Abstract
Self-rated health questions have been proven to be a highly reliable and valid measure of overall health as measured by other indicators in many population groups. It also has been shown to be a very good predictor of mortality, chronic or severe diseases, and the need for services, and is positively correlated with clinical assessments. Genetic factors have been estimated to account for 25 – 64% of the variance in the liability of self-rated health. The aim of the present study was to identify Single Nucleotide Polymorphisms (SNPs) underlying the heritability of self-rated health by conducting a genome-wide association analysis in a large sample of 6,706 Australian individuals aged 18–92. No genome wide significant SNPs associated with self-rated health could be identified, indicating that self-rated health may be influenced by a large number of SNPs with very small effect size. A very large sample will be needed to identify these SNPs.
Keywords: genome-wide association, genes, self-rated health, self-reported health, health
Introduction
Self-rated health questions have been developed with the objective of quantifying an individual’s perception of his or her overall health state. Even though these measures tend to be less sensitive to changes in specific disorders (Beaton & Schemitsch, 2003), it have been proven to be a highly reliable and valid measure of overall health as measured by means of other indicators in different population groups (Lundberg & Manderbacka, 1996). It has also been shown to be a very good predictor of mortality and the need for services (Leinonen, et al., 2005), and is positively correlated with clinical assessments (Romeis, et al., 2000). Furthermore, higher self-rated health has been associated with absence of chronic diseases, severe diseases, disabilities, functional limitations, and with higher physical activity, and better psychosocial wellbeing (Bryant, Beck, & Fairclough, 2000; Idler, 1993; Leinonen, Heikkinen, & Jylha, 2001; Rodin & McAvay, 1992). Typically, self-rated health is based on a single question asking the respondents to rate their current health status. Most individuals rate their health as moderate to good while few would rate their health as bad (Juerges, 2007).
Several twin studies have investigated the heritability of self-rated health (Christensen, Holm, McGue, Corder, & Vaupel, 1999; Harris, Pedersen, McClearn, Plomin, & Nesselroade, 1992; Leinonen, et al., 2005; Lichtenstein & Pedersen, 1995; Romeis, et al., 2000; Silventoinen, Posthuma, Lahelma, Rose, & Kaprio, 2007; Svedberg, Lichtenstein, & Pedersen, 2001) estimating genetic factors to account for 25 – 64 % of the variance in the liability of self-rated somatic health. A large longitudinal study of Finnish twins showed that the heritability of self-rated health was greatest at age 16, at 63% (95% CI: 0.56 – 0.67), declining steadily to age 25 with a heritability of 33% (CI: 0.25 – 0.41) (Silventoinen et al., 2006). The study revealed moderate correlations between the different health ratings at different life stages (r = 0.33–0.61), which were predominately due to genetic factors. The finding of decreasing heritability of self-rated health with age, however, is not confirmed by cross-sectional studies, for example Mosing et al. (2009) found a heritability of 46% in an elderly twin sample (mean age = 61 ± 8.8). As self-rated health has been shown to be for a substantial part due to genetic factors, it would be interesting to explore the genetic variants underlying this trait. The aim of the present study was to identify SNPs underlying the heritability of self-rated health by conducting a genome-wide association (GWA) analysis on a large sample of Australian individuals who have previously rated their health status.
Methods
Participants and measures
Self-rated health data were collected in four twin family studies conducted at the Queensland Institute of Medical Research (QIMR). The two earliest studies (Study 1 and Study 2) were conducted between 1993 and 1995. Study 3 was conducted between 1996 and 2000 and the most recent and largest study (Study 4), was collected between 2001 and 2005 (see Table 1a for more details). All four studies consisted of mailed-out questionnaires assessing health and lifestyle issues as well as demographic information and were approved by the QIMR Human Research Ethics Committee. Content and sampling methods of the four studies have been described in detail elsewhere (Bucholz, et al., 1998; Hansell, et al., 2008; Heath, et al., 1997; Mosing, Gordon, et al., 2009; Mosing, Zietsch, et al., 2009). In Study 3 and 4 (more than 95% of the final sample), self-rated health was assessed with the following item: “How would you describe your general physical health?”, rated on a 4-point Likert scale, “excellent” (1), “good” (2), “fair” (3), “poor” (4). In the other two studies the self-rated health questions were worded slightly differently: “In general, would you say that your physical health now is excellent, good, fair or poor?” (Study 1) and “How would you describe your health at present? – very good, good, fair, poor, very poor” (Study 2). As few individuals rated their health as poor or very poor and for consistency with the self-rated health questions of the other studies, which only had four instead of five categories, the categories poor and very poor of the self-rated health item in Study 2 were collapsed.
Table 1a.
Raw number and percentage (in brackets) of participants drawn from each study. The most recent score of participants who took part in more than one study was used.
Self-rated health and genotype data were available for 6,706 individuals (3,710 females and 2,996 males) from 2,585 independent families aged between 18–92 years (Mean=46; SD=11). Of these, 1,403 (21%) individuals participated in more than one of the studies in which case the most recent rating was used. Test-retest Pearson correlations between the different self-rated health measures ranged between 0.48 and 0.65 and the correlation between the two identically worded items was not higher compared to the questions used in the other two studies. Table 1a shows the number of individuals derived from each study forming the final sample. Finally, in line with previous findings, most individuals rated their health as good while few reported poor health, resulting in a skewed distribution (Table 1b). Therefore, a square root transformation was applied to the final scores and the scores were treated as continuous. Previous behaviour genetic analysis in a subsample of the present sample revealed heritability estimates of 46% with the remaining variance being due to non-shared environmental influences (Mosing, Zietsch, et al., 2009).
Table 1b.
Score distribution of the final sample for self-rated health.
| Excellent | Good | Fair | Poor | Total | |
|---|---|---|---|---|---|
| N (%) | 1837 (27%) | 3640 (54%) | 1053 (16%) | 176 (3%) | 6706 (100%) |
Genotyping, quality control, and imputation procedures
Over more than twenty years a wide range of phenotypic data and DNA samples have been collected as part of the different projects. The DNA samples were collected in accordance with standard protocols and were genotyped using the following Illumina SNP platforms: 317K, HumanCNV370-Quadv3, and Human610-Quad. Quality control (QC) procedures employed are discussed in full detail in Medland et al. (2009). Briefly, checks for ancestry outliers, Mendelian errors, Hardy Weinberg Equilibrium, and Minor Allele Frequency (MAF) were conducted separately for each of the projects and then again for the combined dataset. The final dataset consisted of 269,840 SNPs and was imputed by MACH (Abecasis, unpublished) using the data from the European HapMap 1+2, Release 22 Build 36. SNPs with an imputation quality score (r2) greater than 0.3 were retained resulting in a total of 2,380,486 imputed SNPs. Finally, if only one individual from a monozygotic twin pair had been genotyped, the non-genotyped co-twin was assigned that genotype as well.
Statistical analyses
The best guess genotype at each SNP was tested for association with self-rated health using the family-based association test in Merlin (Chen & Abecasis, 2007) accounting for family relationships. The additive genetic effect was computed by modelling the genotypic mean of the heterozygote (Aa) as the average of the two homozygotes (AA, aa). The generally accepted genome-wide significance level for the association between SNP and phenotype at α = 0.05 is 7.2*10−8 or smaller, correcting for the total number of independent tests (Dudbridge & Gusnanto, 2008), and was also applied in the present study.
Additionally, a gene-based test (VEGAS), feasible for use with GWA data with related individuals (Liu, et al., accepted), was conducted to test whether there are any genes which harbour an excess of associated variants. Details of this procedure are summarized elsewhere (Liu, et al., accepted; Verweij, et al., accepted). In brief, this test explores association on a per-gene basis taking the p-values of all SNPs within 50kb of each gene, as well as linkage disequilibrium (LD) and number of SNPs per gene into account. A p-value below α = 2.8*10−6 was considered to be significant as the gene-based association test included 17,585 genes (0.05/17,585).
Finally, power calculations were conducted by performing association tests on simulated datasets (based on our sample) in Merlin. The simulated datasets maintain the features of the original data in terms of marker informativeness, allele frequency, spacing, missing data patterns, and trait distribution despite replacing the phenotypic values and the genotypes for a randomly selected SNP with a minor allele frequency of 0.25. One-thousand simulated data sets were generated on which association analyses were subsequently performed. Detailed information on the simulation procedure can be found on http://www.sph.umich.edu/csg/abecasis/Merlin/reference/simulation.html. The empirical power estimate is given by the proportion of genome-wide significant association tests detected in the 1000 association analyses. The present sample provides 99% and 50% power to detect SNPs explaining 1% and 0.5% of variance in self-rated health, respectively.
Results
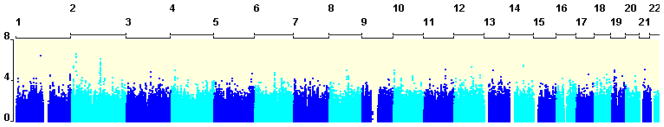
We tested 2,380,486 SNPs for association with self-rated health correcting for age and sex. The Quantile-Quantile (QQ) plot (Figure 1) shows the association between the observed versus the expected (under the null-hypothesis of no association) p-values of the autosomal associations.
Figure 1.
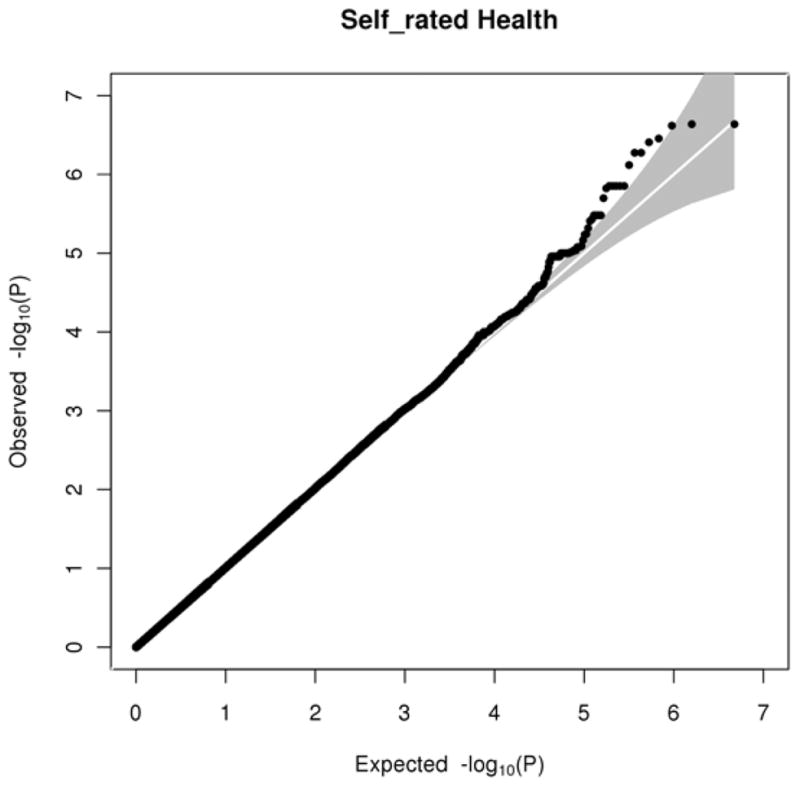
The Q-Q plot shows the association between the observed and expected −log10(P-value) of the autosomal association between SNPs and self-rated health with the grey area representing the 95% confidence interval.
Results of the association analysis (shown in Figure 2) indicate that there are no genome-wide significant (α = 7.2*10−8) association signals, with the smallest p-value of 2.3*10−7 obtained for a SNP (rs17043947) on chromosome 2p24.1. However, though not significant, we found two promising regions on chromosome two (2p24.1 and 2q14.3) with the top hits having p-values of 2.3*10−7 and 7.6*10−7, respectively. Table 2 shows SNPs in the top 50 smallest p-values for self-rated health. Redundant SNPs in high LD (r2 > .70) with a more significant SNP were excluded.
Figure 2.
Manhattan plot showing the results of the genome-wide association analyses for self-rated health with the x-axis showing chromosome numbers and the y-axis the p-value (−log10) of the association signals.
Table 2.
Genetic markers showing strongest association with self-rated health (independent markers within top 50 SNPs).
| Chr | SNP | Base pair location | p-value | SNPs in LD | Minor allele | MA F | Effect size | Closest gene | location |
|---|---|---|---|---|---|---|---|---|---|
| 2 | rs17043947 | 22736987 | 2.3 * 10-7 | 8 | T | .04 | −0.07 | ||
| 1 | rs958798 | 110770223 | 3.5 * 10-7 | T | .17 | 0.03 | KCNC4 | Intronic | |
| 2 | rs17043944 | 22724593 | 5.3 * 10-7 | 7 | T | .02 | −0.10 | ||
| 2 | rs7567389 | 127982645 | 1.5 * 10-6 | 2 | A | .30 | −0.03 | CYP27C1 | Upstream |
| 2 | rs6759460 | 127897011 | 3.3 * 10-6 | 4 | C | .24 | 0.03 | AC110926 | Intronic |
| 14 | rs6573416 | 62518348 | 3.3 * 10-6 | 1 | G | .13 | 0.03 | SYT16 | Intronic |
| 2 | rs2357266 | 9924387 | 3.7 * 10-6 | 1 | G | .35 | −0.02 | Intergenic | |
| 12 | rs300489 | 78485994 | 4.8 * 10-6 | A | .23 | 0.03 | NAV3 | Intronic | |
| 21 | rs7279441 | 24198815 | 8.3 * 10-6 | 1 | G | .14 | 0.03 | Intergenic | |
| 4 | rs17478107 | 16002288 | 9.2 * 10-6 | C | .28 | −0.02 | PROM1 | Intronic | |
| 2 | rs1799810 | 128176040 | 9.2 * 10-6 | T | .41 | −0.02 | |||
| 8 | rs12680321 | 77148730 | 9.8 * 10-6 | T | .20 | −0.03 | Intergenic | ||
| 13 | rs9548119 | 38531581 | 9.3 * 10-6 | 1 | A | .22 | 0.03 | Intergenic | |
| 13 | rs9548119 | 38531581 | 9.3 * 10-6 | 1 | A | .22 | 0.03 | Intergenic | |
| 2 | rs1158867 | 128177377 | 1.0* 10-5 | C | .41 | −0.02 | |||
| 10 | rs11815041 | 1622424 | 1.0 * 10-5 | 5 | G | .49 | 0.02 | ADARB2 | Intronic |
| 19 | rs11085795 | 11988515 | 1.0 * 10-5 | 1 | A | .26 | 0.03 | ZNF439 | Intergenic |
| 11 | rs7120279 | 95720275 | 4.9 * 10-5 | 1 | C | .21 | −0.03 | MAML2 | Intronic |
Note. Independent markers: more than 500kb apart and in LD of r2 < 0.70; SNPs in LD: the number of correlated SNPs that are in the top 50 (nonindependent groups of markers); Chr = Chromosome; MAF = Minor Allele Frequency; Closest gene = name of gene if the SNP is located in a known gene or within 50kb distance from a gene; Base pair locations: obtained from the HapMapI+II (b36r22) CEU legend files; Closest gene to the SNP: obtained from WGA Viewer release 57.
The gene-based test did not reveal significant results (α = 2.8*10−6), with the smallest p-value being 9.0*10−5. Table 3 shows the five genes with the smallest p-values.
Table 3.
Top five genes showing the strongest association with self-rated health.
| Gene | Chromosome | Start position | End position | Number of SNPs in gene | P-value |
|---|---|---|---|---|---|
| ERCC3 | 2 | 127731335 | 127768222 | 74 | 9.0 * 10−5 |
| S100A5 | 1 | 151776246 | 151780865 | 30 | 1.2 * 10−4 |
| PROC | 2 | 127892486 | 127903288 | 75 | 1.5 * 10−4 |
| S100A6 | 1 | 151773699 | 151775341 | 36 | 1.7 * 10−4 |
| S100A4 | 1 | 151782718 | 151784906 | 26 | 1.8 * 10−4 |
Discussion
The present study is the first to perform a genome-wide association analysis on self-rated health. Despite the high power (99%) to detect SNPs accounting for 1% of the variance in self-rated health, no genome-wide significant SNPs were identified. However, though not significant, we found two promising regions on chromosome two. Also, some of the top 50 SNPs were close to genes (e.g. MAML2, PROM1, PROC) broadly associated with a variety of health conditions, such as inflammation, coronary disease, cardio vascular disease, thrombosis, protein C deficiency etc (Reiner, et al., 2008; Trynka, et al., 2009; Wu, et al., 2009). The Protein C (PROC) gene was also in the top 5 genes revealed by the gene-based test. Changes in these genes may have an effect on an individuals’ self-rating of health. As no other study has explored the molecular genetic basis of self-rated health we cannot compare our findings.
Nevertheless, the fact that we did not find a genome-wide significant SNP is not totally unexpected considering that self-rated health is a very broad measure, influenced not only by several general somatic health factors but also by the health status presently experienced at the time of the rating. The concept of self-rated health has also been shown to be strongly associated with mental health, e.g. someone who is depressed may rate their health status as lower than someone who is in a good state of mind. Additionally, particular personality traits may play a role in how an individual rates his or her own health, for example a person scoring high in neuroticism would most likely rate their health slightly worse than a person very low in neuroticism. All these facts indicate that self-rated health is a very broad concept on a phenotypic level and may be genetically even more complicated. We suggest that very many rare variants of small effect size may influence self-rated health and are therefore difficult to detect. The fact that our Q-Q plot (Figure 1) lifts appreciable above the 95% confidence interval also hints at the highly polygenic nature of our trait, self-rated health. A recent paper by Yang et al. (2010) showed that 45% of the variance of human height could be explained considering all SNPs in a study (294,831), as opposed to 5% explained by SNPs detected by the conventional GWAS approach. This indicates that even in a very clear-cut and highly heritable phenotype such as human height, variance is explained by a large number of SNPs with very small effect; too small to be detected in a normal GWAS. The International Schizophrenia Consortium showed that by using the top-half (p-value below 0.5) of the SNPs, they could quite consistently predict Schizophrenia and related disorders (e.g. Bipolar disorder) in other samples, supporting the idea of a polygenic basis to the phenotype (Purcell, et al., 2009). Another study on human height by Lango Allen et al. (submitted) also supports these findings: with a sample of almost 200,000 individuals they show that hundreds of genetic variants influence variance in adult height. This also shows that in order to find genes with such a small effect size a very large sample is needed. Aiming for this, a large consortium has been founded planning to conduct a meta-analysis on self-rated health in the near future, combining several samples in order to possibly confirm the regions of interest found in the present study and find additional genetic variants underlying the variation of self-rated health and possibly even predict self-rated health and related health measures/indicators across different samples as in Purcell et al. (2009).
In summary, no genome wide significant SNPs underlying self-rated health could be identified, indicating that the concept of self-rated health may be influenced by a large number of SNPs with very small effect size. In order to identify these, a very large sample would be needed which only can be accomplished by conducting a meta-analysis combining different samples.
Acknowledgments
We would like to thank the twins and their families registered at the ATR for their participation and QIMR for sample collection. Funding was provided by the Australian NHMRC (241944, 339462, 389927, 389875, 389891, 389892, 389938, 442915, 442981, 496739, 552485, 552498, 613608), the FP-5 GenomEUtwin Project (QLG2-CT-2002-01254), and the U.S. NIH grants (AA07535, AA10248, AA13320, AA13321, AA13326, AA14041, MH66206). MAM and KJHV are supported by ANZ Trustees PhD scholarships in Medical Research. SEM is supported by an NHMRC Sidney Sax fellowship.
References
- Abecasis GR. MACH. unpublished. from http://www.sph.umich.edu/csg/abecasis/MACH/index.html.
- Beaton DE, Schemitsch E. Measures of health-related quality of life and physical function. Clinical Orthopaedics and Related Research. 2003;(413):90–105. doi: 10.1097/01.blo.0000079772.06654.c8. [Review] [DOI] [PubMed] [Google Scholar]
- Bryant LL, Beck A, Fairclough DL. Factors that contribute to positive perceived health in an older population. Journal of Aging and Health. 2000;12(2):169–192. doi: 10.1177/089826430001200202. [Article] [DOI] [PubMed] [Google Scholar]
- Bucholz KK, Heath AC, Madden PAF, Slutske WS, Statham DJ, Dunne MP, et al. Drinking in an older population: Cross-sectional and longitudinal data from the Australian Twin Registry. In: Gomberg ESL, Hegedus AM, Zucker RA, editors. Alcohol problems and aging. Bethesda, MD: National Institutes of Health; 1998. pp. 41–62. [Google Scholar]
- Chen WM, Abecasis GR. Family-based association tests for genomewide association scans. American Journal of Human Genetics. 2007;81(5):913–926. doi: 10.1086/521580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen K, Holm NV, McGue M, Corder L, Vaupel JW. A Danish population-based twin study on general health in the elderly. Journal of Aging and Health. 1999;11(1):49–64. doi: 10.1177/089826439901100103. [DOI] [PubMed] [Google Scholar]
- Dudbridge F, Gusnanto A. Estimation of significance thresholds for genomewide association scans. Genetic Epidemiology. 2008;32(3):227–234. doi: 10.1002/gepi.20297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansell NK, Agrawal A, Whitfield JB, Morley KI, Zhu G, Lind PA, et al. Long-term stability and heritability of telephone interview measures of alcohol consumption and dependence. Twin Research and Human Genetics. 2008;11(3):287–305. doi: 10.1375/twin.11.3.287. [Article] [DOI] [PubMed] [Google Scholar]
- Harris JR, Pedersen NL, McClearn GE, Plomin R, Nesselroade JR. Age-Differences in Genetic and Environmental-Influences for Health from the Swedish Adoption Twin Study of Aging. Journals of Gerontology. 1992;47(3):P213–P220. doi: 10.1093/geronj/47.3.p213. [DOI] [PubMed] [Google Scholar]
- Heath AC, Bucholz KK, Madden PAF, Dinwiddie SH, Slutske WS, Bierut LJ, et al. Genetic and environmental contributions to alcohol dependence risk in a national twin sample: consistency of findings in women and men. Psychological Medicine. 1997;27(6):1381–1396. doi: 10.1017/s0033291797005643. [Article] [DOI] [PubMed] [Google Scholar]
- Idler EL. Age-differences in self-assessments of health - Age-changes, cohort differences, or survivorship. Journals of Gerontology. 1993;48(6):S289–S300. doi: 10.1093/geronj/48.6.s289. [Article] [DOI] [PubMed] [Google Scholar]
- Juerges H. True health vs response styles: Exploring cross-country differences in self-reported health. Health Economics. 2007;16:163–178. doi: 10.1002/hec.1134. [DOI] [PubMed] [Google Scholar]
- Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, et al. Hundreds of variants influence human height and cluster within genomic loci and biological pathways. Nature Genetics submitted. [Google Scholar]
- Leinonen R, Heikkinen E, Jylha M. A pattern of long-term predictors of health ratings among older people. Aging Clinical and Experimental Research. 2001;13(6):454–464. [Article] [PubMed] [Google Scholar]
- Leinonen R, Kaprio J, Jylha M, Tolvanen A, Koskenvuo M, Heikkinen E, et al. Genetic Influences Underlying Self-Rated Health in Older Female Twins. Journal of the American Geriatrics Society. 2005;53(6):1002–1007. doi: 10.1111/j.1532-5415.2005.53319.x. [DOI] [PubMed] [Google Scholar]
- Lichtenstein P, Pedersen NL. Social Relationships, Stressful Life Events, and Self-Reported Physical Health - Genetic and Environmental-Influences. Psychology & Health. 1995;10(4):295–319. [Google Scholar]
- Liu JZ, McRae AF, Nyholt DR, Medland SE, Wray NR, Brown KM, et al. A versatile gene-based test for genome-wide association studies. American Journal of Human Genetics. doi: 10.1016/j.ajhg.2010.06.009. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg O, Manderbacka K. Assessing reliability of a measure of self-rated health. Scandinavian Journal of Social Medicine. 1996;24(3):218–224. doi: 10.1177/140349489602400314. [DOI] [PubMed] [Google Scholar]
- Medland SE, Nyholt DR, Painter JN, McEvoy BP, McRae AF, Zhu G, et al. Common variants in the trichohyalin gene are associated with straight hair in Europeans. American Journal of Human Genetics. 2009;85(5):750–755. doi: 10.1016/j.ajhg.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosing MA, Gordon SD, Medland SE, Statham DJ, Nelson EC, Heath AC, et al. Genetic and environmental influences on the co-morbidity between depression, panic disorder, agoraphobia, and social phobia: A twin study. Depression and Anxiety. 2009;26(11):1004–1011. doi: 10.1002/da.20611. [Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosing MA, Zietsch BP, Shekar SN, Wright MJ, Martin NG. Genetic and environmental influences on optimism and its relationship to mental and self-rated health: A study of aging twins. Behavior Genetics. 2009;39(6):597–604. doi: 10.1007/s10519-009-9287-7. [DOI] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature (London) 2009;460(7256):748–752. doi: 10.1038/nature08185. [Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner AP, Carty CL, Jenny NS, Nievergelt C, Cushman M, Stearns-Kurosawa DJ, et al. PROC, PROCR and PROS1 polymorphisms, plasma anticoagulant phenotypes, and risk of cardiovascular disease and mortality in older adults: the Cardiovascular Health Study. Journal of Thrombosis and Haemostasis. 2008;6(10):1625–1632. doi: 10.1111/j.1538-7836.2008.03118.x. [Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodin J, McAvay G. Determinants of change in perceived health in a longitudinal-study of older adults. Journals of Gerontology. 1992;47(6):P373–P384. doi: 10.1093/geronj/47.6.p373. [Article] [DOI] [PubMed] [Google Scholar]
- Romeis JC, Scherrer JF, Xian H, Eisen SA, Bucholz K, Heath AC, et al. Heritability of self-reported health. Health Services Research. 2000;35(5):995–1010. [PMC free article] [PubMed] [Google Scholar]
- Silventoinen K, Posthuma D, Lahelma E, Rose RJ, Kaprio J. Genetic and environmental factors affecting self-rated health from age 16–25: A longitudinal study of Finnish twins. Behavior Genetics. 2007;37(2):326–333. doi: 10.1007/s10519-006-9096-1. [DOI] [PubMed] [Google Scholar]
- Svedberg P, Lichtenstein P, Pedersen NL. Age and sex differences in genetic and environmental factors for self-rated health: A twin study. Journals of Gerontology Series B-Psychological Sciences and Social Sciences. 2001;56(3):S171–S178. doi: 10.1093/geronb/56.3.s171. [DOI] [PubMed] [Google Scholar]
- Trynka G, Zhernakova A, Romanos J, Franke L, Hunt KA, Turner G, et al. Coeliac disease-associated risk variants in TNFAIP3 and REL implicate altered NF-kappa B signalling. Gut. 2009;58(8):1078–1083. doi: 10.1136/gut.2008.169052. [Article] [DOI] [PubMed] [Google Scholar]
- Verweij KJH, Zietsch BP, Medland SE, Gordon SD, Benyamin B, Nyholt DR, et al. A genome-wide association study of Cloninger’s Temperament scales: Implications for the evolutionary genetics of personality. Biological Psychology. doi: 10.1016/j.biopsycho.2010.07.018. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XF, Spitz MR, Lee JJ, Lippman SM, Ye YQ, Yang HS, et al. Novel Susceptibility Loci for Second Primary Tumors/Recurrence in Head and Neck Cancer Patients: Large-Scale Evaluation of Genetic Variants. Cancer Prevention Research. 2009;2(7):617–624. doi: 10.1158/1940-6207.CAPR-09-0025. [Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, Gordon SD, Henders AK, Nyholt DR, et al. Common SNPs explain a large proportion of the heritability for human height. Nature Genetics. 2010 doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]