Abstract
Networks are increasingly used in biology to represent complex data in uncomplicated symbolic form. However, as biological knowledge is continually evolving, so must those networks representing this knowledge. Capturing and presenting this type of knowledge change over time is particularly challenging due to the intimate manner in which researchers customize those networks they come into contact with. The effective visualization of this knowledge is important as it creates insight into complex systems and stimulates hypothesis generation and biological discovery. Here we highlight how the retention of user customizations, and the collection and visualization of knowledge associated provenance supports effective and productive network exploration. We also present an extension of the Hanalyzer system, ReOrient, which supports network exploration and analysis in the presence of knowledge change.
Introduction
Recently, biologists have been turning to network representations to aid in the interpretation of high-throughput or otherwise complex datasets. Networks are suited to the visualization of biological phenomena due to their ability to represent interactions (edges) between biological entities (nodes), and to illustrate vast amounts of data compactly. Networks allow the visualization of data that is too extensive or complicated to understand in tabular form1, and provide an “approximate model or explanation of the underlying biological process”2. Networks are used to visualize both data (e.g. expression arrays) and existing knowledge (e.g. signaling pathways), and often both together.
The knowledge used to build these networks is not static. The biomedical literature (as represented in PubMed) grew by more than 750,000 articles in the last year. Information in gene-centric databases is growing even faster3. As such, the knowledge relevant to the analysis of a large or complex dataset will likely change during the course of analysis.
Here we present principles and a tool for visualizing networks that facilitates analysis in the presence of knowledge change. In particular we highlight two principles that support the presentation of knowledge change; the retention of user driven customization over time, and the collection and visualization of knowledge associated provenance. The ability to effectively identify and present such knowledge change in a network through the implementation of these principles is crucial for its effectual and continued exploration, and therefore biological discovery. The tool described is an extension of the previously reported Hanalyzer system4,5.
Customization of networks supports the learning process
As humans, our visual systems possess an innate ability to process large amounts of information by identifying patterns and trends viewed in terms of position, shape and color of objects1. Network users unconsciously take advantage of these skills when they devote significant time and effort exploring new networks and customizing them via repositioning and color-coding in order to accentuate their research questions and aims. There are many reasons for this customization. First, despite layout-optimization methods, the default presentation of a network tends to render the knowledge into a dense, highly populated ‘hair-ball’ that fails to consider domain-specific information, and obscures important data6. Second, users bring considerable prior knowledge to the network analysis process. Customization of the network allows them to integrate this knowledge into the network, and also to view the network within the context of their prior knowledge. This transforms the network from a mass of raw information into an organized view of knowledge. Third, adjustments to the network highlight different kinds of information, (i.e. GO annotations or KEGG pathways), which support discovery and hypothesis building. Finally, as researchers continue to gain insights and develop new hypotheses, they encode these in the network with further customizations. The development of personalized versions of a network therefore mirrors the transition of data in the network from raw information into knowledge, while also capturing the unique background of a particular researcher, and what that user learnt and discovered during their analysis7.
However, the learning curve, which must be overcome when exploring and modifying complex networks, can be sizable and requires a significant commitment of time and effort on the part of the user. So it is unsurprising that many life scientists come to the conclusion that the cost of using such networks outweighs any benefit and they have reservations about their use2. Considering this, it is important that once a scientist has made this investment, their efforts are rewarded and they are adequately supported so as to ensure their continued use.
Consistency in presentation supports efficient data exploration and integration
Networks represent static ‘snapshots’ of knowledge at defined points in time, and for a network to remain current updates need to be periodically undertaken. In the case of highly integrated information networks, such as those generated by the Hanalyzer, whole bodies of literature and numerous databases need to be periodically ‘re-read’ in order to keep the knowledgebase current5. Such ‘new release’-type of network updates involve the addition, removal and modification of numerous nodes and edges, and the subsequent loss of user implemented customizations results in massive disorientation and frustration on the part of the user. The generically laid out network bears little similarity to the extensively modified, customized version the scientist was using. In the absence of personalized visual cues, such as spatial organization, to trigger recognition of sections of the network, the researcher once more has to invest a significant amount of time re-imbuing the network with their personalized customizations, before they even start on the rationalization of any changes in knowledge.
The establishment of a common spatial distribution of nodes between networks is important for the rapid permeation of knowledge acquired from, and thus associated with, the previous network to the new representation. By maintaining the same spatial layout when new data is integrated into a network, the user is able to maintain their orientation in information space7. Changes are easier to locate, identify and integrate into the researchers’ understanding of the network.
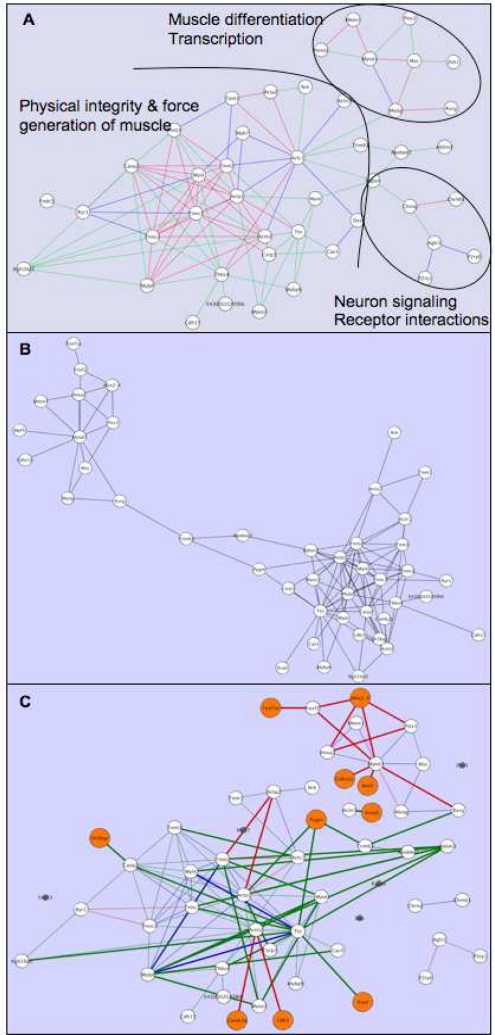
To maintain the spatial orientation of user manipulated networks, we have developed a plug-in ReOrient for Cytoscape8 which tracks node position between different versions of the same network. Using previously reported data5 to demonstrate its functionality, a network which after approximately 48 hours of manual exploration and organization by a researcher was parsed into three clusters of nodes representing tongue muscle differentiation, regulation and initiation of this process by myogenic transcription factors, and synapse development and maintenance5. The manual manipulation of the spatial organization of the network reflected the researchers understanding of the knowledge and further distinguished these clusters from each other (Figure 1A). Subsequently the network was updated and user customizations lost (Figure 1B). However, by applying the ReOrient plug-in, the nodes in the updated were immediately positioned to be consistent with the previous, customized version, and with this new visualization the user was able to quickly identify the absence of the neuron signaling/synapse associated cluster, and the addition of muscle differentiation and transcription cluster nodes to the network (compare Figure 1A & 1C).
Figure 1.
Networks illustrated before and after knowledge update. A) A user customized tongue muscle development network5. Three functional clusters are annotated and edges are colored according the combinatorial metric used to assert them (for details see5). B) The same network as in A, but as automatically generated immediately after update. Note the lack of spatial concordance between the nodes of network A and B. C) The use of the ReOrient plug-in preserves the layout of nodes allowing easy orientation. Provenance provided by the Hanalyzer allows the visualization of knowledge change. New knowledge is represented by enlarged orange nodes and thickened edges, while knowledge loss in the form of nodes which have no longer met a threshold for inclusion in the network are reduced in size and colored grey.
Not all knowledge change is equal
Although maintaining the spatial orientation of a network is crucial for clear representation of knowledge change over time, to understand the implications of the changes which have occurred information about the provenance of each update must be captured and presented in a manner which allows this knowledge evolution to be explored.
Not all change is equal, and some is currently difficult to track. Three types of knowledge change can be observed between network version updates: 1) New knowledge is represented as additional nodes and edges, 2) knowledge reduction, (which occurs when either a threshold for inclusion in the network is no longer met, or due to removal of such knowledge from a data source) is represented as deletion of edges or nodes, and 3) alteration of existing knowledge is represented as modification of an attribute associated to a node or edge. Identifying and reporting how network knowledge has changed over time is critical to the user, to not only to understand how new information can support and develop their current theory, but also to rationalize those discoveries which fail to support assumptions based on previous versions of the network2. The user needs to have these changes in knowledge presented to them, and have them bought to their attention. In current systems the exact reverse is true and the user must actively seek such changes out. Not only are such searches time consuming, but manual searching is also rarely efficient, with details easily missed or over looked.
The Hanalyzer facilitates effective presentation of knowledge change by capturing detailed knowledge provenance. Once available the user is able to leverage this information to easily identify and track the flux of knowledge in/out/within a network (Figure 1C). Such alterations can obviously have both supporting and undermining effects to currently held hypotheses (and associated research efforts) and so needs to be presented to the user urgently.
Identifying the presence of new knowledge is relatively simple task. New database entries are logged and date stamped, and therefore easy to parse. This new information is highly desirable to the user as it represents knowledge development, expansion and gain. The main concern here, is being able to quickly identify this new information within the network and understand where and how it fits into the larger picture. Knowledge reduction is often overlooked in the desire to be exposed to all that is new. However, considering the volume of inaccurate information housed in biological databases which will gradually be corrected it is prudent to track it 9. Removal of information is not directly reported by databases and identifying this type of knowledge change requires a user to notice when a detail disappears. Such manual checking obviously is untenable when dealing with large networks comprising 1000+ nodes. Tracking modifications to pre-existing knowledge is more complicated. Information that has been modified (i.e. a new gene added to an OMIM entry, a newly observed phenotype in a previously documented mouse model) is valuable because it represents subtle changes in the state of current knowledge upon which theories have been built. Parsing the precise nature of this type of change is challenging however, as entry updates may be date stamped, but the details of the change not noted. Applying our CommonAttributes plug-in5 the user is able to drill into node and edge attributes and retrieve the details of such knowledge modifications.
Support for tracking knowledge change over time
Scientists are increasingly turning to networks to aid in their interpretation and investigation of highly-complex data, however they recognize that using networks can require significant amounts of their valuable time which can be a barrier to use2. Here we have outlined how through consideration and maintenance of a users highly personalized interaction with a network, knowledge change can be incorporated in a manner that is efficient and supportive to hypothesis generation and biological discovery. Effective visualization of knowledge improves insight, which leads to formulation of better questions and hypotheses, which are the real key to discovery7.
Previously, users may not have interacted so intimately with networks. However, the depth and complexity of data presented by recent 3R systems invites significant exploration, and as such personalization5. Such features as provenance capture and spatial consistency, as supported by the Hanalyzer, and plug-ins CommonAttributes and ReOrient, ensures that not only is the knowledge content of a network current, but also that customizations provided by the researcher remains in the representation.
Adapting the network to highlight new nodes and edges bought these new features to the attention of the user, while consistent spatial organization between versions allowed the user to integrate this new data into their pre-existing understanding of the knowledge captured and represented here. What is key, is that the identification of important features within the network really must come from the user7. As exploration proceeds and new updated networks are released, what is deemed important may shift, and such knowledge evolution should be easily tracked and traced.
Availability
All software mentioned in this manuscript is available as open source software via SourceForge at hanalyzer.sourceforge.net.
Acknowledgments
H. Johnson for manuscript development. NIH grants R01LM008111, R01LM009254 and R01GM083649 to LH, T15LM009451 supported RS. Fulbright-AstraZeneca Fellowship funded HT.
References
- 1.Tao Y, Liu Y, Friedman C, Lussier YA. Information visualization techniques in bioinformatics during the postgenomic era. Drug Discovery Today: BIOSILICO. 2004;2(6):237. doi: 10.1016/S1741-8364(04)02423-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saraiya P, North C, Duca KA. Visualizing biological pathways: requirements analysis, systems evolution and research agenda. Information Visualization. 2005;1 [Google Scholar]
- 3.Galperin MY.The Molecular Biology Database Collection: 2008 update Nucleic Acids Res 36(Database issue), D22008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tipney H, et al. Leveraging existing biological knowledge in the identification of candidate genes for facial dysmorphology BMC BioinformaticsAccepted for publication 2008 [DOI] [PMC free article] [PubMed]
- 5.Leach S, et al. 3R Systems for Biomedical Discovery Acceleration, with Applications to Craniofacial Development PLoS Computational BiologySubmitted 2008 [DOI] [PMC free article] [PubMed]
- 6.Suderman M, Hallett M. Tools for visually exploring biological networks. Bioinformatics. 2007;23(20):2651. doi: 10.1093/bioinformatics/btm401. [DOI] [PubMed] [Google Scholar]
- 7.Thomas J, et al. Discovering Knowledge Through Visual Analysis. Journal of Universal Computer Science. 2001;7(6):517. [Google Scholar]
- 8.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13(11):2498. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagy A, et al. Identification and correction of abnormal, incomplete and mispredicted proteins in public databases. BMC Bioinformatics. 2008;9:353. doi: 10.1186/1471-2105-9-353. [DOI] [PMC free article] [PubMed] [Google Scholar]