Abstract
Integrating and relating images with clinical and molecular data is a crucial activity in translational research, but challenging because the information in images is not explicit in standard computer-accessible formats. We have developed an ontology-based representation of the semantic contents of radiology images called AIM (Annotation and Image Markup). AIM specifies the quantitative and qualitative content that researchers extract from images. The AIM ontology enables semantic image annotation and markup, specifying the entities and relations necessary to describe images. AIM annotations, represented as instances in the ontology, enable key use cases for images in translational research such as disease status assessment, query, and inter-observer variation analysis. AIM will enable ontology-based query and mining of images, and integration of images with data in other ontology-annotated bioinformatics databases. Our ultimate goal is to enable researchers to link images with related scientific data so they can learn the biological and physiological significance of the image content.
Introduction
Relating images to pathophysiological, clinical, and molecular data is a critical activity to advance translational research. Rapid advances in radiological imaging modalities are now providing a rich set of “imaging biomarkers” that have potential utility for understanding disease biology and for predicting responses to therapeutic interventions.1, 2 Relating images to other data is challenging however, because the key image content relevant to the biological or disease process (“image metadata”) is not explicit; image metadata currently extracted by researchers is recorded in ad-hoc ways and disconnected from the images.
The informatics methods currently used to manage and integrate non-imaging biomedical data are difficult to apply to images. The first challenge is that images contain rich content that is not explicit and not accessible to machines. Images contain implicit knowledge about anatomy and abnormal structure (“semantic content”) that is deduced by the viewer of the pixel data. This knowledge not recorded in a structured manner nor directly linked to the image. Thus images cannot be easily searched for their semantic content (e.g., find all images containing liver).
A second challenge for managing and integrating imaging is that the terminology and syntax for describing images varies, with no widely-adopted standards, hindering interoperability and ability to share image data. Schemes for annotating images have been proposed in non-medical domains,3, 4 but no standard schemes for describing medical image contents—the imaging observations, the anatomy, and the pathology—are currently in use.
A final challenge for managing and integrating images with other data is that the lack of localization of all pertinent image metadata. Image metadata is fragmented; patient and study information resides in certain fields of the DICOM header, image annotations are in graphical overlays of presentation state objects, and image interpretations are recorded in unstructured radiology reports. Thus, it is difficult to query images based on their content.
We describe our approach to tackling the above challenges to achieve semantic access to image data: Annotation and Image Markup (AIM).* AIM, a project of the caBIG Imaging Workspace, makes all the key semantic content of images explicit. It includes an ontology to represent the entities and relations pertinent to imaging. Through these methods, AIM will permit linking imaging, clinical, and molecular data. Ultimately, image warehouses will be minable with the potential to facilitate integrative translational research. In this paper, we describe AIM, use cases, and a prototype implementation.
Methods
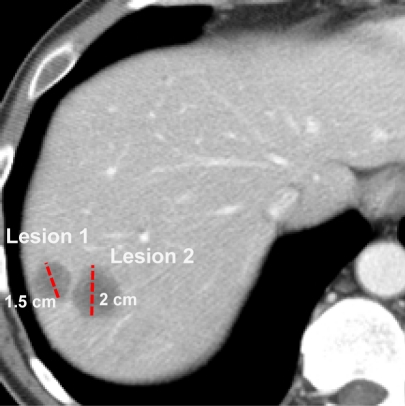
In AIM, we distinguish between image annotation and markup. Image annotations are explanatory or descriptive information, generated by humans or machines, directly related to the content of an image (generally non-graphical, such as abnormalities seen in images, measurements, and anatomic or coordinate locations). Image markup refers to graphical symbols that are associated with an image and optionally with one or more annotations in the image (Figure 1). Accordingly, the key information content about an image lies in the annotation; the markup is simply a graphical presentation of the annotation. In current image display programs, the semantics of image markups are inferred by human readers, but they are not directly machine-accessible (Figure 1). These image annotations are recorded in image graphics, the DICOM header, or in text reports—a form that is not easily accessed and integrated.
Figure 1.
Image markups for recording semantic information. CT scan showing two liver lesions with image markup to indicate the longest diameter of each lesion (dashed line). The lesions are also named (“Lesion 1” and “Lesion 2”) so that the corresponding lesions can be identified and re-assessed in images on follow-up studies. This information is only recorded as a graphic overlay; the semantics of these markups is not explicit for computational processing.
Use cases
To drive the requirements and to evaluate the potential utility of AIM, we developed representative use cases for medical image annotation and markup in the cancer research domain. We derived these use cases by reviewing the use of image information in clinical trials from the American College of Radiology Imaging Network (ACRIN), the Quality Assurance and Review Center (QARC), and the Radiation Therapy Oncology Group (RTOG). We also reviewed queries made to the National Cancer Imaging Archive (NCIA) to survey the scope of information sought in images.
1. Tracking imaging biomarkers in clinical trials:
Objective criteria for measuring response to cancer treatment are critical in cancer research and practice. The Response Evaluation Criteria in Solid Tumors (RECIST)5 criteria quantify treatment response based on information in images. RECIST evaluates response by assessing lesions in baseline and follow-up imaging studies. RECIST requires defining a set of target lesions to be tracked over time. The sum of the longest diameters (SLD) of target lesions needs to be calculated in order to apply RECIST crirteria. In this use case, AIM would provide the necessary information to recognize lesion identity and to track lesion measurements across serial imaging studies.
2. Query for images in imaging databases and on semantic Web:
Clinicians and researchers need to search for images to guide care and to formulate new hypotheses. In this use case, users can query images according to anatomy, visual observations, or other metadata associated with images. These queries cannot be done at present because image metadata are currently stored only in the DICOM header, in image graphical overlays, or in the radiology report.
3. Multi-reader imaging studies:
Interpreting images can be a subjective task, with variation among readers. An important use case is to enable multiple readers to interpret the same image and to analyze inter-reader variation in their observations. This task requires associating information about the reader with each piece of image information they contribute.
Review of existing standards
There are a number of existing efforts relevant to describing images and annotating them. We reviewed the specifications of DICOM, HL-7, W3C, MPEG7 and SVG to harmonize this work with AIM. The goal was to identify entities, relations, and/or value domains pertaining to images and annotations and markup. We also reviewed specification of measurements and calculations in images.
Ontology and Information Model
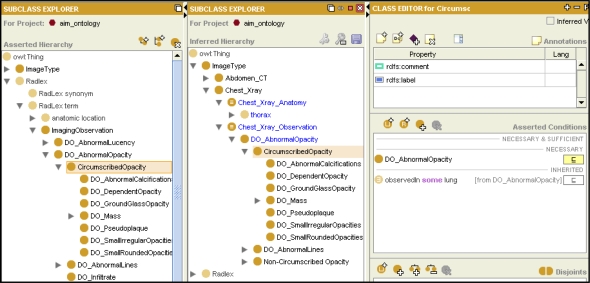
We built an ontology and an information model for image annotation and markup. The AIM ontology is in OWL-DL and represents the entities and relations needed to describe medical images and their contents. The AIM ontology includes RadLex, an ontology of radiology.6 RadLex provides terms for anatomic structures that can been seen in images (such as “liver” and “lung”) and the observations made by radiologists about images (such as “opacity” and “density” of structures in images). In addition to RadLex, the AIM ontology includes classes describing geometry—shapes and the spatial regions that can be visualized in images—as well as other image metadata (Figure 2).
Figure 2.
AIM Ontology. The AIM ontology (left panel) includes a taxonomy of anatomy and imaging observations. Assertions on classes (right panel) record knowledge about the anatomic regions that will be visible in particular types of images (for example, the screenshot shows an assertion that abnormal opacity in images is observed in the lung), as well as the imaging observations that will occur in those anatomic regions. The ontology also contains classes representing imaging procedures such as chest X-ray (middle panel). The ontology enables query of images according to anatomy or findings at different levels of granularity, enabled by OWL classification. For example, the ontology indicates that thoracic anatomy and abnormal opacity occur in chest X-rays.
The AIM ontology provides a knowledge model of imaging, relating imaging observations to other image metadata necessary for the use cases. For example, the ontology represents the fact that certain image observations are visualized in particular anatomic regions (Figure 2). This information is needed for use cases related to image query. For example, the ontology can be used to recognize that chest X-rays should be retrieved if the user is interested in images containing thoracic anatomy or findings of abnormal opacity. Such reasoning is enabled by applying an OWL-DL classifier to the ontology and examining the inferred taxonomy of classes representing types of images (Figure 2).
We also created an information model (“AIM schema”)—a standard syntax for creating and storing instances of AIM image annotations. The AIM schema was originally created in UML, and subsequently translated to XML schema (an XSD file). The AIM schema can be viewed in Protege as an ontology. Individual image annotations are instances in the AIM ontology (or XML files valid against the AIM XSD). AIM schema provides a syntax to describe the rich metadata associated with images. Since image features are localized to regions in the image, AIM schema also specifies spatial regions in images and their geometry. For example, lines have a length and an angle with respect to a reference line. Thus, “line” and “angle” are entities in the AIM schema.
In the AIM schema, there are two types of annotation: ImageAnnotation (annotation on an image) and AnnotationOnAnnotation (annotation on an annotation). Image annotations include information about the image as well as their semantic contents (anatomy, imaging observations, etc). Annotation on annotations provide an annotation abstraction layer, permitting users to make statements about groups of any pre-existing annotations. Such annotations enable our use cases for multi-reader image evaluations and making statements about change in disease status across a series of images.
Prototype Implementation and Evaluation
We created a prototype implementation of AIM to evaluate its sufficiency to meet the needs of our use cases—an image annotation tool that collects information about images and stores it as AIM XML. This tool7 extracts information from the DICOM header and from graphical objects that users create as they annotate the images. We created AIM annotations on images obtained from a study where RECIST was used to assess disease status, and we evaluated the ability of AIM to represent the key image content and to enable the image-based analyses required for that study. We also evaluated the AIM ontology and information model for its sufficiency to meet the requirements our three key use cases for image annotation.
Results
In reviewing the existing practices related to image annotation, we found a variety of approaches, varying in semantic expressiveness, breadth, and depth. Among DICOM, HL-7, W3C, MPEG7 and SVG, there were many useful entities which we adopted (and harmonized in AIM) relating to geometry, coordinates, and pixels. None of existing standards described individual image regions (the focus is annotating the entire image), and none incorporated controlled terminology for annotation.
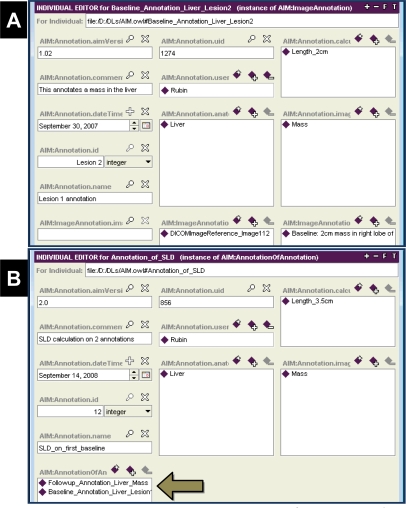
An AIM annotation is created as an XML file that is compliant with the AIM schema file (XSD). That file can be transformed into a format that can be viewed in Protege (Figure 3) or serialized to standard formats such as DICOM-SR or HL7 Clinical Document Architecture. The AIM annotation file can be created by image annotation tools that support the AIM format, such as the one we recently developed.7
Figure 3.
Annotations in AIM. Two types of AIM annotations are shown as instances of the information model in Protege. Both of these example annotations enable RECIST scoring. (A) An AIM ImageAnnotation describing a lesion seen on a baseline image, including a lesion identifier (“Lesion 2”) and measurement (2 cm). (B) An AIM AnnotationOnAnnotation describing the sum of linear dimensions of the measurable disease, which are measured on two separate ImageAnnotations (arrow).
The AIM ontology and information model provided the semantic expressiveness required to fulfill the annotation requirements of our use cases, summarized here by use case.
1. Tracking imaging biomarkers in clinical trials:
In this use case, the goal is to track measurable lesions in serial studies and to calculate the SLD composite measure of lesions. AIM provided the necessary information to recognize lesion identity and to integrate information across lesions necessary to compute SLD. Measurable lesions were identified by unique labels (e.g., “Lesion 2”) and measured at each study time point, recording this information as AIM ImageAnnotations (Figure 3A). The SLD calculation could be automated by applying a summation calculation to the AIM annotations of measurable lesions at each time point (Figure 3B). The summation is represented by creating an AIM AnnotationOnAnnotation, referencing the two ImageAnnotations that describe the measurements of each measurable lesion.
2. Query for images in imaging databases and on semantic Web:
AIM provided a human-readable and machine-interpretable infrastructure enabling query of images according to anatomy, visual observations, and other metadata associated with images. The AIM schema makes each metadata field explicit in the ImageAnnotation (Figure 3A). Each of these fields is accessible for query, so complex queries can be formulated. The AIM XML can be published on the semantic Web and made accessible for querying online radiological images to which they are linked.
The ontology-based annotations enabled the use of the AIM ontology for query at varying levels of granularity. Such queries could be constructed by exploiting the taxonomic structure of AIM ontology, particularly for anatomy and imaging observations, such as “find all images showing a circumscribed opacity.” The ontology would enable these queries to return images annotated with abnormal calcifications, dependent opacities, and other subclasses of circumscribed opacity (Figure 2). The ontology also enabled queries that exploit logical definitions of classes, such as for types of images according to the anatomy or findings they contain. For example, the ontology would permit a query service searching for abnormal thoracic anatomy and abnormal opacities to recognize it should look for images of chest X-rays (Figure 2).
3. Multi-reader imaging studies:
A common use case in clinical research is where multiple readers interpret the same image and the researchers wish to analyze inter-reader variation in the results. AIM enabled this task by associating information about the reader with each element of image metadata that readers contribute. In addition, the AnnotationOnAnnotation could be used to describe whether or not multiple readers agree in their interpretations.
Discussion
Images are currently not exploited well in research because their contents are not easily accessed by machines (Figure 1). AIM addresses the challenges of managing and integrating images with other data by making the semantic content of images explicit using an ontology and an information model (Figure 3). The approach is similar to that being undertaken in other communities developing minimal information standards, such as MIAME8 and the Open Microscopy Environment,9 an effort to define information standards for microscopy images. To our knowledge, AIM is the only effort to create information standards for radiological images.
AIM provides several benefits to translational research. First, AIM is a single format for image metadata, unifying the diverse image metadata that are currently fragmented in different locations. AIM will thus permit researchers to share images and to build minable image repositories. A second benefit is that AIM enables complex queries exploiting knowledge in the ontology; for example, an image search engine using AIM could recognize that searches for thoracic anatomy and abnormal opacity should look at images that are chest X-rays (Figure 2). A third benefit is that AIM enables identifying and tracking lesions and their measurements, a common need in research and clinical practice. In the future, AIM annotations could drive automated calculation of assessment criteria such as RECIST (Figure 3). A final benefit is that AIM unifies all image metadata and encodes its content with ontologies, which will permit the semantic content of radiology images to be integrated with that of other types of images in the future, such as pathology and molecular images. AIM is a caBIG project with the potential for widespread adoption throughout cancer centers, enabling many institutions to share and interoperate with their image data.
We anticipate that an initial use of AIM in translational research will be to standardize the way image metadata are stored and queried. Since AIM describes the key image features that are commonly studied in translational studies, researchers will be able to build databases integrating radiology images with pathology and molecular data. For example, consider a clinical trial for a new drug in which imaging was used to assess response to treatment. Since AIM describes each lesion—its name, location, size, etc, it will be possible to mine databases of images acquired in clinical trials to compare different criteria used to assess treatment response. It will also be possible to link the image-based features of each lesion to other disease characteristics, such as pathology, molecular, and clinical data to identify novel imaging biomarkers of disease and treatment response.
A potential limitation of our work is that it was driven by particular use cases. However, our use cases were carefully selected to be representative of common clinical research activities involving imaging. A second potential limitation is that it could be too ambitious to expect researchers to provide the full spectrum of rich information in AIM annotations. Since much of the information in AIM can be extracted from the DICOM header and from graphical elements users draw on images, some of the labor in creating AIM documents could be streamlined by “intelligent” annotation tools.7 Such tools could capture the structured AIM information while the researcher works with the image in the manner they are currently accustomed (Figure 1).
We are currently undertaking research studies in which imaging data is recorded in AIM, and we are conducting assessment studies with our tools. Ultimately, we expect that making the semantic contents computationally-accessible will catalyze translational research by helping researchers to integrate, explore, and make discoveries with the rapidly accumulating wealth of biomedical images and related information.
Acknowledgments
This work is supported by a grant from the National Cancer Institute (NCI) through the cancer Biomedical Informatics Grid (caBIG) Imaging Workspace, subcontract from Booz-Allen & Hamilton, Inc.
Footnotes
References
- 1.Guha C, Alfieri A, Blaufox MD, Kalnicki S. Tumor biology-guided radiotherapy treatment planning: gross tumor volume versus functional tumor volume. Semin Nucl Med. 2008 Mar;38(2):105–13. doi: 10.1053/j.semnuclmed.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 2.Tardivon AA, Ollivier L, El Khoury C, Thibault F. Monitoring therapeutic efficacy in breast carcinomas. Eur Radiol. 2006 Nov;16(11):2549–58. doi: 10.1007/s00330-006-0317-z. [DOI] [PubMed] [Google Scholar]
- 3.Khan L. Standards for image annotation using Semantic Web. Computer Standards & Interfaces. 2007 Feb;29(2):196–204. [Google Scholar]
- 4.Petridis K, Bloehdorn S, Saathoff C, Simou N, Dasiopoulou S, Tzouvaras V, et al. Knowledge representation and semantic annotation of multimedia content. Iee Proceedings-Vision Image and Signal Processing. 2006 Jun;153(3):255–62. [Google Scholar]
- 5.Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, et al. New guidelines to evaluate the response to treatment in solid tumors European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000 Feb 2;92(3):205–16. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 6.Langlotz CP. RadLex: a new method for indexing online educational materials. Radiographics. 2006 Nov–Dec;26(6):1595–7. doi: 10.1148/rg.266065168. [DOI] [PubMed] [Google Scholar]
- 7.Rubin DL, Rodriguez C, Shah P, Beaulieu C. iPad: Semantic Annotation and Markup of Radiological Images. Proc AMIA Symp. 2008. (in press) [PMC free article] [PubMed]
- 8.Whetzel PL, Parkinson H, Causton HC, Fan L, Fostel J, Fragoso G, et al. The MGED Ontology: a resource for semantics-based description of microarray experiments. Bioinformatics. 2006 Apr 1;22(7):866–73. doi: 10.1093/bioinformatics/btl005. [DOI] [PubMed] [Google Scholar]
- 9.Goldberg IG, Allan C, Burel JM, Creager D, Falconi A, Hochheiser H, et al. The Open Microscopy Environment (OME) Data Model and XML file: open tools for informatics and quantitative analysis in biological imaging. Genome Biol. 2005;6(5):R47. doi: 10.1186/gb-2005-6-5-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]