Abstract
Prostate cancer initiation and progression are uniquely dependent on the androgen receptor (AR). Even when the cancer progresses to a castration-resistant stage, AR signaling remains active via a variety of mechanisms. In the present study, we showed that NF-κB/p52 can activate the AR, resulting in increased transactivation of AR-responsive genes, such as PSA and NKX3.1, in a ligand-independent manner. NF-κB2/p52 enhances nuclear translocation and activation of AR by interacting with its NH2-terminal domain and enhances the recruitment of coactivators such as p300 to the promoters of AR-dependent genes. These results were confirmed in three different prostate cancer cell lines: LAPC-4 (wild-type AR), LNCaP (mutant AR), and C4-2 (castration resistant). Transfection of p52 into LAPC-4 and LNCaP cells (which express low levels of p52) showed increased activation of the endogenous AR. Downregulation of endogenous p52 in C4-2 cells resulted in abrogation of AR constitutive activation. Comparison of the relative effects of p52 and p65 (RelA) showed that p52, but not p65, could activate the AR. Collectively, these findings, together with previous reports that the levels of NF-κB2/p52 are elevated in prostate cancer cells and that active NF-κB2/p52 promotes prostate cancer cell growth in vitro and in vivo, suggest that NF-κB2/p52 may play a critical role in the progression of castration-resistant prostate cancer.
Introduction
As the growth of prostate cancer cells depends on the presence of androgens, almost all patients respond initially to androgen ablation. However, virtually every patient will relapse due to the growth of castration-resistant cancer cells. Androgen receptor (AR) activation plays an important role in prostate carcinogenesis and progression. Ligand-dependent activation leads to the binding of AR to androgen response elements (ARE). Castration-resistant prostate cancer (CRPC) cells often continue to express androgen-responsive genes, such as prostate-specific antigen (PSA), and often express AR (1, 2), suggesting that the AR becomes activated by an androgen-independent mechanism in AR-positive CRPC cells. The AR can also be activated in the absence of or very low levels of androgen by cross talk with other signaling pathways (3–9). Mutations, deletions, and amplification of the AR gene or alterations of the interactions between AR and some of its coregulators may increase tumor cell sensitivity to very low levels of androgen or allow it to respond to other steroids or even antiandrogens (10–14). In addition, levels of intraprostatic androgens are at concentrations sufficient to activate the AR and stimulate tumor growth (15–18). Thus, abnormal activation of the AR plays a major role in the development of castration resistance.
The NF-κB family comprises five proteins—RelA/p65, NF-κB1/p50, c-Rel, RelB, and NF-κB2/p52—which have been identified as important mediators in the oncogenesis of many cancers. The classic NF-κB pathway involving the p65/p50 heterodimer has been well studied and shown to be constitutively activated in several cancers, including prostate. The noncanonical NF-κB pathway involves the processing of p100 to NF-κB2/p52 via the recruitment of NF-κB–inducing kinase and subsequent activation of IκB kinase α. The processing of p100 to p52 is a tightly controlled event in many cells and tissues (19–22). The proteolytic processing of p100 to p52 can be activated by lymphotoxin β (23, 24), B-cell–activating factor (25, 26), CD40 ligand (24, 27), and its cis-acting domain (22). The functional significance of p100 processing has been confirmed by genetic evidence from humans and mice (28). Constitutive production of p52 in p100 knock-in mice causes marked hyperplasia in lymphocytes and liver, leading to early postnatal death (27). Overproduction of p52 in the mammary gland in p100 transgenic mice disrupted normal ductal development and led to hyperplastic growth (29). Overproduction of p52 has been observed in several solid tumors, including breast and prostate cancers (30, 31). We previously showed that p100 is processed to p52 by activated signal transducer and activator of transcription 3 (Stat3) in prostate cancer cells (32). Further studies showed that NF-κB2/p52 induces castration-resistant growth in LNCaP cells by inhibiting cell cycle arrest and apoptotic cell death induced by androgen deprivation (33). This was accompanied by continued expression and activation of the AR, which suggests that p52 may activate AR during the progression of CRPC. Our studies also show that several genes involved in cell growth, proliferation, cell movement, etc. are potential targets of NF-κB2/p52 (34).
In this study, we examined the mechanism of AR activation by p52. We show that AR is activated by p52 via recruiting the AR and coactivators such as p300 to the promoters of AR-responsive genes, resulting in their increased transactivation even in androgen-depleted conditions.
Materials and Methods
Cells, reagents, and antibodies
LAPC-4, LNCaP, C4-2, and PC-3 prostate cancer cells were obtained from the American Type Culture Collection and cultured in RPMI 1640 containing either 10% complete fetal bovine serum (FBS) or 10% charcoal-dextran–stripped FBS (CS-FBS) and penicillin/streptomycin. LNCaP passage numbers <20 were used throughout the study. NF-κB2/p52 (K-27), AR (441; mouse monoclonal), hemagglutinin (HA), actin, and tubulin antibodies were purchased from Santa Cruz Biotechnology. Anti-Flag-M2 antibodies were purchased from Sigma Chemical Co. All other reagents were of analytic grade and obtained from local suppliers.
Western blot analysis
Cells were lysed in high salt buffer as described earlier (33). Western blots were probed with the indicated primary antibodies, and the chemiluminescence was detected by ECL (Amersham) after incubation with the appropriate horseradish peroxidase–conjugated secondary antibodies.
Measurement of PSA
PSA levels were measured in the culture supernatants using ELISA (United Biotech, Inc.) according to the manufacturer’s instructions and as described previously (35).
Luciferase assays
LAPC-4 and LNCaP cells were transfected with pGL3-ARE-Luc, pGL3-PSA6.0-Luc, pGL3-AREI/II-Luc, and pGL3-AREIII-Luc reporters along with p52, p65, p100, AR, p300 plasmids, and AR and p300 siRNAs as indicated in the figures (see Supplementary Materials and Methods for details). Cell lysates were subjected to luciferase assays with the Luciferase Assay System (Promega).
Coimmunoprecipitation
Equal amounts of LNCaP or C4-2 cell lysates were immunoprecipitated with 1 μg of anti-AR, anti-Flag, anti-p52, or anti-HA antibodies (as indicated in the figures) overnight. The precipitated proteins were analyzed by Western blotting with the indicated antibodies.
Chromatin immunoprecipitation assay
LAPC-4 and LNCaP cells were transfected with the indicated plasmids, and 1 nmol/L dihydrotestosterone (DHT) was added 24 h after transfection. DNA-protein complexes were isolated, and chromatin immunoprecipitation (ChIP) assays were performed as described earlier using primers spanning either the proximal or the distal enhancer AREs of the PSA promoter or ARE sites in the NKX3.1 promoter (−3013 to −2995; refs. 36, 37). Isotype-matched IgG was used as control (primer sequences in Supplementary Materials and Methods).
Immunofluorescence
LNCaP (grown in CS-FBS for 3 d to reduce background AR activation) and PC-3 cells were transfected with the indicated plasmids or treated with 1 nmol/L DHT and used to assess the extent of AR nuclear translocation by immunofluorescence (see Supplementary Materials and Methods for details).
Real-time quantitative reverse transcription-PCR
LNCaP and LAPC-4 cells were transfected with the indicated plasmids, and total RNAs were extracted using Trizol reagent (Invitrogen). cDNAs were prepared after digestion with RN-ase-free RQ1 DNase (Promega). The cDNAs were subjected to real-time reverse transcription-PCR (RT-PCR) using SYBR Green iQ Supermix (Bio-Rad) according to the manufacturer’s instructions. Each reaction was normalized by coamplification of actin. Triplicates of samples were run on default settings of the real-time PCR machine (Bio-Rad) as described earlier (primer sequences in Supplementary Materials and Methods; ref. 38).
Statistical analysis
Data are shown as the mean ± SD. Multiple group comparison was performed by one-way ANOVA followed by the Scheffe procedure for comparison of means. P < 0.05 was considered significant.
Results
NF-κB2/p52 enhances PSA expression
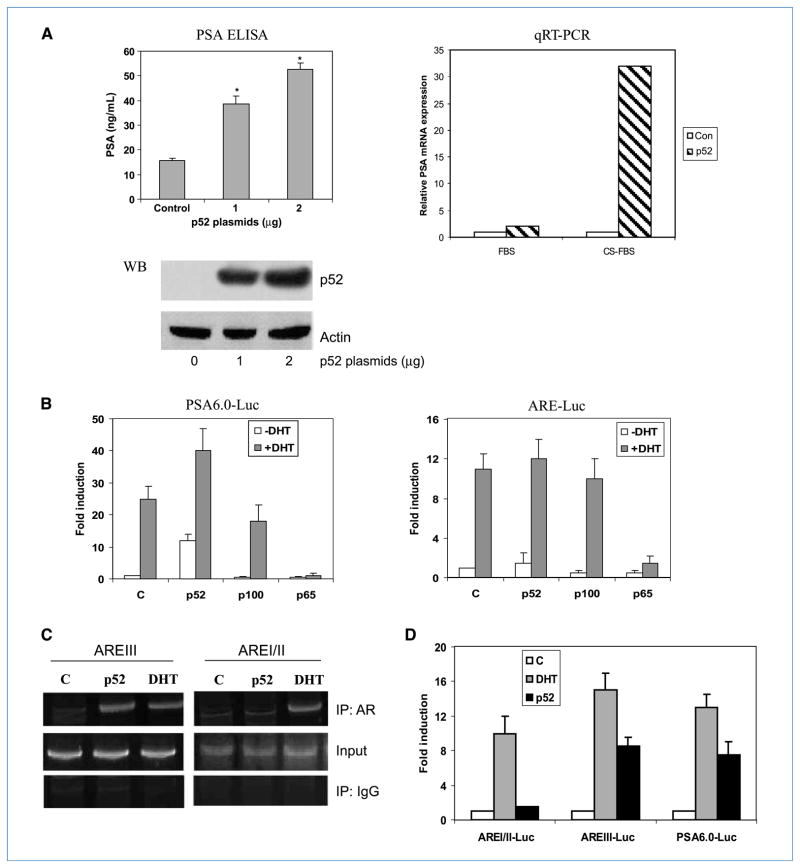
Our previous studies found that LNCaP cells overexpressing NF-κB2/p52 were able to form rapidly growing tumors when injected s.c. into castrated nude mice. This was accompanied by continued expression and activation of the AR as well as production of PSA in castrated animals (33). To verify that p52 enhances PSA expression in vitro, LNCaP cells transfected with varying amounts of p52-expressing plasmids were grown in CS-FBS. Secretion of PSA was assayed in the supernatants by ELISA after 48 hours. The values were normalized to cell numbers. Expression of p52 induced a dose-dependent increase in PSA expression in the absence of androgen in LNCaP cells. This result was also confirmed by real-time RT-PCR to measure endogenous PSA mRNA expression. Expression of p52 enhanced PSA mRNA expression ~30-fold in CS-FBS (Fig. 1A). These observations confirm the in vivo finding that mice bearing LNCaP tumors expressing p52 have elevated PSA levels.
Figure 1.
A, p52 enhances PSA expression. LNCaP cells were transfected with 1 to 2 μg of p52 plasmid in CS-FBS for 2 d. PSA ELISA: PSA secretion was measured by ELISA in supernatants. Columns, mean of triplicate samples; bars, SD. *, P < 0.05. Western blot (WB): cell lysates were analyzed by immunoblotting using p52 antibodies. Actin was the internal control. qRT-PCR: total RNAs from p52-transfected and vector control–transfected LNCaP cells were analyzed by qRT-PCR for PSA mRNA expression. p52-induced PSA mRNA levels are expressed relative to their respective controls in FBS and CS-FBS. B, p52 transactivates PSA promoter. LNCaP cells in CS-FBS were cotransfected with pGL3-PSA6.0-Luc or ARE-Luc and plasmids expressing p52, p100, or p65. Cells were cultured in CS-FBS (−DHT) or 1 nmol/L DHT (+DHT) for 48 h. Luciferase activities were measured. Columns, mean of triplicate samples; bars, SD. Three independent experiments were performed. C, p52 recruits AR preferentially to the distal enhancer of PSA promoter. Androgen-deprived LNCaP cells were transfected with plasmids expressing p52 and vector ± 1 nmol/L DHT. ChIP assays were performed using AR antibodies. p52-activated AR is recruited only to AREIII, whereas DHT-activated AR is recruited to both AREIII and AREI/II. D, androgen-deprived LNCaP cells were cotransfected with p52 and pGL3-AREI/II-Luc, pGL3-AREIII-Luc, or pGL3-PSA-Luc ± 1 nmol/L DHT for 48 h. Luciferase activities were measured. Columns, mean of triplicate samples; bars, SD. Three independent experiments were performed.
NF-κB2/p52 induces transactivation of PSA promoter
We used PSA promoter as a model to examine the effects of p52 on AR signaling, as it is the best-characterized androgen-responsive promoter. LNCaP cells were cotransfected with PSA enhancer-promoter-luciferase (pGL3-PSA-6.0-Luc) and plasmids carrying p52, p65, p100, or vector control. As shown in Fig. 1B, PSA promoter activity was increased 11-fold by overexpression of p52 compared with the vector control in CS-FBS. However, p100 decreased PSA promoter activity by ~2-fold compared with the vector control. In contrast to the activation of AR by p52, overexpression of p65 significantly reduced DHT-induced transactivation of ARE-Luc and pGL3-PSA-6.0-Luc reporters, consistent with previous reports that NF-κB–p65 and AR exhibit mutual transcriptional antagonism (39, 40). To our surprise, p52 did not induce ARE-Luc activity, which can be activated by DHT (Fig. 1B), suggesting that p52-activated AR is distinct from androgen-bound AR.
NF-κB2/p52–activated AR is preferentially recruited to the PSA distal enhancer
The PSA promoter in the PSA-Luc construct contains two sets of AREs: AREI/II (the proximal enhancer) located at −0.1 kb and AREIII (the distal enhancer) located at ~−6 kb. The ARE-Luc construct contains only AREI/II (the proximal enhancer). To determine whether p52 modulates recruitment of AR to the PSA promoter, ChIP assays were performed to examine the recruitment of AR by p52 and DHT. As shown in Fig. 1C, p52-activated AR binds only to the distal enhancer (AREIII), whereas DHT-bound AR is recruited to both ARE sites. To test whether the differential recruitment of AR to AREs by p52 translates to differential AR activation, we performed luciferase assays using reporters containing the enhancer element (AREIII-Luc), the proximal AREs (AREI/II-Luc), or the full-length PSA promoter (PSA-6.0-Luc). We found that p52 induced the activity of AREIII-Luc and the full-length promoter but not AREI/II-Luc. As expected, DHT stimulates the activity of AREI/II-Luc, AREIII-Luc, and PSA-6.0-Luc (Fig. 1D). These data showed that p52-activated AR is preferentially recruited to the PSA distal enhancer.
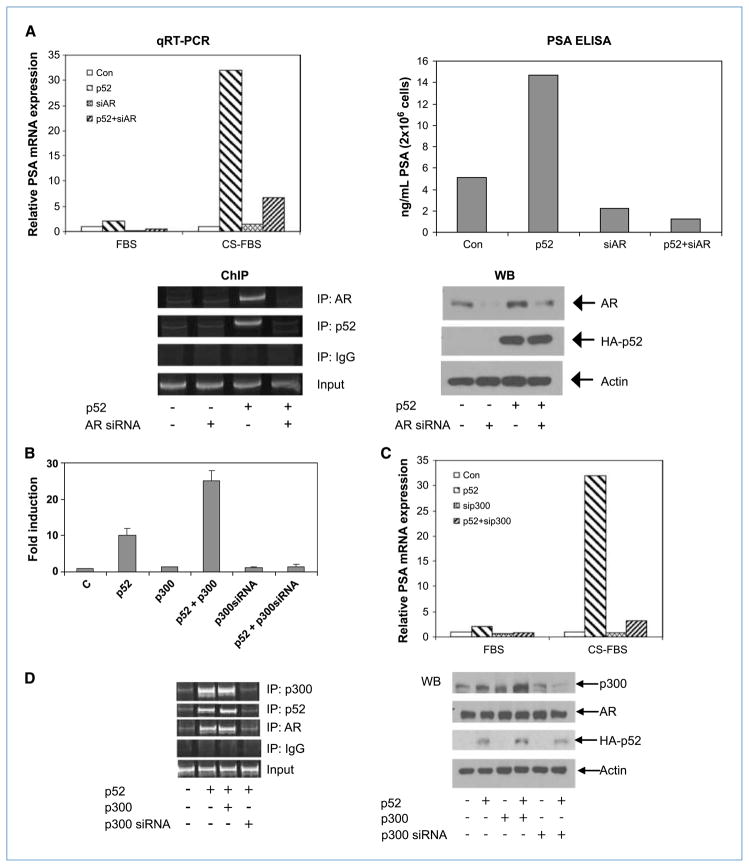
NF-κB2/p52–induced PSA expression is dependent on AR expression
Previous studies showed that NF-κB–p65: p50 complex binds directly to the distal core enhancer region in the PSA promoter and enhances PSA expression (41). So, we examined whether p52 binding to the AREIII element in the PSA enhancer is AR dependent or independent. Total RNAs were isolated from LNCaP cells transfected with plasmids encoding p52 and AR siRNA, and PSA mRNA expression was analyzed by quantitative RT-PCR (qRT-PCR). We found that p52 enhanced PSA mRNA expression ~30-fold in CS-FBS, which was almost completely abolished by down-regulation of AR. This was also confirmed using ELISA, where p52 induced the secretion of PSA by LNCaP cells in CS-FBS, which was abolished by downregulation of AR (Fig. 2A). To confirm that the recruitment of p52 to AREIII is AR dependent, we performed ChIP assay with lysates from LNCaP cells transfected with p52 and AR siRNA in CS-FBS. The results showed that recruitment of p52 to the AREIII site was completely AR dependent (Fig. 2A, ChIP), which was consistent with the above results. Lysates from these cells were analyzed by immunoblotting with anti-AR antibodies to confirm that AR expression was downregulated >70% in AR siRNA–transfected cells (Fig. 2A, WB). These findings clearly showed that the p52-enhanced PSA expression is AR dependent.
Figure 2.
p52-enhanced PSA expression is dependent on AR. LNCaP cells were transfected with plasmids expressing p52 or AR siRNA in FBS or CS-FBS. A, qRT-PCR: real-time PCR analysis was performed for PSA mRNA and results were expressed as fold change in relative mRNA expression. PSA ELISA: PSA protein expression was measured by ELISA in the supernatants. ChIP: ChIP assay was performed using AR, p52, or IgG antibodies. Recruitment of p52 to AREIII in the PSA promoter was abolished in the absence of AR. WB: expression levels of AR and p52 in the above experiments. B, p300 enhances PSA transactivation by p52. Androgen-deprived LNCaP cells were transfected with pGL3-PSA-Luc and plasmids expressing p52, p300, or p300 siRNA in CS-FBS for 48 h. Luciferase activities were measured. Columns, mean of triplicate samples; bars, SD. Three independent experiments were performed. C, p300 siRNA abolishes p52-induced increase in PSA mRNA. LNCaP cells transfected with plasmids expressing p52 or p300 siRNA were analyzed by qRT-PCR for PSA mRNA. D, p52 enhances p300 recruitment to the distal enhancer of PSA promoter. ChIP assay was performed using p300, p52, AR, or IgG antibodies and AREIII primers. WB: expression levels of HA-p52, AR, and p300 in the above experiments.
NF-κB2/p52 enhances the recruitment of p300 to the PSA distal enhancer
Optimal activation of AR requires the recruitment of coregulators, including p300, SRC-1, or TIF-2, to the AREs. To test whether p52 enhances the recruitment of these coregulators, we transfected the pGL3-PSA-6.0-Luc reporter along with p52- and p300-expressing plasmids or p300 siRNA into LNCaP cells in CS-FBS. Interestingly, pGL3-PSA-6.0-Luc was synergistically activated in the presence of p52 and p300 (>20-fold; Fig. 2B). p300 siRNA was able to reduce p52-induced transcriptional activity of AR. These results were confirmed using ChIP assays by transfecting p52 and p300 plasmids into LNCaP cells to test the recruitment of AR and p300 to AREIII. Overexpression of p52 enhanced the recruitment of p300 to AREIII, which was abolished by p300 siRNA (Fig. 2C). We also tested the effect of p300 siRNA on p52-mediated PSA mRNA expression using qRT-PCR. The results clearly showed that p300 siRNA almost completely abolished the p52-enhanced PSA expression (Fig. 2D). These results suggest that p52 may play an important role in the recruitment of coactivators such as p300 to AREs, thereby increasing the DNA-binding and, consequently, the transactivating ability of AR. LNCaP cells possess a functional but mutant AR. To test whether wild-type AR (Wt-AR) responds similarly to p52 expression, the experiments were repeated in LAPC-4 cells, which possess Wt-AR with similar results (Supplementary Fig. S1). These findings confirm that mutant AR in LNCaP cells as well as Wt-AR in LAPC-4 cells responds to activation by p52 in a similar manner.
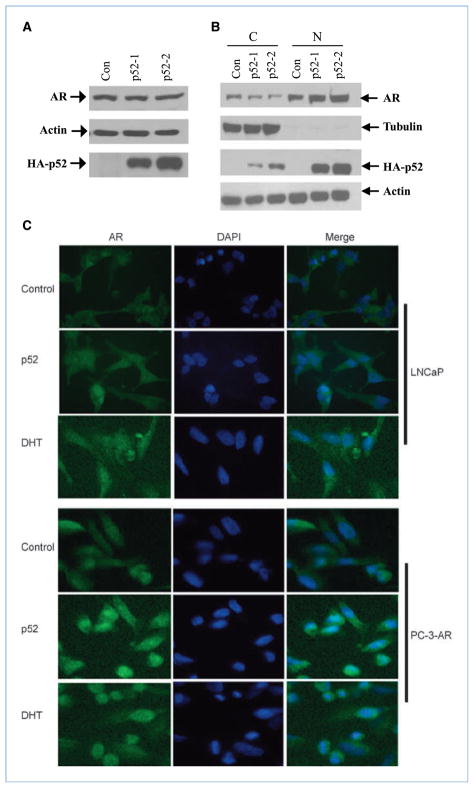
NF-κB2/p52 enhances the nuclear translocation of AR
To test whether the above effects were due to an increase in p52-induced AR expression, LNCaP cells were grown in CS-FBS for 2 days and transfected with p52-expressing plasmids. Whole-cell extracts were analyzed with anti-AR antibodies. The results showed that p52 did not enhance the expression of AR (Fig. 3A). The AR typically translocates from the cytoplasm to the nucleus on activation. To examine whether p52 enhances nuclear translocation of the AR protein in the presence or absence of low levels of androgen, nuclear extracts from p52-transfected LNCaP cells grown in CS-FBS were analyzed by immunoblotting with anti-AR antibodies. The level of nuclear translocation of AR under conditions of androgen deprivation was very low, which was enhanced with the expression of p52 (Fig. 3B). To further confirm these results, LNCaP cells transfected with p52 or control were processed for immunofluorescent staining with AR antibodies. AR was confined mainly to the cytoplasmic compartment in control cells in CS-FBS, whereas stronger nuclear staining of AR was observed in p52-transfected cells, although some AR was still seen in the cytoplasm (Fig. 3C, top). To verify that these results were not due to the T877A mutation present in the AR in LNCaP cells, we cotransfected AR-negative PC-3 cells with Wt-AR and p52 followed by immunofluorescent staining with AR antibodies. AR expression was mostly seen in the cytoplasmic compartment in control cells, whereas it was mostly located in the nucleus in p52-transfected cells (Fig. 3C, bottom). These results suggested that p52 may facilitate the nuclear translocation of AR during androgen deprivation.
Figure 3.
p52 induces nuclear translocation of AR. Androgen-deprived LNCaP cells were transfected with HA-p52 and control vector. Whole-cell (A), cytoplasmic (C), and nuclear (N) proteins (B) were immunoblotted with AR, actin, tubulin, or HA antibodies. Actin and tubulin were used as loading controls for whole-cell and cytoplasmic proteins, respectively. C, top, androgen-deprived LNCaP cells were transfected with p52 plasmid (1 or 2 μg) or control vector or treated with 1 nmol/L DHT for 24 h; bottom, PC-3 cells were transfected with Wt-AR and HA-p52 in CS-FBS for 24 h. Cells were processed for immunofluorescent staining of AR, and nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI).
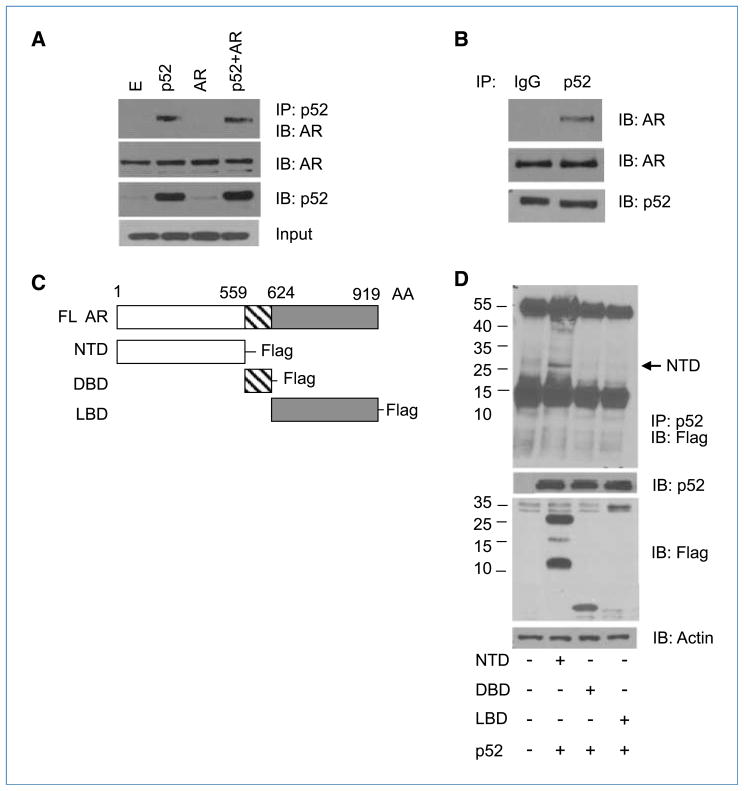
NF-κB2/p52 interacts with the NH2-terminal domain of AR
The above results suggested a physical interaction between the AR and p52. To determine whether p52 can interact with the AR, we transfected HA-p52 into LNCaP cells along with Wt-AR. Nuclear fractions were immunoprecipitated with anti-p52 antibodies, and the resulting eluates were analyzed by immunoblotting with anti-AR antibodies. We observed that p52 was able to coimmunoprecipitate with AR in the nuclear extracts (Fig. 4A). Immunoprecipitation with anti-p52 antibodies did not pull down detectable levels of AR in p52-nontransfected cells due to low levels of p52 in LNCaP cells (33). Coimmunoprecipitation of endogenous p52 with endogenous AR was also observed in C4-2 cells (Fig. 4B). These results suggest that p52 physically interacts with AR.
Figure 4.
p52 interacts with the NTD of AR. A, p52 immunoprecipitates with AR. Nuclear extracts from LNCaP cells were immunoprecipitated with p52 antibodies and probed for AR. Bottom, expression of AR or p52 in nuclear extracts. B, endogenous p52 immunoprecipitates with AR in C4-2 cells. Nuclear extracts from C4-2 cells were immunoprecipitated (IP) with p52 or IgG antibodies and immunoblotted (IB) with AR antibodies. C, schematic representation of the functional domains of AR [919 amino acids (AA)]. D, p52 associates with the NTD of AR. Extracts from LNCaP cells expressing Flag-tagged DBD, LBD, and NTD and p52 were immunoprecipitated with p52 antibodies and probed with anti-Flag antibodies. Bottom, immunoblotting with p52, Flag, and actin antibodies in cell extracts.
To test which domain of the AR interacts with p52, we cloned the three domains of AR [NH2-terminal transactivation domain (NTD), DNA-binding domain (DBD), and ligand-binding domain (LBD)] into Flag-tagged expression vectors by PCR amplification (Fig. 4C; Supplementary Fig. S2A; Supplementary Results). LNCaP cells were transfected with HA-tagged p52 and Flag-tagged AR-NTD/DBD/LBD–expressing plasmids and immunoprecipitated with anti-p52 antibodies and immunoblotted with anti-Flag antibodies (Fig. 4D) or immunoprecipitated with anti-Flag antibodies and probed with anti-HA antibodies (Supplementary Fig. S2B). These experiments revealed that the NTD of AR coprecipitates with p52, suggesting that p52 may interact with the NTD of AR.
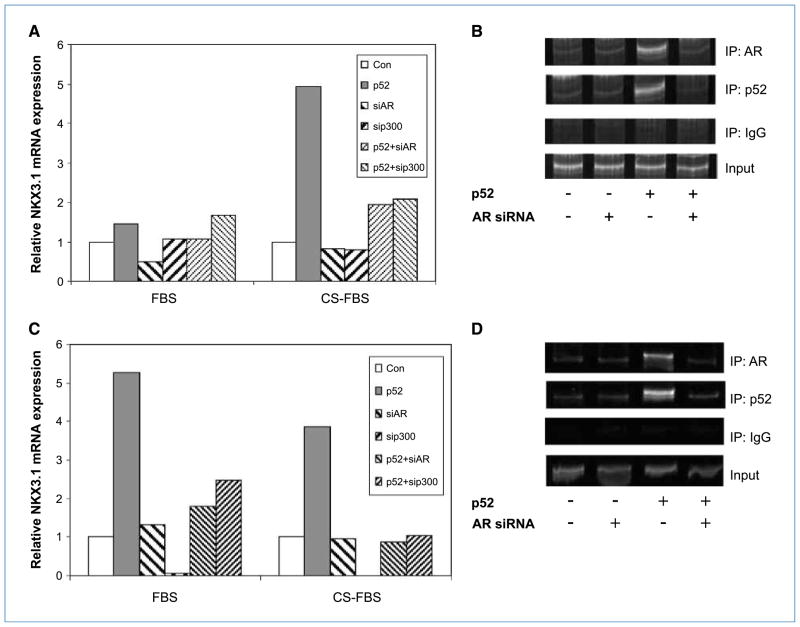
NF-κB2/p52 induces recruitment of AR to AREs in NKX3.1 promoter
The above results showed that NF-κB2/p52 activates transcription of the androgen-responsive gene PSA under conditions of androgen deprivation. To determine whether this effect is specific to PSA, we examined the effect of p52 expression on another typical AR target gene, NKX3.1. LNCaP and LAPC-4 cells were transfected with plasmids expressing p52, AR siRNA, or p300 siRNA in CS-FBS. Total RNAs were analyzed by qRT-PCR for NKX3.1 mRNA. The results showed that p52 induced ~4–5-fold increase in NKX3.1 mRNA, which was abolished in the presence of AR or p300 siRNAs (Fig. 5A and C). ChIP assays were performed to confirm whether the observed induction of NKX3.1 transcription was mediated by recruitment of AR to AREs in its promoter. Results showed that p52 induced the recruitment of AR to AREs in NKX3.1 promoter in CS-FBS. Recruitment of p52 to the AREs was also observed, which was abolished by coexpression of AR siRNA in both LNCaP and LAPC-4 cells (Fig. 5B and D). The above findings showed that the effect of NF-κB2/p52 in activation of AR is not restricted to PSA.
Figure 5.
Expression of the AR-regulated gene NKX3.1 is enhanced by p52 in an AR-dependent manner. LNCaP cells were transfected with plasmids expressing p52 (1 μg), or p300 and AR siRNAs in FBS or CS-FBS. A, total RNAs were analyzed by qRT-PCR for NKX3.1 mRNA. p52 induced the expression of NKX3.1 mRNA, which was abolished by downregulation of AR. B, ChIP assays were performed with AR, p52, or IgG antibodies. p52 enhanced the recruitment of AR to NKX3.1 promoter, whereas recruitment of p52 was AR dependent. LAPC-4 cells were transfected with plasmids expressing p52, or p300 or AR siRNAs in FBS or CS-FBS. C, total RNAs were analyzed by qRT-PCR for NKX3.1 mRNA. p52-induced expression of NKX3.1 mRNA was abolished by downregulation of AR. D, ChIP assays were performed with AR, p52, or IgG antibodies. p52 induced the recruitment of AR to NKX3.1 promoter, whereas recruitment of p52 was AR dependent.
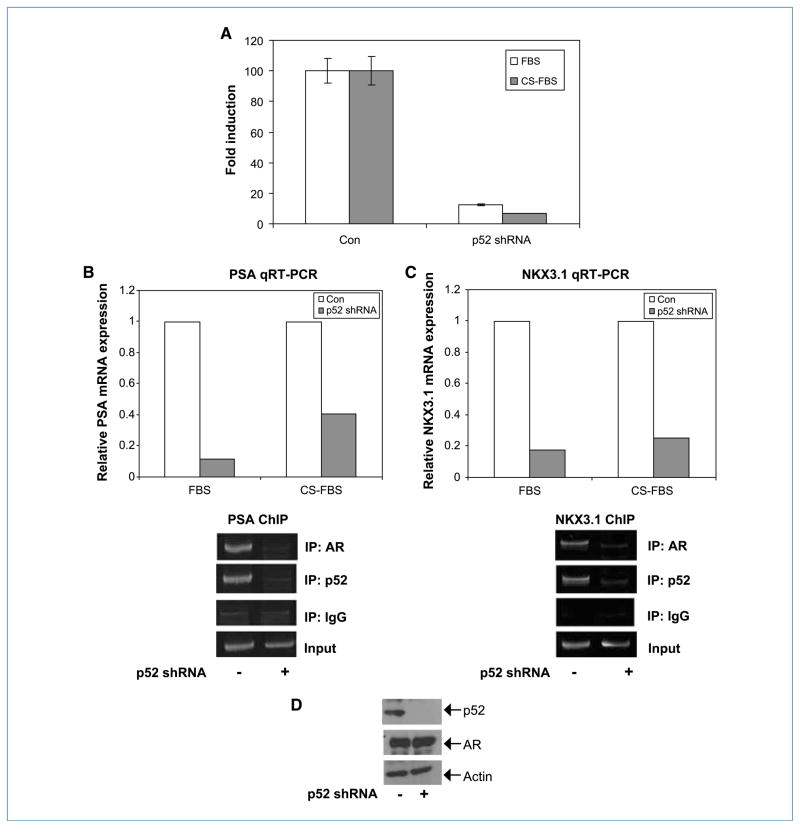
Downregulation of NF-κB2/p52 in C4-2 cells abolishes AR activation
C4-2 cells, a castration-resistant subline of LNCaP, exhibit high endogenous levels of p52 and constitutive activation of the AR compared with LNCaP. To test whether blocking endogenous p52 expression affects constitutive activation of AR in C4-2 cells, we transfected p52 and control short hairpin RNAs (shRNA) along with PSA-6.0-Luc reporter into C4-2 cells in FBS and CS-FBS. Luciferase activities were measured after downregulation of p52 expression was confirmed. The results showed that downregulation of p52 reduced transactivation of the PSA promoter-reporter by AR by ~80% (Fig. 6A). Total RNAs from the cells were also analyzed by qRT-PCR for PSA and NKX3.1 mRNAs. We found that downregulation of endogenous p52 expression resulted in 60% to 80% decrease in the transcript levels of both PSA and NKX3.1 (Fig. 6B and C, qRT-PCR). To test whether downregulation of p52 affects recruitment of AR to the promoters of PSA and NKX3.1 genes, ChIP assays were performed with extracts from C4-2 cells treated with p52 shRNA. Knockdown of p52 abolished the recruitment of AR to these promoters almost completely (Fig. 6B and C, ChIP). Expression levels of AR and p52 were verified by immunoblotting (Fig. 6D). These findings showed that downregulation of endogenous p52 expression reduced AR activity in cells expressing high levels of both proteins.
Figure 6.
Downregulation of endogenous p52 reduces AR activity in C4-2 cells. A, C4-2 cells were cotransfected with p52 or control shRNAs and pGL3-PSA-Luc in FBS or CS-FBS, and luciferase activity was measured after 48 h. Columns, mean; bars, SD. Downregulation of endogenous p52 expression in C4-2 cells reduced the transactivation of PSA promoter. C4-2 cells were transfected with p52 or control shRNAs. B, PSA qRT-PCR: total RNAs were analyzed by qRT-PCR for PSA mRNA. Results are expressed as fold change in expression. PSA ChIP: extracts were analyzed by ChIP assay using PSA-AREIII primers. Recruitment of AR to AREIII was abolished when endogenous p52 expression was downregulated. C, NKX3.1 qRT-PCR: cDNAs were also analyzed for the expression of NKX3.1 mRNA by qRT-PCR. Results are expressed as fold change in expression. NKX3.1 ChIP: extracts were analyzed by ChIP assay using primers against NKX3.1-ARE site. Constitutive recruitment of AR to the ARE was dependent on endogenous p52 expression in C4-2 cells. D, immunoblotting of above cell lysates with p52 and AR antibodies.
Comparison of relative effects of p52 and p65 on AR
To determine whether the above observed effects of p52 on AR activation were due to a p52-induced increase in AR expression and to compare the relative effects of p52 and p65 on AR expression, we analyzed AR protein and mRNA levels in LNCaP and LAPC-4 cells transfected with p52 or p65 by immunoblotting and qRT-PCR. Although we observed 1.5- to 2-fold induction of AR mRNA levels by p52, no significant induction in protein levels was detected by overexpression of p52 or p65 (Supplementary Fig. S3A and C). However, p52, but not p65, induced AR nuclear translocation (Fig. 3; Supplementary Fig. S3B).
To compare the effects of p52 and p65 on AR-regulated genes, we analyzed expression levels of PSA and NKX3.1 in LNCaP and LAPC-4 cells expressing p52 or p65 in the presence or absence of androgen. p52, and not p65, induced significant increases in expression levels of PSA and NKX3.1 (Supplementary Fig. S4A–C) and transactivated PSA promoter-driven luciferase reporter (Supplementary Fig. S4D). These findings showed that p52-induced AR activation is not due to an increase in AR expression.
Discussion
Prostate cancer is the most common type of cancer in American men, and failure of androgen deprivation therapy leads to castration-resistant disease progression. Accumulating evidence suggests that ligand-independent activation of AR and development of apoptosis-resistant cells contribute to CRPC. AR activation in CRPC may occur by a variety of mechanisms that alter the sensitivity or specificity of AR. Previous studies showed that overexpression of NF-κB2/p52 induced castration-resistant growth of androgen-sensitive LNCaP cells in vivo. In this study, we investigated the potential role of p52 in activation of AR and show that p52 activates AR by interacting with its NH2-terminal domain. p52 not only recapitulates DHT-mediated AR activation but also has a unique action in that p52-activated AR is preferentially recruited to the distal enhancer of the PSA promoter and is assembled into transcriptional complexes with coactivators such as p300 to optimize target gene transcription under androgen deprivation. These findings were confirmed in cell lines that possess Wt-AR (LAPC-4) as well as mutant AR (LNCaP). Downregulation of p52 in C4-2 cells, which constitutively express high levels of p52, led to abrogation of the constitutive activation of AR under conditions of androgen deprivation. These results suggest that NF-κB2/p52 plays a major role in aberrant activation of the AR in prostate cancer cells after androgen ablation.
The classic pathway of NF-κB is constitutively activated in prostate cancer, whereas the number of reports on activation of the alternative pathway is limited (31–33). We have reported that activated Stat3 may be involved in the processing of NF-κB2/p100 in prostate cancer (32) and that overexpression of p52 induces androgen-independent growth of androgen-dependent LNCaP cells in vivo (33). We used several different approaches and provide direct evidence that p52 increases AR activation and transactivation of AR-responsive genes, may interact with the AR directly via its NTD, and enhances recruitment of coactivators such as p300 to the AREs of AR-responsive promoters. Previous studies have shown that p65 (RelA) induces expression and activation of the AR (42, 43), whereas our results show that p52 did not induce AR protein expression but induced nuclear translocation and enhanced transactivation of AR-responsive genes. The present study thus reveals a hitherto unknown positive link between the AR and the noncanonical NF-κB pathway.
It is interesting to note that p52 recruits AR preferentially to the distal, but not to the proximal, promoter region of the PSA gene, whereas DHT recruits AR to both distal and proximal enhancers of the PSA gene. These observations suggest that p52-activated AR may be conformationally different from DHT-activated AR and that it is likely to require other transcriptional mediators to “help” stabilize binding and potentiate transcription, as shown by results that p52 enhances recruitment of coactivators such as p300 to AR-responsive promoters and facilitates transactivation of androgen-responsive genes. Our finding is echoed by several recent studies (44, 45), which showed that the AR activated by aberrant signals such as neuropeptide and epidermal growth factor is recruited preferentially to AREI/II. It was shown that the modified AR efficiently translocates into the nucleus and assembles coactivator complexes different from those of DHT-bound AR, similar to our findings. These results are significant in showing that the AR activated by aberrant signals may have a conformation different from the DHT-bound one and could account for the different transcriptional outputs and hence phenotypes of hormone-sensitive versus CRPC.
In summary, we showed that NF-κB/p52 activates the AR and enhances nuclear translocation and activation of AR by interacting with its NTD. p52 enhances the recruitment of coactivators such as p300 to the promoters of AR-dependent genes, resulting in increased transactivation of AR-responsive genes in androgen-deprived conditions. Our findings, together with previous reports that the levels of p52 are elevated in prostate cancer cells and that NF-κB2/p52 promotes prostate cancer cell growth in vitro and in vivo, suggest that p52 may play a critical role in the progression of CRPC.
Supplementary Material
Acknowledgments
We thank Dr. Chinghai Kao (University of Indiana) for pGL3-PSA-6.0-Luc and Dr. Chawnshang Chang (University of Rochester) for Wt-AR.
Grant Support
NIH grants CA118887 and CA109441 and Department of Defense grant PC080538.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Hobisch A, Culig Z, Radmayr C, Bartsch G, Klocker H, Hittmair A. Distant metastases from prostatic carcinoma express androgen receptor protein. Cancer Res. 1995;55:3068–72. [PubMed] [Google Scholar]
- 2.van der Kwast TH, Schalken J, Ruizeveld de Winter JA, et al. Androgen receptors in endocrine-therapy-resistant human prostate cancer. Int J Cancer. 1991;48:189–93. doi: 10.1002/ijc.2910480206. [DOI] [PubMed] [Google Scholar]
- 3.de Miguel F, Lee S, Onate S, Gao A. Stat3 enhances transactivation of steroid hormone receptors. Nucl Recept. 2003;1:3–9. doi: 10.1186/1478-1336-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abreu-Martin MT, Chari A, Palladino AA, Craft NA, Sawyers CL. Mitogen-activated protein kinase kinase kinase 1 activates androgen receptor-dependent transcription and apoptosis in prostate cancer. Mol Cell Biol. 1999;19:5143–54. doi: 10.1128/mcb.19.7.5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sadar MD. Androgen-independent induction of prostate-specific antigen gene expression via cross-talk between the androgen receptor and protein kinase A signal transduction pathways. J Biol Chem. 1999;274:7777–83. doi: 10.1074/jbc.274.12.7777. [DOI] [PubMed] [Google Scholar]
- 6.Chen T, Wang LH, Farrar WL. Interleukin 6 activates androgen receptor-mediated gene expression through a signal transducer and activator of transcription 3-dependent pathway in LNCaP prostate cancer cells. Cancer Res. 2000;60:2132–5. [PubMed] [Google Scholar]
- 7.Wen Y, Hu MC-T, Makino K, et al. HER-2/neu promotes androgen-independent survival and growth of prostate cancer cells through the Akt pathway. Cancer Res. 2000;60:6841–5. [PubMed] [Google Scholar]
- 8.Hobisch A, Eder IE, Putz T, et al. Interleukin-6 regulates prostate-specific protein expression in prostate carcinoma cells by activation of the androgen receptor. Cancer Res. 1998;58:4640–5. [PubMed] [Google Scholar]
- 9.DeMiguel F, Lee SO, Lou W, et al. Stat3 enhances the growth of LNCaP human prostate cancer cells in intact and castrated male nude mice. Prostate. 2002;52:123–9. doi: 10.1002/pros.10110. [DOI] [PubMed] [Google Scholar]
- 10.Tilley WD, Buchanan G, Hickey TE, Bentel JM. Mutations in the androgen receptor gene are associated with progression of human prostate cancer to androgen independence. Clin Cancer Res. 1996;2:277–85. [PubMed] [Google Scholar]
- 11.Linja MJ, Savinainen KJ, Saramaki OR, Tammela TL, Vessella RL, Visakorpi T. Amplification and overexpression of androgen receptor gene in hormone-refractory prostate cancer. Cancer Res. 2001;61:3550–5. [PubMed] [Google Scholar]
- 12.Dehm SM, Schmidt LJ, Heemers HV, Vessella RL, Tindall DJ. Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res. 2008;68:5469–77. doi: 10.1158/0008-5472.CAN-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu R, Dunn TA, Wei S, et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res. 2009;69:16–22. doi: 10.1158/0008-5472.CAN-08-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yeh S, Miyamoto H, Shima H, Chang C. From estrogen to androgen receptor: a new pathway for sex hormones in prostate. Proc Natl Acad Sci U S A. 1998;95:5527–32. doi: 10.1073/pnas.95.10.5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohler JL, Gregory CW, Ford OH, III, et al. The androgen axis in recurrent prostate cancer. Clin Cancer Res. 2004;10:440–8. doi: 10.1158/1078-0432.ccr-1146-03. [DOI] [PubMed] [Google Scholar]
- 16.Montgomery RB, Mostaghel EA, Vessella R, et al. Maintenance of intratumoral androgens in metastatic prostate cancer: a mechanism for castration-resistant tumor growth. Cancer Res. 2008;68:4447–54. doi: 10.1158/0008-5472.CAN-08-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marks LS, Mostaghel EA, Nelson PS. Prostate tissue androgens: history and current clinical relevance. Urology. 2008;72:247–54. doi: 10.1016/j.urology.2008.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mostaghel EA, Nelson PS. Intracrine androgen metabolism in prostate cancer progression: mechanisms of castration resistance and therapeutic implications. Best Pract Res Clin Endocrinol Metab. 2008;22:243–58. doi: 10.1016/j.beem.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao G, Harhaj EW, Sun SC. NF-κB-inducing kinase regulates the processing of NF-κB2 p100. Mol Cell. 2001;7:401–9. doi: 10.1016/s1097-2765(01)00187-3. [DOI] [PubMed] [Google Scholar]
- 20.Xiao G, Fong A, Sun SC. Induction of p100 processing by NF-κB-inducing kinase involves docking IκB kinase ακ(IKKα) to p100 and IKKα-mediated phosphorylation. J Biol Chem. 2004;279:30099–105. doi: 10.1074/jbc.M401428200. [DOI] [PubMed] [Google Scholar]
- 21.Qing G, Xiao G. Essential role of IκB kinase ακ in the constitutive processing of NF-κB2 p100. J Biol Chem. 2005;280:9765–8. doi: 10.1074/jbc.C400502200. [DOI] [PubMed] [Google Scholar]
- 22.Qing G, Qu Z, Xiao G. Regulation of NF-κB2 p100 processing by its cis-acting domain. J Biol Chem. 2005;280:18–27. doi: 10.1074/jbc.M406619200. [DOI] [PubMed] [Google Scholar]
- 23.Dejardin E, Droin NM, Delhase M, et al. The lymphotoxin-βκ receptor induces different patterns of gene expression via two NF-κB pathways. Immunity. 2002;17:525–35. doi: 10.1016/s1074-7613(02)00423-5. [DOI] [PubMed] [Google Scholar]
- 24.Coope HJ, Atkinson PG, Huhse B, et al. CD40 regulates the processing of NF-κB2 p100 to p52. EMBO J. 2002;21:5375–85. doi: 10.1093/emboj/cdf542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caamano JH, Rizzo CA, Durham SK, et al. Nuclear factor (NF)-κB2 (p100/p52) is required for normal splenic microarchitecture and B cell-mediated immune responses. J Exp Med. 1998;187:185–96. doi: 10.1084/jem.187.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Franzoso G, Carlson L, Poljak L, et al. Mice deficient in nuclear factor (NF)-κB/p52 present with defects in humoral responses, germinal center reactions, and splenic microarchitecture. J Exp Med. 1998;187:147–59. doi: 10.1084/jem.187.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishikawa H, Carrasco D, Claudio E, Ryseck RP, Bravo R. Gastric hyperplasia and increased proliferative responses of lymphocytes in mice lacking the COOH-terminal ankyrin domain of NF-κB2. J Exp Med. 1997;186:999–1014. doi: 10.1084/jem.186.7.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao G, Rabson AB, Young W, Qing G, Qu Z. Alternative pathways of NF-κB activation: a double-edged sword in health and disease. Cytokine Growth Factor Rev. 2006;17:281–93. doi: 10.1016/j.cytogfr.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 29.Connelly L, Robinson-Benion C, Chont M, et al. A transgenic model reveals important roles for the NF-κB alternative pathway (p100/p52) in mammary development and links to tumorigenesis. J Biol Chem. 2007;282:10028–35. doi: 10.1074/jbc.M611300200. [DOI] [PubMed] [Google Scholar]
- 30.Cogswell PC, Guttridge DC, Funkhouser WK, Baldwin AS., Jr Selective activation of NF-κB subunits in human breast cancer: potential roles for NF-κB2/p52 and for Bcl-3. Oncogene. 2000;19:1123–31. doi: 10.1038/sj.onc.1203412. [DOI] [PubMed] [Google Scholar]
- 31.Lessard L, Karakiewicz PI, Bellon-Gagnon P, et al. Nuclear localization of nuclear factor-κB p65 in primary prostate tumors is highly predictive of pelvic lymph node metastases. Clin Cancer Res. 2006;12:5741–5. doi: 10.1158/1078-0432.CCR-06-0330. [DOI] [PubMed] [Google Scholar]
- 32.Nadiminty N, Lou W, Lee SO, Lin X, Trump DL, Gao AC. Stat3 activation of NF-κB p100 processing involves CBP/p300-mediated acetylation. Proc Natl Acad Sci U S A. 2006;103:7264–9. doi: 10.1073/pnas.0509808103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nadiminty N, Chun JY, Lou W, Lin X, Gao AC. NF-κB2/p52 enhances androgen-independent growth of human LNCaP cells via protection from apoptotic cell death and cell cycle arrest induced by androgen-deprivation. Prostate. 2008;68:1725–33. doi: 10.1002/pros.20839. [DOI] [PubMed] [Google Scholar]
- 34.Nadiminty N, Dutt S, Tepper C, Gao AC. Microarray analysis reveals potential target genes of NF-kB2/p52 in LNCaP prostate cancer cells. Prostate. 2010;70(3):276–87. doi: 10.1002/pros.21062. [DOI] [PubMed] [Google Scholar]
- 35.Lee SO, Chun JY, Nadiminty N, Lou W, Gao AC. Interleukin-6 undergoes transition from growth inhibitor associated with neuroendocrine differentiation to stimulator accompanied by androgen receptor activation during LNCaP prostate cancer cell progression. Prostate. 2007;67:764–73. doi: 10.1002/pros.20553. [DOI] [PubMed] [Google Scholar]
- 36.Yoon H-G, Wong J. The corepressors silencing mediator of retinoid and thyroid hormone receptor and nuclear receptor corepressor are involved in agonist- and antagonist-regulated transcription by androgen receptor. Mol Endocrinol. 2006;20:1048–60. doi: 10.1210/me.2005-0324. [DOI] [PubMed] [Google Scholar]
- 37.Chun JY, Nadiminty N, Lee SO, Onate S, Lou W, Gao AC. Mechanisms of selenium downregulation of androgen receptor signaling in prostate cancer. Mol Cancer Ther. 2006;5:913–8. doi: 10.1158/1535-7163.MCT-05-0389. [DOI] [PubMed] [Google Scholar]
- 38.Chun JY, Nadiminty N, Dutt S, et al. Interleukin-6 regulates androgen synthesis in prostate cancer cells. Clin Cancer Res. 2009;15:4815–22. doi: 10.1158/1078-0432.CCR-09-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palvimo JJ, Reinikainen P, Ikonen T, Kallio PJ, Moilanen A, Janne OA. Mutual transcriptional interference between RelA and androgen receptor. J Biol Chem. 1996;271:24151–6. doi: 10.1074/jbc.271.39.24151. [DOI] [PubMed] [Google Scholar]
- 40.Cinar B, Yeung F, Konaka H, et al. Identification of a negative regulatory cis-element in the enhancer core region of the prostate-specific antigen promoter: implications for intersection of androgen receptor and nuclear factor-κB signalling in prostate cancer cells. Biochem J. 2004;379:421–31. doi: 10.1042/BJ20031661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen CD, Sawyers CL. NF-κB activates prostate-specific antigen expression and is upregulated in androgen-independent prostate cancer. Mol Cell Biol. 2002;22:2862–70. doi: 10.1128/MCB.22.8.2862-2870.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin RJ, Lho Y, Connelly L, et al. The nuclear factor-κB pathway controls the progression of prostate cancer to androgen-independent growth. Cancer Res. 2008;68:6762–9. doi: 10.1158/0008-5472.CAN-08-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang L, Altuwaijri S, Deng F, et al. NF-κB regulates androgen receptor expression and prostate cancer growth. Am J Pathol. 2009;175:489–99. doi: 10.2353/ajpath.2009.080727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang JC, Ok JH, Busby JE, Borowsky AD, Kung HJ, Evans CP. Aberrant activation of androgen receptor in a new neuropeptide-autocrine model of androgen-insensitive prostate cancer. Cancer Res. 2009;69:151–60. doi: 10.1158/0008-5472.CAN-08-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Desai SJ, Ma AH, Tepper CG, Chen HW, Kung HJ. Inappropriate activation of the androgen receptor by nonsteroids: involvement of the Src kinase pathway and its therapeutic implications. Cancer Res. 2006;66:10449–59. doi: 10.1158/0008-5472.CAN-06-2582. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.