institutions have either proposed alternative methods for knowledge management, or have not addressed this issue as part of their current efforts. Some have proposed to use an approach based upon the Cyc project19, and others are taking advantage of the resources provided by the caBIG initiative. The Cyc project is attempting to assemble a comprehensive ontology and knowledge base of everyday knowledge, with the goal of enabling knowledge-based applications to perform human-like reasoning. Current criticisms of this work include scalability issues, and the lack of breadth and depth of its content. As part of the caBIG initiative, the NCI Center for Bioinformatics (NCICB) has built the cancer Common Ontologic Representation Environment (caCORE)18, which provides the MDA-based infrastructure necessary to create interoperable biomedical information systems. Class models can be developed in either Enterprise Architect or ArgoUML, and semantically annotated using a combination of caCORE tools, including the cancer Data Standards Repository (caDSR) and the Semantic Integration Workbench (SIW). However, the caDSR is centrally maintained by NCICB and all of these tools are cancer-specific. For CTSA institutions, these two aspects of the caBIG initiative introduce issues of scalability and generalizability.
Though our proposed framework aims to address such issues, there are limitations to the methodologies currently used to implement it. Primarily, each of the tools utilized throughout the framework only addresses a subset of the overall necessary functionalities. The result of using such a disjoint set of tools is reliance upon a human-mediated workflow, which limits the scalability and extensibility of the approach. Though human intervention is necessary in any KE process, our next steps include the extension of the proposed framework to integrate all utilized KE tools into a seamless semi-automated workflow. This will allow for maximal scalability and extensibility, which are key elements to the CCTS Knowledge Management Initiative (KMI). We are currently in the process of evaluating various tools that could be utilized during each phase of the proposed KE framework. Alternatives to and extensions of the previously described Apelon DTS10 and Protégé Ontology Editor and Knowledge Acquisition System9 include the Protégé Prompt Tab20, LexGrid Editor21, and the SIW17. The Prompt tab supports the management of multiple ontologies in Protege, and enables the comparison of versions of the same ontology, movement of frames between projects, merging of multiple ontologies, and extraction of ontological subsets. The LexGrid Editor is an Eclipse-based open source tool for authoring, viewing and maintaining lexical resources that conform to a formal terminology model. Resources can be developed locally or viewed in context of a networked ‘grid’ of terminologies. The SIW is a tool that allows for the mapping of class model elements to metadata concepts defined in the NCI Thesaurus and, where possible, DEs already registered in the caDSR. Some of these tools, such as the LexGrid and Protégé editors have been integrated in the Eclipse workbench22, and our goal is to utilize this platform to further enable the development of a seamless semi-automated KE pipeline that addresses all four phases of our proposed framework.
Conclusion
Our objective in designing this agile framework, which builds upon prevailing KE and semantic interoperability methods and best practices, is to enable the rapid development of reference information models comprised of ontology-anchored conceptual knowledge and locally relevant vocabularies to provide for the widespread interoperability of CCTS information systems both internally and with the translational research community at-large. We believe that the described KE approach satisfies such an aim and provides a step towards addressing open research questions surrounding the design of translational research information systems capable of being both locally relevant and globally interoperable.
Acknowledgments
This work was supported in part by the OSU CCTS (NCRR, 1U54RR024384-01A1).
References
- 1.Translational Research Re-engineering the Clinical Research Enterprise [cited 2008 Aug 25]; Available from: http://nihroadmap.nih.gov/clinicalresearch [Google Scholar]
- 2.Zerhouni EA. Translational research: moving discovery to practice. Clin Pharmacol Ther. 2007;81(1):126–8. doi: 10.1038/sj.clpt.6100029. [DOI] [PubMed] [Google Scholar]
- 3.Shaw MLG, Gaines BR. The synthesis of knowledge engineering and software engineering in Advanced Information Systems Engineering. Springer Berlin; Heidelberg: 1992. pp. 208–220. [Google Scholar]
- 4.Studer R, Benjamins VR, Fensel D. Knowledge engineering: principles and methods. Data & Knowledge Engineering. 1998;25(1–2):161–197. [Google Scholar]
- 5.Choi N, Song I-Y, Han H. A survey on ontology mapping. ACM SIGMOD Record. 2006;35(3):34–41. [Google Scholar]
- 6.Payne PR, et al. Conceptual knowledge acquisition in biomedicine: A methodological review. J Biomed Inform. 2007;40(5):582–602. doi: 10.1016/j.jbi.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schreiber G, et al. Knowledge Engineering and Management: The CommonKADS Methodology. MIT Press; 2000. [Google Scholar]
- 8.Fensel D, et al. The Unified Problem-solving Method Development Language UPML. Knowledge and Information Systems. 2002;5(1):83–131. [Google Scholar]
- 9.http://protege.stanford.edu
- 10.http://apelon-dts.sourceforge.net/
- 11.OMG Model Driven Architecture [cited 2009 Jan 29]; Available from: http://www.omg.org/mda/
- 12.https://cabig.nci.nih.gov/ [Google Scholar]
- 13.Fridsma DB, et al. The BRIDG project: a technical report. J Am Med Inform Assoc. 2008;15(2):130–7. doi: 10.1197/jamia.M2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.http://ncicb.nci.nih.gov/core/EVS
- 15.Din FM, Sperzel D. Terminology Mapping Guide, in Mapping Process and Quality Assurance. Apelon, Inc; 2008. [Google Scholar]
- 16.Chronic Lymphocytic Leukemia Research Consortium [cited 2009 Jan 29]; Available from: http://cll.ucsd.edu/
- 17.Komatsoulis GA, et al. caCORE version 3: Implementation of a model driven, service-oriented architecture for semantic interoperability. J Biomed Inform. 2008 Feb;41(1):106–23. doi: 10.1016/j.jbi.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.https://cabig.nci.nih.gov/tools/catissuesuite [Google Scholar]
- 19.Cycorp. [cited 2009 Jan 29]; Available from: http://www.cyc.com/
- 20.Noy NF, Musen MA.PROMPT: Algorithm and Tool for Automated Ontology Merging and Alignment Proceedings of the 17th National Conference on Artificial Intelligence and 12th Conference on Innovative Applications of Artificial Intelligence2000. AAAI Press / The MIT Press. [Google Scholar]
- 21.http://informatics.mayo.edu/LexGrid
- 22.www.eclipse.org