Abstract
Epstein–Barr virus (EBV) evolved an episomal system for maintaining life-long, latent infection of human B lymphocytes. Circular episomes engineered from EBV components required for this latent form of infection have the capacity to persist in most types of replicating mammalian cells without DNA integration and the pitfalls of insertional mutagenesis. EBV episomes are typically transduced using low-efficiency methods. Here we present a method for efficient delivery of EBV episomes to nuclei of hepatocytes in living mice using a helper-dependent adenoviral vector and Cre-mediated recombination in vivo to generate circular EBV episomes following infection. Cre is transiently expressed from a hepatocyte-specific promoter so that vector generation and transgene expression are tissue specific. We show long-term persistence of the circularized vector DNA and expression of a reporter gene in hepatocytes of immunocompetent mice.
Keywords: transduction, stable transformation, viral vector, eukaryotic plasmid
Of several genes expressed by Epstein–Barr virus (EBV) in latently infected B cells, only EBV nuclear antigen 1 (EBNA-1) is required to confer persistence in mitotic cells to a plasmid bearing the EBV oriP region.1–4 EBNA-1 is a sequence-specific DNA-binding protein that binds to multiple sites in oriP. A dyad symmetry (DS) of inverted EBNA-1 binding sites at one end of oriP functions as an origin of DNA replication in primate cells.4 EBNA-1 bound to the DS recruits cellular DNA pre-replication complex components that result in one firing of the origin per S phase, maintaining EBV episomes at a constant number per cell.4 Binding to 21–30 bp imperfect direct repeats (the family of repeats, or FR) that make up most of oriP ensures proper episomal segregation of EBV episomes during mitosis. EBNA-1 bound to the FR interacts with metaphase chromosomes, effectively tethering daughter episomes generated during S phase to host daughter chromosomes. This EBNA-1-mediated tethering confers segregation of the episomes to each daughter cell during mitosis, ensuring nuclear retention in replicating cells.4,5 A plasmid containing both oriP and an EBNA-1 expression cassette can persist in replicating cultured human cells with ~95% episome retention per cell cycle without selection.2,4
Although circular plasmids containing the EBV oriP and an EBNA-1 expression cassette are maintained in replicating primate and canine cultured cells, they are not efficiently maintained in cultured rodent cells.2,4 However, when the region of DS is replaced by a large fragment of human DNA (~20 kb), maintenance in replicating cultured rodent fibroblasts is restored, probably because the region functions as a replication origin.3 The combination of mammalian genomic DNA as an origin and the FR region to ensure segregation to daughter cells during mitosis allows efficient episome maintenance in murine cells.6 Conventional delivery of EBV episomes has used physical means of gene transfer, such as hydrodynamic injection, and electroporation with varying degrees of efficiency that falls off rapidly with episomes of >10 kb.7
In the system presented here, EBV episomes are delivered to cell nuclei by a helper-dependent adenovirus vector (HDAd). These vectors are deleted for all adenovirus genes and can contain up to 39 kb of DNA.8 However, the DNA introduced by HDAds is a double-stranded linear molecule. In the system presented, the DNA to be maintained as an EBV episome is present on the HDAd DNA flanked by LoxP sites. Circularization into a functional EBV episome is achieved by transient Cre expression. In HDAd vectors, nonadenovirus DNA replaces viral DNA, except for the adenovirus inverted terminal repeats (ITR) required as the initiation sites of adenovirus DNA replication, and the ψ region near one end, required for viral packaging.9 The absence of even low-level adenovirus protein expression inherent in conventional partially deleted adenovirus (first-generation Ad vectors) leads to substantially lower cytotoxicity in target cells and less immune-mediated targeting of transduced cells.8,10
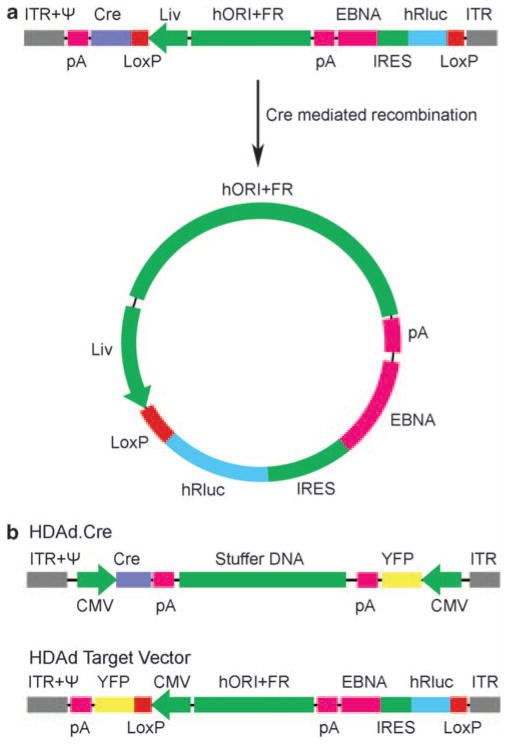
We conducted our initial in vivo experiments in mouse liver as adenovirus-5-based vectors injected into the tail vein readily infect hepatocytes through clotting factor X receptors.11,12 The design of the ‘single-virus HDAd–EBV vector’ is summarized in Figure 1a. The ~29 kb vector genome contains a Cre coding region transcribed from a hepatocyte specific promoter/enhancer.13 It also contains an ‘EBV episome target’ region flanked by parallel LoxP sites (Figure 1a) so that Cre-mediated site-specific recombination circularizes the intervening DNA, leaving a single copy of the LoxP sequence (the vector sequence ‘arms’ flanking the target sequence are left as a linear DNA linked by a single LoxP site; not shown). The vector is designed so that Cre-mediated recombination both circularizes the DNA and places the hepatocyte-specific promoter/enhancer upstream of an expression cassette for a reporter gene to monitor transduced cells, Renilla luciferase.14 The resulting transcript, and hence, luciferase activity, is expressed only following Cre-mediated recombination of the target sequence within the vector. Cre-mediated recombination also prevents further Cre expression by separating the hepatocyte-specific promoter from the Cre coding region (Figure 1a).
Figure 1.
Diagrams of helper-dependent adenovirus-Epstein-Barr virus hybrid vectors (HDAd–EBV vectors). (a) Single-virus vector, HDAd-EBV.1x.hRluc and its conversion through Cre-mediated site-specific recombination to an EBV episome. The vector consists of the Ad5 inverted terminal repeats (ITR); cis-acting packaging element (ψ); an expression cassette (transcription to the left) consisting of the hepatocyte-specific promoter (Liv), Cre-recombinase recognition sequence (LoxP), coding region for Cre and the bovine growth hormone polyadenylation signal (pA); a 19 kb fragment from human chromosome 10 that functions as an origin of replication; the EBV family of repeats (FR); a bicistronic promoterless expression cassette (transcription to the left) with the reporter transgene, humanized Renilla luciferase (hRluc), an internal ribosomal entry site (IRES), coding region for Epstein–Barr virus nuclear antigen-1 (EBNA-1) and an SV40 polyadenylation signal. This is followed by a second LoxP site and ITR. Cre-mediated recombination at the parallel LoxP sites generates the ~27 kb circular EBV episome as diagrammed. (b) Components of the two-virus HDAd–EBV vector system.16 HDAd.Cre (above) contains two expression cassettes, one for Cre, and the other for yellow fluorescence protein (YFP) to aid in quantitating vector concentration, both driven by the HCMV-IE promoter, with an intervening stuffer sequence of human chromosomal DNA. Below is the target vector, HDAd-EBV.hRluc, which also contained a YFP expression cassette. Cre-mediated recombination of the target vector generates an EBV episome identical to that generated by the single-virus vector shown in (a), except that the hepatocyte-specific enhancer/promoter is replaced by the HCMV-IE promoter.
The transcript generated from the circularized target DNA by the hepatocyte-specific promoter also contains an internal ribosome entry site from murine encephalo-myocarditis virus that promotes translation of EBNA-1,15 required for maintenance of the EBV episome. The target DNA also contains the EBV FR to promote efficient episome segregation during mitosis and a 19 kb fragment of human genomic DNA from chromosome 10, termed ‘hORI,’ to confer DNA replication origin function.3 Consequently, the ~27 kb circular DNA molecule generated in vivo following vector infection contains all the components required for maintenance as an EBV episome in mammalian cells.
It is essential to limit Cre-recombinase expression during the generation and expansion of these types of vectors as recombination between LoxP sites flanking the EBV episome target sequence prevents propagation of the vector. The ‘two-virus’ HDAd–EBV vector system developed earlier14,16 prevents Cre-mediated recombination at LoxP sites during vector construction and expansion because the Cre expression cassette and the EBV episome target sequence are carried by two HDAd vectors that are constructed and expanded separately (Figure 1b). The ‘single-virus’ design diagrammed in Figure 1a sought to overcome the problem of precocious vector recombination during construction and propagation by using a highly cell-type-specific promoter that should not be expressed at significant level in 293 cells where HDAd vectors are constructed and propagated. We used a promoter/enhancer construct reported to be highly hepatocyte specific consisting of the human α1 antitrypsin promoter region with four upstream ApoE enhancer tandem repeats.13 Reduced promoter silencing17 and decreased immunogenicity elicited from transgene products18 are additional desirable aspects of the use of a cell-type-specific promoter in place of the strong, ubiquitously expressed human cytomegalovirus (HCMV) immediate-early promoter used in our ‘proof-of-principle’ two-virus vector system (Figure 1b).14
Preliminary transient transfection assays confirmed high specificity of the ApoE/α1 antitrypsin promoter in HepG2 cells, a hepatocellular carcinoma cell line,19 compared to 293 cells, and we found that it was possible to generate and propagate the vector DNA diagrammed in Figure 1a (see Supplementary Table 1 for titers of vector stocks used in these studies). Because the ~29 kb HDAd-EBV.1x.hRluc vector contains LoxP sites, it was generated and propagated using a helper virus with its packaging sequence flanked by FRT sites in 293 cells constitutively expressing the Flp site-specific recombinase.20 During the initial transfection of vector DNA prepared from Escherichia coli and in early passages of the vector, 293-Flp cells were also transduced with siRNA directed against the Cre coding region to further inhibit Cre expression during vector propagation (see Supplementary Materials for details).
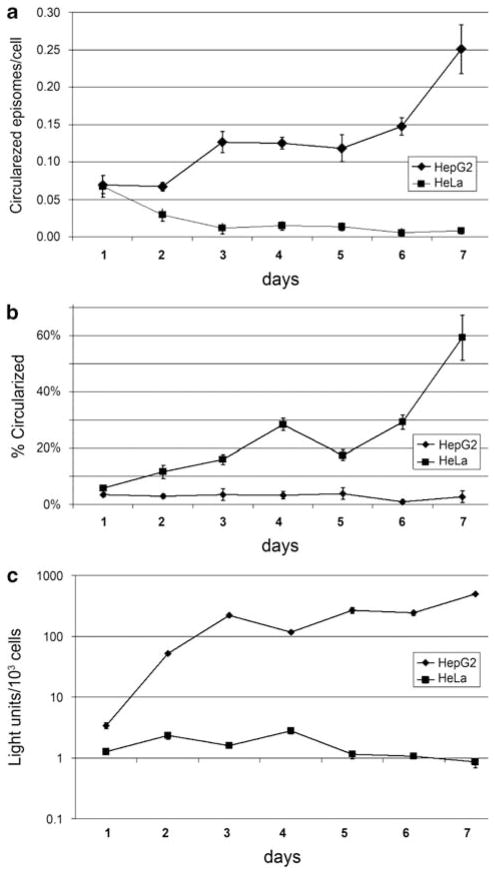
The ability to circularize the target DNA between the two LoxP sites in the single-virus vector and the dependence of this process on tissue-specific Cre expression were analyzed first in cultured cells. HepG2 cells derived from a human hepatocellular carcinoma and HeLa cells derived from a cervical carcinoma were infected with the single-virus vector HDAd-EBV.1x.hR-luc (Figure 1a). Because no promoter lies upstream of the Renilla luciferase coding region until Cre-mediated recombination occurs (Figure 1a), luciferase expression is a functional assay of Cre-mediated recombination, which places the hepatocyte-specific promoter upstream of the luciferase coding region. Circularized target DNA was assayed by real-time PCR (qPCR) using an amplicon spanning the junction formed after Cre-mediated site-specific recombination at the parallel LoxP sites, detecting only recombined vector. This was compared with the number of total vector DNA molecules quantitated by qPCR using primers against the EBNA1 gene. Luciferase DNA and circularized target DNA were assayed daily for 1 week after infection and maintenance of cells in low (2%) calf serum to minimize cell replication (Figure 2).
Figure 2.
Generation and stability of EBV episomes by Cre-mediated recombination in cultured cells. (a) HepG2 and HeLa cells were infected with HDAd-EBV.1x.hRluc at an MOI 2 IGU per cell and incubated in 2% calf serum containing media for 7 days. Cells were harvested daily and circularized vectors were assayed in isolated total nuclear DNA by quantitative real-time PCR (qPCR) with primers that detect circularized episomes specifically. Cell counts were determined by qPCR detecting the endogenous EEF2 gene. Error bars signify probable uncertainty calculated from standard deviation of triplicate qPCR samples. (b) Percentage of circularized vectors determined in HepG2 and HeLa cells over time. The same samples were subjected to qPCR to detect total vectors by targeting the EBNA sequence, contained in both circularized and unrecombined vectors. Percentage was determined by dividing detected circular vectors over total vectors. Error bars signify probable uncertainty calculated from standard deviation of triplicate qPCR samples, or the range of duplicates used in the last two samples for EBNA qPCR in HepG2 cells. (c) Luciferase assay results for extracts prepared at the indicated days post infection.
The fraction of target vector DNA circularized increased with time in HepG2 cells, reaching ~60% of total molecules 7 days after infection (Figure 2b). No increase in circularized target DNA was observed in HeLa cells. Luciferase activity increased more than two orders of magnitude in HepG2 cells compared with the low level initially expressed in HeLa cells, which fell by 7 days after infection (Figure 2c). This high degree of cell-type-specific luciferase expression results from two consequences of the cell-type specificity of the hepatocyte-specific promoter: (1) circularization of the target DNA that places the promoter in front of the luciferase coding region was much greater in HepG2 cells than in HeLa cells, probably because of much higher transient Cre expression from the liver-specific promoter in HepG2 cells (Figure 2a); (2) once circularized target DNA is generated by Cre-mediated recombination, transcription from the hepatocyte-specific promoter is much higher in HepG2 cells than in HeLa cells.13 Having observed cell-type-specific generation of the EBV episome and cell-type-specific expression of the reporter luciferase gene in cultured cells, we proceeded with experiments in mice.
Our initial experiments to analyze the efficiency of episome formation and maintenance of transgene expression in hepatocytes in an intact animal were performed in immune compromised Foxn1nu Balb/c (‘nude’) mice. These initial experiments were performed in nude mice to separate analysis of the efficiency of EBV episome formation and introduction into nuclei of hepatocytes in intact animals from potential consequences of immune-mediated responses to gene expression from the episomes. Nude mice were pretreated by tail vein administration of mouse IgG to reduce Kupffer cell function,21 and then administered a single tail vein injection of 3.2 × 108 infectious genome units (IGU)14 HDAd-EBV.hRluc. For comparison, nude mice were also administered 5 × 109 IGU each of the HDAd vectors of the two-virus system diagrammed in Figure 1b. A high dose of vectors is required in the two-vector system to ensure that most hepatocytes are co-infected with both HDAd vectors. Circularization of the target vector in the two-virus system generates the same circular EBV episomal DNA as diagrammed in Figure 1a, except that the hepatocyte-specific promoter/enhancer is replaced with the HCMV-IE promoter/enhancer.14
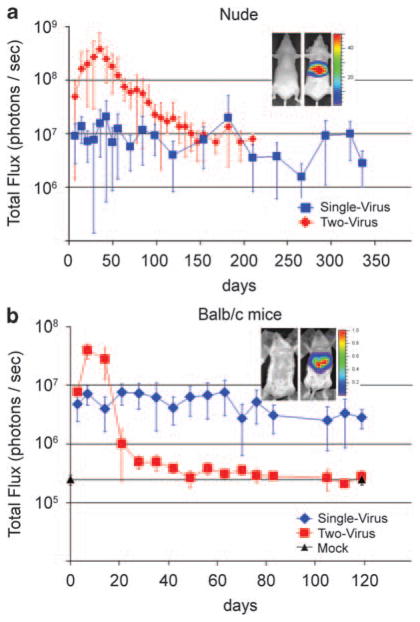
Luciferase expression in the livers of treated mice was assayed at approximately weekly intervals by measuring transdermal bioluminescence after tail vein injection of the luciferase substrate14 (Figure 3a). For the two-virus vector system, luciferase activity peaked at 35 days post-vector administration and then diminished over the next 100 days until it reached a steady level ~3% of the peak level (Figure 3a). This experiment was terminated at 210 days post-vector administration because luciferase expression had remained at this low level for 10 weeks.
Figure 3.
Persistence of transgene expression in vivo assayed by bioluminescence imaging of luciferase activity in the liver of the same mice over time. (a, red) A single tail vein injection of 3.2 × 108 IGU of the single-virus vector HDAd-EBV.1x.hRluc (Figure 1a) was administered to nine nude mice. Of these, one died at day 98, two at day 154 and two at day 238, from causes unrelated to vector administration; four mice remained at the final time point of 336 days post-vector administration. At each time point indicated, all remaining mice were assayed for luciferase expression in the liver by bioluminescence following tail vein administration of the Renilla luciferase substrate coelenterazine, as described.14 (Inset shows bioluminescence images of a control (left) and a transduced (right) animal. Color scale 1, 50 × 106 photons per sec.) (blue) A second group of 16 mice were administered 5 × 109 each of the two HDAds of the two-vector system (Figure 1b). Mice (number at each time point given in Supplementary Table 1) were assayed for bioluminescence from the region of the liver at the indicated times after injection. (b, red) Eight immunocompetent Balb/c mice were administered a single tail vein injection of 7.2 × 108 IGU of the single-virus vector HDAd-EBV.1x.hRluc, or (blue) six Balb/c mice were administered 7.2 × 108 IGU each of the two-virus vector system. Luciferase activity in the region of the liver was quantitated by bioluminescence at the indicated days post-vector administration.
In contrast to the two-virus system, the single-virus vector generated luciferase activity with less than one vector per hepatocyte. qPCR detected ~0.2 circularized vector copies per cell in total liver DNA isolated from mice 322 days after vector administration. Although the level of luciferase activity in livers of mice treated with the single-vector system was lower than in mice treated with the two-vector system, luciferase expression from the single-vector system remained at high level for a prolonged period following a peak at 42 days after vector administration (Figure 3a). Luciferase activity was ~50% of this peak signal at 300 and 330 days post-vector administration, with an average of 46% of maximum luciferase activity throughout the 330-day period. The fluctuation in luciferase bioluminescence of several fold observed over time in the same mice was due, in part, to the complexity of the in vivo assay. Nonetheless, luciferase expression from the single-virus vector clearly persisted longer than for the two-virus vector. The higher peak level of luciferase activity in the two-vector system was probably a consequence of both the higher number of EBV episomes per hepatocyte introduced using the two-vector system and the initial high activity of the HCMV-IE promoter promoting expression of Renilla luciferase in the two-vector system. As mentioned earlier, the two-vector system requires a high MOI of vector to ensure that hepatocytes are co-infected by both the Cre-expressing and the target vectors.14 At the high MOIs used, an average of 20–40 EBV episomes were generated per hepatocyte.14 Although the amount of single-vector administered was ~6% of the target vector used in the two-vector system (3.2 × 108 IGU single-virus vector versus 5 × 109 IGU each of the Cre vector and the target vector in the two-virus system), the single-vector system generated only approximately 0.5–1% as many EBV episomes per hepatocyte (approximately 0.2/20–40 copies per hepatocyte14). This probably occurred because transduction of hepatocytes by tail vein injection of adenovirus vectors does not scale linearly with increasing vector dose, but rather is more efficient at higher vector doses such as those used with the two-vector system.21
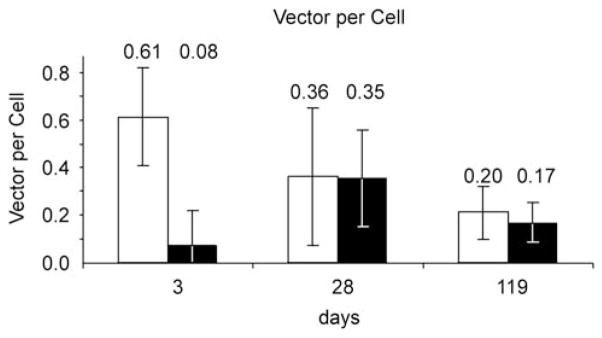
The stability of transgene expression in nude mice prompted us to analyze transgene expression and vector DNA stability in immunocompetent mice. In an initial pilot experiment, one immunocompetent C57Bl/6 mouse expressed a stable level of luciferase for 323 days (Supplementary Figure S1). A larger experiment was set up in immunocompetent Balb/c mice, considered to be less tolerant to adenovirus 5 antigens than C57Bl/6 mice.22 Eight Balb/c mice were administered a single tail vein injection of 7.2 × 108 IGU of the single-virus vector HDAd-EBV.1x.hRluc. Total liver DNA was isolated from two mice at both 3 and 28 days after vector administration, and from four mice at the end of the experiment at 119 days. qPCR with specific primer sets was used to assay total vector DNA as well as target DNA recombined at the LoxP sites (Figure 4). At day 3, ~10% of the vector DNA had undergone recombination at the LoxP sites, similar to the extent of recombination observed in 3 days in cultured HepG2 (Figure 2a). But at days 28 and 119, >85% of detected vector target DNA was circularized at the LoxP site. A similar high percentage of circularized target vector was observed in hepatocytes using the two-virus vector in living mice at 9 days and greater after vector administration.14 Total vector DNA in the liver was relatively stable; the average level measured dropped by only a factor of ~3 between 3 and 119 days. Total vector DNA introduced into livers of nude mice by the two-vector system was also stable for 50 weeks.14 An average of 0.35 and 0.17 EBV episomes per hepatocyte were observed at 28 and 119 days after vector administration, although these numbers were not statistically different. We observed a similar level of EBV episome per hepatocyte in Balb/c mice as in nude mice, even though we administered approximately two times more vector to the Balb/c mice. This small discrepancy is probably within the error limits of the methods used, but may have resulted from a lower efficiency of hepatocyte transduction in immunocompetent Balb/c mice compared with immunodeficient nude mice. Likewise, Balb/c mice administered 7.2 × 108 IGU of each of the vectors of the two-vector system contained 0.72 episomes per hepatocyte at 119 days post-vector administration, compared with 20–40 episomes per hepatocyte observed in nude mice after tail vein injection of 5 × 109 IGU per cell of each component of the two-vector system.14 If vector delivery increased linearly with vector dose, we would have expected approximately 3–6 episomes per hepatocyte. The observation of only ~0.7 episomes per hepatocyte was probably a consequence of the higher efficiency of vector delivery at the higher vector doses used in the earlier study.14,21 These results indicate that circularized EBV episomes generated by both the single-vector and the two-vector systems were relatively stable in hepatocytes of immunocompetent Balb/c mice, as we observed earlier for the two-virus system in immunodeficient nude mice.14
Figure 4.
Circularization and maintenance of EBV episomes in vivo. Balb/C mice were injected with 7.2 × 108 IGU of HDAd-EBV.1x.hRluc. Two mice were killed at 3 and 28 days, and four at 119 days post-vector administration. Liver samples were assayed by qPCR to determine vector molecules per cell, normalizing to the mouse Eef2 gene.14 Light bars show total vector; dark bars show only circularized EBV target DNA. Error bars signify the range of assays from two mice at days 2 and 28, and the standard deviation from four mice at day 119.
Luciferase expression from the single-virus vector remained fairly stable in the Balb/c mice, dropping only ~50% in 119 days (Figure 3b). In contrast, luciferase expression from the two-vector system peaked at approximately fivefold higher level than observed with the single-vector system at days 7–14 after administration, despite administration of equivalent doses of target vector in the two-vector system compared to the single vector (7.2 × 108 IGU for each). This may be due to higher initial activity of the HCMV-IE promoter compared to the liver-specific promoter. However, this initial high level of transgene expression dropped precipitously by day 21 and reached near background levels by day 44 (Figure 3b). Because vector DNA was not lost from the animals during the time luciferase expression dropped in both immuno-compromised and immunocompetent animals treated with the two-virus system, the drop in luciferase activity was probably not due to cell-mediated immune elimination of transduced hepatocytes. Rather, loss of luciferase expression was likely due to silencing of the HCMV-IE promoter over time, a phenomenon that has been observed in several cell types,23 including hepatocytes.24 The observation that inhibition of luciferase expression showed much faster kinetics in the immunocompetent Balb/c mice compared with the immunodeficient nude mice suggests that some aspect of the immune response can contribute to the shutoff of transcription from an HDAd vector in the absence of DNA loss.25 During dissection of liver tissue from transduced nude and Balb/c mice, no evidence of tumors or macroscopic liver pathology was evident, as was the case in our earlier studies with the two-vector system in nude mice.14 In the earlier study, confocal microscopy of liver tissue from mice 6 days after a dose of vector that transduced 98% of hepatocytes revealed normal histology.14
These results show the efficient delivery of EBV episomes to nuclei in the tissue of an intact animal using a single HDAd vector. A highly cell-type-specific promoter allowed the generation and propagation of the vector outlined in Figure 1a because of sufficiently low expression in 293-Flp cells20 used to generate the vector. It also resulted in stable transgene expression in immunocompetent animals. This system may be useful for the efficient transduction of alternative animal tissues using appropriate cell-type-specific promoters. Further development of this vector system may allow gene therapy of replicating cells in human patients without the risk of insertional mutagenesis by the vector.
Supplementary Material
Acknowledgments
This work was supported by USPHS Grant R37 CA025235 and the Eli and Edythe Broad Center of JSG was partially supported by the Ruth L Kirchstein National Research Service Award GM007185, and SDG by UCLA Training Grant in Genetic Mechanisms T32-GM07104.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Tsurumi T, Fujita M, Kudoh A. Latent and lytic Epstein–Barr virus replication strategies. Rev Med Virol. 2005;15:3–15. doi: 10.1002/rmv.441. [DOI] [PubMed] [Google Scholar]
- 2.Yates J, Warren N, Sugden B. Stable replication of plasmid derived from Epstein–Barr virus in various mammalian cells. Nature. 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- 3.Krysan PJ, Calos MP. Epstein–Barr virus-based vectors that replicate in rodent cells. Gene. 1993;136:137–143. doi: 10.1016/0378-1119(93)90457-e. [DOI] [PubMed] [Google Scholar]
- 4.Lindner SE, Sugden B. The plasmid replicon of Epstein–Barr virus: mechanistic insights into efficient, licensed, extrachromosomal replication in human cells. Plasmid. 2007;58:1–12. doi: 10.1016/j.plasmid.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nanbo A, Sugden A, Sugden B. The coupling of synthesis and partitioning of EBV’s plasmid replicon is revealed in live cells. EMBO J. 2007;26:4252–4262. doi: 10.1038/sj.emboj.7601853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wohlgemuth JG, Kang SH, Bulboaca GH, Nawotka KA, Calos MP. Long-term gene expression from autonomously replicating vectors in mammalian cells. Gene Therapy. 1996;3:503–512. [PubMed] [Google Scholar]
- 7.Stoll SM, Sclimenti CR, Baba EJ, Meuse L, Kay MA, Calos MP. Epstein–Barr virus/human vector provides high-level, long-term expression of alpha1-antitrypsin in mice. Mol Ther. 2001;4:122–129. doi: 10.1006/mthe.2001.0429. [DOI] [PubMed] [Google Scholar]
- 8.Segura MM, Alba R, Bosch A, Chillon M. Advances in helper-dependent adenoviral vector research. Curr Gene Ther. 2008;8:222–235. doi: 10.2174/156652308785160647. [DOI] [PubMed] [Google Scholar]
- 9.Parks RJ, Chen L, Anton M, Sankar U, Rudnicki MA, Graham FL. A helper-dependent adenovirus vector system: removal of helper virus by Cre-mediated excision of the viral packaging signal. Proc Natl Acad Sci USA. 1996;93:13565–13570. doi: 10.1073/pnas.93.24.13565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brunetti-Pierri N, Ng P. Progress and prospects: gene therapy for genetic diseases with helper-dependent adenoviral vectors. Gene Therapy. 2008;15:553–560. doi: 10.1038/gt.2008.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waddington SN, McVey JH, Bhella D, Parker AL, Barker K, Atoda H, et al. Adenovirus serotype 5 hexon mediates liver gene transfer. Cell. 2008;132:397–409. doi: 10.1016/j.cell.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 12.Lowenstein PR. With a little help from my f(X)riends!: the basis of Ad5-mediated transduction of the liver revealed. Mol Ther. 2008;16:1004–1006. doi: 10.1038/mt.2008.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okuyama T, Huber RM, Bowling W, Pearline R, Kennedy SC, Flye MW, et al. Liver-directed gene therapy: a retroviral vector with a complete LTR and the ApoE enhancer-alpha 1-antitrypsin promoter dramatically increases expression of human alpha 1-antitrypsin in vivo. Hum Gene Ther. 1996;7:637–645. doi: 10.1089/hum.1996.7.5-637. [DOI] [PubMed] [Google Scholar]
- 14.Gallaher SD, Gil JS, Dorigo O, Berk AJ. Robust in vivo transduction of a genetically stable Epstein–Barr virus episome to hepatocytes in mice by a hybrid viral vector. J Virol. 2009;83:3249–3257. doi: 10.1128/JVI.01721-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan BT, Wu L, Berk AJ. An adenovirus-Epstein-Barr virus hybrid vector that stably transforms cultured cells with high efficiency. J Virol. 1999;73:7582–7589. doi: 10.1128/jvi.73.9.7582-7589.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dorigo O, Gil JS, Gallaher SD, Tan BT, Castro MG, Lowenstein PR, et al. Development of a novel helper-dependent adenovirus-Epstein-Barr virus hybrid system for the stable transformation of mammalian cells. J Virol. 2004;78:6556–6566. doi: 10.1128/JVI.78.12.6556-6566.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Al-Dosari M, Zhang G, Knapp JE, Liu D. Evaluation of viral and mammalian promoters for driving transgene expression in mouse liver. Biochem Biophys Res Commun. 2006;339:673–678. doi: 10.1016/j.bbrc.2005.11.063. [DOI] [PubMed] [Google Scholar]
- 18.Pastore L, Morral N, Zhou H, Garcia R, Parks RJ, Kochanek S, et al. Use of a liver-specific promoter reduces immune response to the transgene in adenoviral vectors. Hum Gene Ther. 1999;10:1773–1781. doi: 10.1089/10430349950017455. [DOI] [PubMed] [Google Scholar]
- 19.Martin GR, Evans MJ. Differentiation of clonal lines of teratocarcinoma cells: formation of embryoid bodies in vitro. Proc Natl Acad Sci USA. 1975;72:1441–1445. doi: 10.1073/pnas.72.4.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Umana P, Gerdes CA, Stone D, Davis JR, Ward D, Castro MG, et al. Efficient FLPe recombinase enables scalable production of helper-dependent adenoviral vectors with negligible helper-virus contamination. Nat Biotechnol. 2001;19:582–585. doi: 10.1038/89349. [DOI] [PubMed] [Google Scholar]
- 21.Tao N, Gao GP, Parr M, Johnston J, Baradet T, Wilson JM, et al. Sequestration of adenoviral vector by Kupffer cells leads to a nonlinear dose response of transduction in liver. Mol Ther. 2001;3:28–35. doi: 10.1006/mthe.2000.0227. [DOI] [PubMed] [Google Scholar]
- 22.Barr D, Tubb J, Ferguson D, Scaria A, Lieber A, Wilson C, et al. Strain related variations in adenovirally mediated transgene expression from mouse hepatocytes in vivo: comparisons between immunocompetent and immunodeficient inbred strains. Gene Therapy. 1995;2:151–155. [PubMed] [Google Scholar]
- 23.Brooks AR, Harkins RN, Wang P, Qian HS, Liu P, Rubanyi GM. Transcriptional silencing is associated with extensive methylation of the CMV promoter following adenoviral gene delivery to muscle. J Gene Med. 2004;6:395–404. doi: 10.1002/jgm.516. [DOI] [PubMed] [Google Scholar]
- 24.Loser P, Jennings GS, Strauss M, Sandig V. Reactivation of the previously silenced cytomegalovirus major immediate-early promoter in the mouse liver: involvement of NFkappaB. J Virol. 1998;72:180–190. doi: 10.1128/jvi.72.1.180-190.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lowenstein PR, Kroeger K, Castro MG. Immunology of neurological gene therapy: how T cells modulate viral vector-mediated therapeutic transgene expression through immunological synapses. Neurotherapeutics. 2007;4:715–724. doi: 10.1016/j.nurt.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.