Abstract
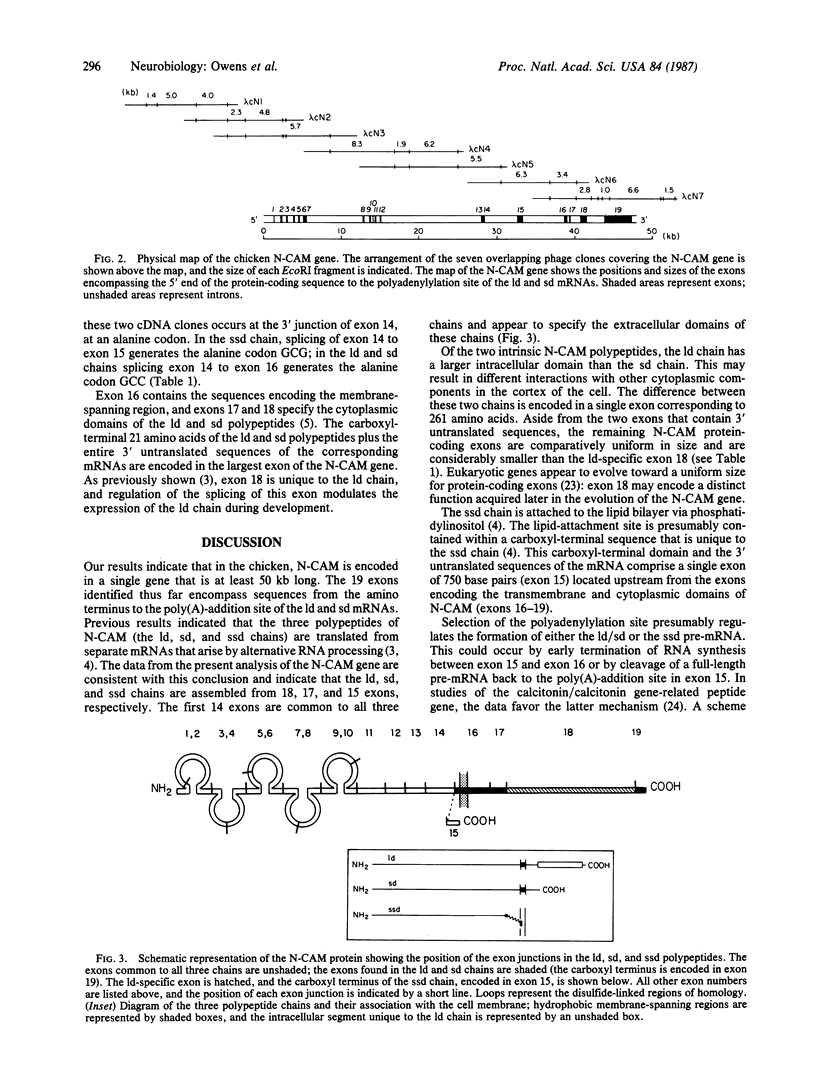
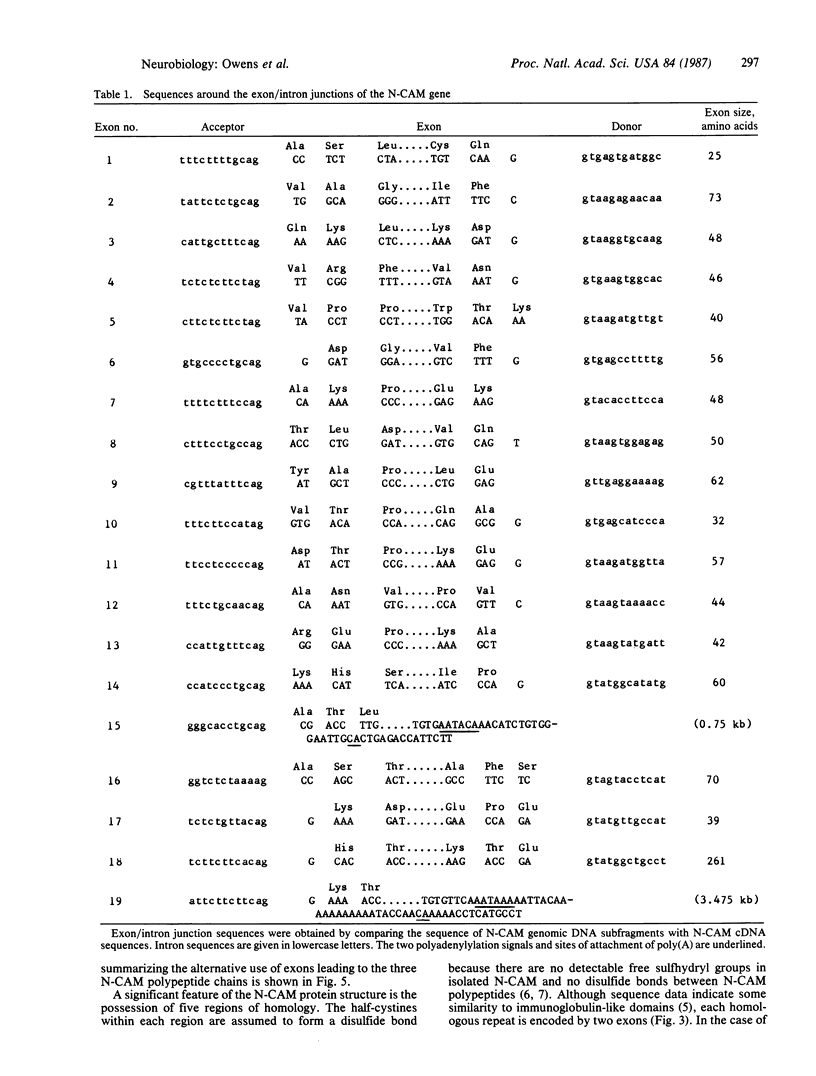
The neural cell adhesion molecule, N-CAM, is expressed as at least three polypeptide chain, (ld, sd, and ssd chains) specified by a single gene and derived by alternative splicing and polyadenylation-site selection during RNA processing. We describe here the characterization of seven overlapping genomic phage clones reactive with N-CAM cDNA, indicating that the chicken N-CAM gene is more than 50 kilobases long. Analysis of the gene shows that there are at least 19 exons and that the coding sequences for the ld, sd, and ssd chains are assembled from 18, 17, and 15 exons, respectively. The first 14 exons appear to be common to all three chains and encode the amino-terminal portion of N-CAM, which contains five tandem homologous repeats resembling those seen in the immunoglobulin gene superfamily. In contrast to other genes containing such domains, each of these segments in N-CAM is specified by two exons. The carboxyl-terminal portion of each N-CAM chain is different as a result of the alternative use of exons. A single exon encodes the carboxyl-terminal 26 amino acids of the ssd chain and the 3' untranslated region of its mRNA, ending with a poly(A)-addition site. Two exons encode the transmembrane and cytoplasmic sequences common to the ld and sd chains, and another exon encodes the additional 261 amino acids found in the cytoplasmic domain of the ld chain. The carboxyl-terminal 21 amino acids common to the ld and sd chains and the 3' untranslated region common to their mRNAs are encoded by a single large exon of 3475 base pairs that ends with a second poly(A)-addition site. Sequences from the 13-kilobase intron that separates the exons encoding the amino-terminal and carboxyl-terminal regions of the molecule hybridize to a 2-kilobase poly(A)+ RNA transcript of unknown identity. This description of the chicken N-CAM gene provides a basis for determining the mechanisms that regulate the differential expression of the N-CAM polypeptide chains during development.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amara S. G., Evans R. M., Rosenfeld M. G. Calcitonin/calcitonin gene-related peptide transcription unit: tissue-specific expression involves selective use of alternative polyadenylation sites. Mol Cell Biol. 1984 Oct;4(10):2151–2160. doi: 10.1128/mcb.4.10.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Blin N., Stafford D. W. A general method for isolation of high molecular weight DNA from eukaryotes. Nucleic Acids Res. 1976 Sep;3(9):2303–2308. doi: 10.1093/nar/3.9.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgois A. Evidence for an ancestral immunoglobulin gene coding for half a domain. Immunochemistry. 1975 Nov;12(11):873–876. doi: 10.1016/0019-2791(75)90244-x. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Choi O. R., Trainor C., Graf T., Beug H., Engel J. D. A single amino acid substitution in v-erbB confers a thermolabile phenotype to ts167 avian erythroblastosis virus-transformed erythroid cells. Mol Cell Biol. 1986 May;6(5):1751–1759. doi: 10.1128/mcb.6.5.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham B. A., Hoffman S., Rutishauser U., Hemperly J. J., Edelman G. M. Molecular topography of the neural cell adhesion molecule N-CAM: surface orientation and location of sialic acid-rich and binding regions. Proc Natl Acad Sci U S A. 1983 May;80(10):3116–3120. doi: 10.1073/pnas.80.10.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Eustachio P., Owens G. C., Edelman G. M., Cunningham B. A. Chromosomal location of the gene encoding the neural cell adhesion molecule (N-CAM) in the mouse. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7631–7635. doi: 10.1073/pnas.82.22.7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelman G. M. Cell adhesion and the molecular processes of morphogenesis. Annu Rev Biochem. 1985;54:135–169. doi: 10.1146/annurev.bi.54.070185.001031. [DOI] [PubMed] [Google Scholar]
- Edelman G. M. Expression of cell adhesion molecules during embryogenesis and regeneration. Exp Cell Res. 1985 Nov;161(1):1–16. doi: 10.1016/0014-4827(85)90485-9. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Goridis C., Hirn M., Santoni M. J., Gennarini G., Deagostini-Bazin H., Jordan B. R., Kiefer M., Steinmetz M. Isolation of mouse N-CAM-related cDNA: detection and cloning using monoclonal antibodies. EMBO J. 1985 Mar;4(3):631–635. doi: 10.1002/j.1460-2075.1985.tb03676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemperly J. J., Edelman G. M., Cunningham B. A. cDNA clones of the neural cell adhesion molecule (N-CAM) lacking a membrane-spanning region consistent with evidence for membrane attachment via a phosphatidylinositol intermediate. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9822–9826. doi: 10.1073/pnas.83.24.9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemperly J. J., Murray B. A., Edelman G. M., Cunningham B. A. Sequence of a cDNA clone encoding the polysialic acid-rich and cytoplasmic domains of the neural cell adhesion molecule N-CAM. Proc Natl Acad Sci U S A. 1986 May;83(9):3037–3041. doi: 10.1073/pnas.83.9.3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman S., Sorkin B. C., White P. C., Brackenbury R., Mailhammer R., Rutishauser U., Cunningham B. A., Edelman G. M. Chemical characterization of a neural cell adhesion molecule purified from embryonic brain membranes. J Biol Chem. 1982 Jul 10;257(13):7720–7729. [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray B. A., Hemperly J. J., Gallin W. J., MacGregor J. S., Edelman G. M., Cunningham B. A. Isolation of cDNA clones for the chicken neural cell adhesion molecule (N-CAM). Proc Natl Acad Sci U S A. 1984 Sep;81(17):5584–5588. doi: 10.1073/pnas.81.17.5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray B. A., Owens G. C., Prediger E. A., Crossin K. L., Cunningham B. A., Edelman G. M. Cell surface modulation of the neural cell adhesion molecule resulting from alternative mRNA splicing in a tissue-specific developmental sequence. J Cell Biol. 1986 Oct;103(4):1431–1439. doi: 10.1083/jcb.103.4.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naora H., Deacon N. J. Relationship between the total size of exons and introns in protein-coding genes of higher eukaryotes. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6196–6200. doi: 10.1073/pnas.79.20.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen C., Mattei M. G., Mattei J. F., Santoni M. J., Goridis C., Jordan B. R. Localization of the human NCAM gene to band q23 of chromosome 11: the third gene coding for a cell interaction molecule mapped to the distal portion of the long arm of chromosome 11. J Cell Biol. 1986 Mar;102(3):711–715. doi: 10.1083/jcb.102.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K. R., Alberts B. M., Benzinger R., Lawhorne L., Treiber G. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology. 1970 Mar;40(3):734–744. doi: 10.1016/0042-6822(70)90218-7. [DOI] [PubMed] [Google Scholar]