Atg1-mediated myosin II activation regulates autophagosome formation during starvation-induced autophagy
In Drosophila, the autophagy kinase Atg1 phosphorylates the MLCK-like kinase spaghetti-squash activator (Sqa). In turn, Sqa-mediated regulation of myosin II activity is critical for starvation-induced autophagosome formation. A similar pathway likely operates in mammalian cells.
Keywords: Atg1, Atg9, autophagy, myosin II, ZIPK
Abstract
Autophagy is a membrane-mediated degradation process of macromolecule recycling. Although the formation of double-membrane degradation vesicles (autophagosomes) is known to have a central role in autophagy, the mechanism underlying this process remains elusive. The serine/threonine kinase Atg1 has a key role in the induction of autophagy. In this study, we show that overexpression of Drosophila Atg1 promotes the phosphorylation-dependent activation of the actin-associated motor protein myosin II. A novel myosin light chain kinase (MLCK)-like protein, Spaghetti-squash activator (Sqa), was identified as a link between Atg1 and actomyosin activation. Sqa interacts with Atg1 through its kinase domain and is a substrate of Atg1. Significantly, myosin II inhibition or depletion of Sqa compromised the formation of autophagosomes under starvation conditions. In mammalian cells, we found that the Sqa mammalian homologue zipper-interacting protein kinase (ZIPK) and myosin II had a critical role in the regulation of starvation-induced autophagy and mammalian Atg9 (mAtg9) trafficking when cells were deprived of nutrients. Our findings provide evidence of a link between Atg1 and the control of Atg9-mediated autophagosome formation through the myosin II motor protein.
Introduction
Autophagy is a highly conserved catabolic process in which long-lived proteins, RNA, and organelles are degraded in lysosomes. It occurs at a relatively low level during normal growth conditions but can be strongly induced under such environmental stress as nutrient starvation, hypoxia, and oxidative stress. Once autophagy is induced, cytoplasmic components are engulfed within specialized double-membrane vesicles known as autophagosomes (Mizushima, 2007). These vesicles subsequently fuse with lysosomes for degradation and recycling. Although much of our knowledge on autophagy was first obtained from morphological observation in mammalian cells, the molecular components of this process were largely identified in yeast (Klionsky, 2007). Through the genetic screening of yeast, about 30 autophagy-related (ATG) genes have been identified (Yorimitsu and Klionsky, 2005; Suzuki and Ohsumi, 2007). A subset of these Atg proteins assembles into different protein complexes, including the Atg1 protein kinase complex, the autophagy-specific phosphatidylinositol 3-kinase (PI3K) complex, the Atg8 and Atg12 ubiquitin-like conjugation systems, and Atg9 recycling system (Xie and Klionsky, 2007), and is essential for the formation of autophagosomes. Interestingly, many of these ‘core' Atg proteins are also found in mammals, suggesting the autophagic mechanism has been conserved.
One major question about autophagy is where and how autophagosomes emerge. In yeast, the pre-autophagosomal structure (PAS) is thought to be the organizing centre for formation of sequestering vesicles (Reggiori and Klionsky, 2005; Xie and Klionsky, 2007). It seems that for this formation to begin, the PAS must be targeted by Atg proteins. The molecular mechanism underlying this process is not known, though the Atg1 complex may be involved in the recruitment of Atg proteins to PAS (Kawamata et al, 2008). The serine/threonine protein kinase Atg1 acts as an important link between the nutrient-sensing target of rapamycin (TOR) kinase signalling and autophagy. It has been shown that Atg1 associates with Atg13 and Atg17 in response to TOR regulation (Kabeya et al, 2005). Moreover, Atg1 is involved in the recycling of Atg9 between PAS and multiple peripheral structures (Reggiori et al, 2004), which may contribute to cycling of autophagosome membrane. In mammals, two Atg1 homologues have been identified: the UNC-51-like kinases, Ulk1 and Ulk2 (Kuroyanagi et al, 1998; Yan et al, 1999). It has been reported that both Ulk1 and Ulk2 can relocalize to mammalian PAS (called isolation membrane/phagophore) when cells are starved of nutrients and the formation of autophagosomes is inhibited when either Ulk1 or Ulk2 is depleted (Hara et al, 2008). These findings suggest a functional redundancy between these proteins. Like yeast Atg1, Ulk1 has been found to regulate the trafficking of mammalian Atg9 (mAtg9) from the trans-Golgi network (TGN) to forming autophagosomes under starvation conditions (Young et al, 2006). More recent studies have shown that Ulk1 also interacts with mammalian Atg13 (mAtg13), the scaffolding protein FIP200 (a functional analogue of yeast Atg17), and Atg101 (Hara et al, 2008; Jung et al, 2009; Hosokawa et al, 2009a, 2009b). These proteins form a large, stable complex and are essential for autophagosome formation. Apart from the Ulk1-mAtg13-FIP200 complex, it is unclear what other Ulk1 downstream signals are generated to trigger autophagy.
In a recent study, we identified an interaction between Atg1 and the cytoskeletal protein paxillin (Chen et al, 2008). We also found that overexpression of Drosophila Atg1 induced autophagy and aberrant actin structures, suggesting a link between Atg1 and actin cytoskeleton in autophagy. As actin filaments form cables that serve as tracks guiding the movement of various cargos (Lanzetti, 2007), it may be that the actin cytoskeleton functions as structural support or as a track assisting the movement of autophagic components to PAS. In fact, in mammalian cells treated with actin-depolymerizing drugs such as cytochalasins B and D, the formation of autophagosomes has been found to be impaired (Aplin et al, 1992; Seglen et al, 1996). In yeast, actin filaments are reported to be essential for selective types of autophagy and the movement of Atg9 between the peripheral sites and the PAS (Reggiori et al, 2005; Monastyrska et al, 2008). However, the precise role that Atg1 has in the regulation of actin-dependent Atg9 cycling and autophagosome formation remains unknown.
In this study, we investigated the relationship between Atg1 and actin cytoskeleton and their possible involvement in autophagy. We discovered in Drosophila a new substrate of Atg1, Sqa, a myosin light chain kinase (MLCK)-like kinase. Depletion of Sqa, its mammalian homolog zipper-interacting protein kinase (ZIPK), and the inactivation of myosin II compromised starvation-induced autophagy in both Drosophila and mammalian cells. In addition, depletion of ZIPK and inhibition of myosin II markedly inhibited the redistribution of mAtg9 from TGN to the peripheral pool in response to nutrient deprivation. Our study not only identifies a novel role of Atg1 in the regulation of actomyosin activation, but also provides some insight into the mechanism underlying Atg1 regulation of Atg9 trafficking during the early stages of autophagosome formation.
Results
Overexpression of Atg1 induces myosin II activation
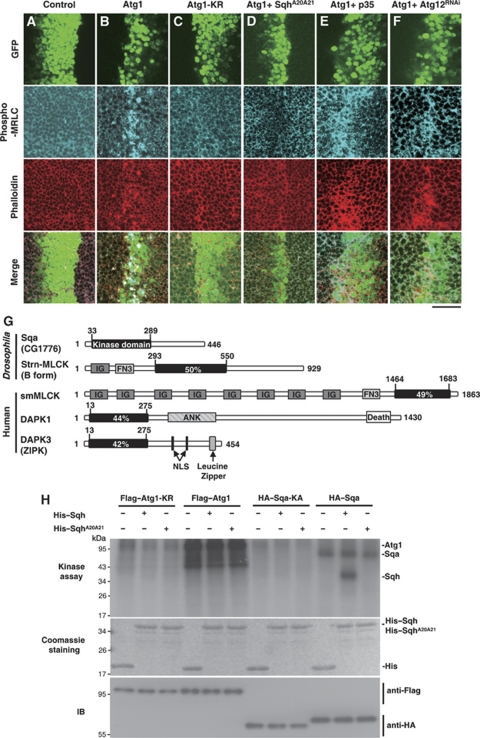
In one of our previous studies, as well as in one by Scott et al (2007), overexpression of Drosophila Atg1 was found to increase autophagy and result in apoptotic cell death (Chen et al, 2008). We also found that high levels of Atg1 led to abnormal cell morphology and F-actin reorganization (Figure 1B), as has been found previously (Chen et al, 2008). This suggests an increase in actomyosin contractility. As myosin II activity is regulated by the phosphorylating state of the myosin regulatory light chain (MRLC), we used a phospho-specific MRLC antibody that would recognize the pSer21 of Drosophila MRLC homolog Spaghetti squash (Sqh) to assess the myosin II activity of wing imaginal discs expressing Atg1 (Matsumura et al, 1998). Overexpression of Atg1 in the developing wing with ptc-GAL4 driver resulted in a dramatic increase level of phospho-MRLC and F-actin accumulation in GFP-marked Atg1-expressing cells, but not in ptc-GAL4 controls or in cells expressing the kinase-deficient Atg1, Atg1-KR (Figure 1A–C). Furthermore, the Atg1-induced MRLC phosphorylation could be suppressed by co-expressing the non-phosphorylatable form of Sqh, SqhA20A21 (Figure 1D) (Jordan and Karess, 1997). Taken together, these data suggested that Atg1 mediated the activation of actomyosin in a kinase-dependent manner. In addition to wing imaginal discs, we also found a robust increase in phopho-MRLC in response to Atg1 expression in fat-body cells (Supplementary Figure S1A) and in cultured S2R+ cells (Supplementary Figure S1B), suggesting that the Atg1-induced actomyosin activation is not tissue specific.
Figure 1.
Atg1 induces myosin II activation and spaghetti-squash activator (Sqa) identification. (A–F) Atg1-induced myosin II activation depends on the kinase activity of Atg1. Third-instar wing imaginal discs from ptc-GAL4 UAS-GFP controls or flies expressing indicated transgenes were stained with phospho-MRLC (blue) and TRITC-labelled phalloidin (red). Low level of phospho-MRLC staining was observed in controls (A) and cells overexpressing kinase-deficient Atg1-KR (C), whereas a robust increase in phospho-MRLC was found in cells overexpressing Atg1 (B). Co-expression of Atg1 and spaghetti-squash (SqhA20A21) exhibited low level of phospho-MRLC staining (D). Atg1-induced high level of phospho-MRLC staining was not suppressed by expression of caspase inhibitor p35 (E) or by depletion of Atg12 (Atg12RNAi) (F). Bar, 20 μm. (G) Schematic diagram of Drosophila and mammalian MLCK family. The quantities within the kinase domains indicate the degree of amino acid identity to the kinase domain of Sqa (CG1776). Ank, ankyrin repeats; Death, death domain; Fn, fibronectin domain; Ig, immunoglobulin domain; NLS, nuclear localization signal. (H) Sqa, but not Atg1, directly phosphorylated Sqh in vitro. Flag–Atg1, Flag–Atg1-KR, HA–Sqa and HA–Sqa-KA were immunoprecipated from lysate of transfected cells and incubated in an in vitro kinase reaction mixture containing [γ-32P]ATP and bacterially expressed recombinant wild-type Sqh or SqhA20A21. As shown on the autoradiogram (top panel), wild-type but not the kinase-deficient Atg1 (Atg1-KR) and Sqa (Sqa-KA) was autophosphorylated. No phosphorylation was seen with SqhA20A21. The equal input of His-fusion proteins is shown on the Coomassie staining. Anti-Flag and anti-HA immunoblottings (IBs) were used as controls to quantify the amount of proteins precipitated.
We next examined whether the increased MRLC phosphorylation in Atg1-expressing cells was caused by Atg1-induced cell death or autophagy. We found that neither expression of the caspase inhibitor p35, nor RNAi-mediated downregulation of Atg12 suppressed the Atg1-induced MRLC phosphorylation (Figure 1E and F), indicating that Atg1-induced myosin II activation occurred through a caspase and autophagy-independent process. We further investigated whether actomyosin activation had a mediating role in Atg1-induced cell death and autophagy. We found the Atg1-induced caspase activation and autophagosome formation to be significantly suppressed by co-expressing SqhA20A21 (Supplementary Figure S2), suggesting that myosin II activation is required for Atg1-induced autophagy and cell death.
CG1776 encodes a novel MLCK-like protein
To investigate the possibility of a direct relationship between Atg1 and Sqh, we first tested whether Sqh could be a direct phosphorylation target of Atg1. Using His-tagged Sqh as a substrate in our in vitro kinase analysis, we did not find Atg1 to directly phosphorylate Sqh (Figure 1H). Therefore, it might be possible that Atg1 induces MRLC phosphorylation through myosin II activators, as phosphorylation of MRLC has been reported to be regulated by a spectrum of stimulating kinases, including MLCK, Rho-associated protein kinase (ROK), and the death-associated protein kinase (DAPK) family proteins (Vicente-Manzanares et al, 2009).
In Drosophila, Stretchin-Mlck (Strn-Mlck) is the MLCK most closely related to the vertebrate smooth muscle/non-muscle MLCKs (sm/nmMLCK) and skeletal MLCKs (skMLCK). In a BLAST homology search against the BDGP database using the kinase domain of Strn-Mlck, we found a new MLCK-like protein, CG1776. The kinase domain of CG1776 is 50 and 49% identical with that of Strn-Mlck and smMLCKs, respectively (Figure 1G). However, unlike Strn-Mlck and smMLCK, CG1776 does not contain a Ca2+/calmodulin regulatory domain. We also found the kinase domain of CG1776 to be highly homologous to that of the DAPK family proteins such as DAPK1 and DAPK3/ZIPK (Figure 1G). Interestingly, a recent study has implicated a role of CG1776 in Drosophila S6 kinase (S6K) phosphorylation (Findlay et al, 2007). As Atg1 has been found to inhibit TOR/S6K-dependent cell growth via a negative feedback regulation (Lee et al, 2007; Scott et al, 2007), we wanted to study the relationship between Atg1 and CG1776. We renamed CG1776 as sqa for the following reasons. First, we performed in vitro kinase assays using Sqh as a substrate to assess whether the sequence homology of Sqa and MLCK extends to their biochemical activity. We found that wild-type Sqa could phosphorylate itself and Sqh (Figure 1H), but the catalytically inactive form, Sqa-KA, could not. Second, when Sqa was clonally expressed in wing imaginal discs using the flip-out GAL4 system (Ito et al, 1997), a dramatic increase of Sqh phosphorylation was detected in GFP-positive Sqa-expressing cells (Figure 3C). Thus, both our biochemical and immunostaining analyses indicated that Sqa is a bona fide MLCK-like protein capable of activating Sqh.
Sqa genetically interacts with Atg1
To determine whether Sqa is involved in the Atg1-mediated activation of myosin II, we examined the effects of RNAi-mediated inhibition of Sqa on the Atg1-induced wing defects. Overexpression of Atg1 with ptc-GAL4 resulted in disappearance of anterior cross-veins in adult wings (Figure 2B), as was found in a previous study (Chen et al, 2008). Importantly, we found that ablation of Sqa expression by Sqa-RNAi strongly reversed the Atg1-induced disappearance of the cross-veins (Figure 2G). Moreover, similar to that of Atg1, overexpression of Sqa in wing discs using ptc-GAL4 resulted in an anterior cross-vein missing phenotype (Figure 2C). The Sqa-induced wing vein defects could be suppressed by co-expressing either Sqa-RNAi or SqhA20A21 (Figure 2I and J), suggesting the observed phenotype is Sqa specific and depends on myosin II activation. However, epistasis analysis revealed that depletion of Atg1 did not suppress the Sqa-induced wing vein defects (Figure 2K), suggesting that Sqa might function as a downstream target of Atg1. To determine whether Sqa is required for Atg1-induced activation of myosin II, we investigated whether ablation of Sqa expression could suppress the activation of myosin II and the disorganization of actin cytoskeleton induced by Atg1. Using anti-phospho-MRLC antibody in our immunofluoresence analysis, we found a marked decrease in MRLC phosphorylation and normal actin organization in the regions where Atg1 and Sqa-RNAi were co-expressed (Supplementary Figure S3A and B). Furthermore, consistent with our findings in Supplementary Figure S2, we found that inhibition of myosin II activation by depletion of Sqa strongly suppressed the Atg1-induced cell death and autophagy (Supplementary Figure S3C–F). These results together indicate that Sqa functions as an important downstream target of Atg1 in the regulation of myosin II activation and is required for Atg1-induced autophagy and cell death.
Figure 2.
Genetic and biochemical interactions between Atg1 and spaghetti-squash activator (Sqa). (A–K) Genetic interactions between Atg1, Sqa, and spaghetti-squash (Sqh). Compared with the ptc-GAL4 controls (A), expression of Atg1 or Sqa by ptc-GAL4 resulted in missing anterior cross-vein phenotypes (B, C). However, RNAi-mediated downregulation of Atg1 (Atg1RNAi) or Sqa (SqaRNAi) did not cause wing vein defects (D, E). Depletion of Atg1 and Sqa suppressed Atg1 and Sqa-induced wing vein defects, respectively (F, I). Atg1-induced wing defects were modulated by depletion of Sqa (G) or by co-expression of SqhA20A21 (H). Nevertheless, Sqa-induced wing vein defects were suppressed by co-expression of SqhA20A21 (J) but not Atg1RNAi (K). (L) Schematic presentation of the domain structures of Sqa and deletion mutants. (M–N) Atg1 physically interacted with Sqa. (M) 293T cells were transfected with HA-tagged Sqa WT (wild-type) or KA, together with Flag-tagged Atg1 WT or KR. The cells were lysed 48 h after transfection and immunoprecipitated (IP) with anti-Flag antibodies. The immunoprecipitated proteins and the total cell lysates (TCL) were analysed by immunoblotting (IB) with antibodies as indicated. (N) 293T cells transfected with Flag–Atg1-KR and various HA-tagged Sqa contracts were subjected to immunoprecipitations with anti-HA antibody. The immunoprecipitated proteins and the total cell lysates were analysed by immunoblotting with antibodies as indicated. (O) Atg1 directly phosphorylated Sqa in vitro. Flag-tagged Atg1 WT or KR immunoprecipated from lysate of transfected cells was used to phosphorylate bacterially expressed recombinant Sqa-K1, Sqa-K2, and Sqa-C in an in vitro kinase assay. The lower panels represent equal input of His-fusion proteins and Atg1 immunoprecipitates.
Sqa is a substrate of Atg1
To further elucidate the interaction between Atg1 and Sqa, we investigated whether Sqa physically interacted with Atg1. HEK 293T cells were transfected with Flag-tagged wild-type Atg1 together with HA-tagged Sqa. Immunoblotting of the anti-Flag immunoprecipates from cell lysates revealed no co-precipitation between wild-type Atg1 and Sqa (Figure 2M). It has been reported that protein kinases generally interact with substrates transiently and with relatively low affinity (Manning and Cantley, 2002). To overcome this problem for our binding assay, we employed a kinase substrate-trapping approach in which the catalytically inactive kinase variants formed a stable interaction with their substrates (Deminoff et al, 2006). We found that Sqa specifically co-immunoprecipitated with the kinase inactive form of Atg1, Atg1-KR, but not with the wild-type Atg1 (Figure 2M). Conversely, the kinase inactive form of Sqa, Sqa-KA, did not interact with wild-type Atg1. These results indicated that Sqa may be a substrate of Atg1. Next, to determine the region of Sqa responsible for its interaction with Atg1, two HA-tagged Sqa mutants, HA–Sqa-K and HA–Sqa-C (Figure 2L), were constructed and tested for their ability to pull down Flag-tagged Atg1-KR. Co-immunoprecipitation assays revealed that Atg1-KR specifically interacted with Sqa-K, which contains the N-terminal kinase domain (amino acids 1–301), but not with Sqa-C, which contains only the C-terminal region (amino acids 291–446) (Figure 2N). Taken together, these results suggest that Sqa is an Atg1-interacting protein, and the interaction is mediated through the kinase domain of Sqa.
Both our genetic and biochemical data suggested that Sqa might be a direct substrate of Atg1. To find out, we performed in vitro kinase assays using Atg1 isolated from 293T transfectants and various recombinant Sqa mutants as substrates. As shown in Figure 2O, Atg1 phosphorylated the kinase domain region of Sqa, Sqa-K1 (amino acids 1–189) and Sqa-K2 (amino acids 190–301), but not the C-terminal region of Sqa, Sqa-C, suggesting that there are multiple Atg1 phosphorylation sites within the N-terminal kinase domain of Sqa. Furthermore, the kinase-inactive Atg1-KR was not able to phosphorylate either region of the Sqa (Figure 2O). Therefore, consistent with the binding assay, our data strongly suggest that Sqa is a direct substrate of Atg1.
Phosphorylation of Sqa at Thr-279 is required for Atg1-mediated myosin II activation
It has been shown that MLCK activity is regulated by multisite phosphorylation through various protein kinases, including cAMP-dependent protein kinase (PKA), protein kinase C, and CaM kinase II (Stull et al, 1993; Gallagher et al, 1997). However, these kinases phosphorylate MLCK at sites in the Ca2+/calmodulin regulatory domain, which is not found in Sqa. A recent study by Graves et al (2005) revealed that phosphorylation of Thr-180, Thr-225, and Thr-265 in the kinase domain of the ZIPK (also known as DAPK3) is essential for full enzymatic activity to occur. Our sequence analysis showed Sqa to be highly homologous to ZIPK and to exhibit three corresponding phosphorylation sites at Thr-194, Thr-239, and Thr-279 (Supplementary Figure S4A). We further determined the Atg1 phosphorylation sites of Sqa using mass spectrometry-based analyses. Notably, MS/MS analysis revealed a single phosphorylation site at Thr-279 in a peptide corresponding to residues 273–285 of Sqa (Supplementary Figure S4B). We thus mutated Thr-194, Thr-239, and Thr-279 of Sqa-K2 to alanine and assayed their ability to be substrates for Atg1 in an in vitro kinase assay. Compared with the wild-type Sqa-K2, the substitution of Ala for Thr-279, but not for Thr-194 or Thr-239 strongly attenuated the phosphorylation of Sqa-K2 by Atg1 (Figure 3A). On the basis of these findings, it is likely that Thr-279 of Sqa is a major phosphorylation site of Atg1 and an additional site(s) may be present within the stretch of residues spanning from amino acids 1 to 301.
Figure 3.
Phosphorylation of spaghetti-squash activator (Sqa) at Thr-279 is required for Atg1-mediated myosin II activation. (A) Characterization of Atg1-dependent phosphorylation sites on Sqa. 293T cells transfected with Flag-tagged Atg1 or Atg1-KR were subjected to immunoprecipitation with anti-Flag antibodies, followed by in vitro kinase assays with bacterially expressed Sqa-K2, Sqa-K2-T194A, Sqa-K2-T239A, and Sqa-K2-T279A as substrates. Atg1 but not Atg1-KR was autophosphorylated (top panel). Relative phosphorylation levels of substrates were quantified. Data are represented as mean±s.e. of triplicates. (B–D) Phosphorylation of Sqa at Thr-279 is critical for the kinase activity of Sqa. (B) HA-tagged Sqa or Sqa-T279A immunoprecipated from lysate of transfected cells was used to phosphorylate bacterially expressed recombinant spaghetti-squash (Sqh) WT and A20A21, in an in vitro kinase assay. (C) Clonal expression of Sqa but not Sqa-T279A (GFP-positive cells) in the larval wing imaginal discs resulted in a marked increase in phospho-MRLC staining (blue) and actin reorganization (red). Bar, 20 μm. (D) The larval fat body of denoted genotypes were dissected, lysed, and subjected to western blot analysis using anti-phospho-MRLC and anti-Sqh antibodies. For quantification, the levels of MRLC phosphorylation in each genotype were measured with ImageJ and normalized to the Sqh levels. Data are expressed as a fold change compared with the wild-type controls. Each value represents mean±s.e. from triplicate experiments. See Supplementary data for genotypes.
To assess the importance of Thr-279 on the function of Sqa, we generated the T279A mutation in the full-length Sqa and determined its enzymatic activity both in vivo and in vitro. Compared with the wild-type Sqa, T279A mutant showed markedly reduced autophosphorylation in an in vitro kinase assay (Figure 3B). Consistent with the autophosphorylation data, T279A mutant dramatically reduced the catalytic activity of Sqa in phosphorylating Sqh in vitro (Figure 3B). Furthermore, both immunofluorescence and immunoblotting analyses showed that T279A mutant failed to stimulate MRLC phosphorylation in vivo (Figure 3C and D). Notably, co-expression of T279A mutant also reduced Sqa-induced wing vein defects (Supplementary Figure S5E). These findings indicated that T279A mutant could behave as a dominant negative mutation. We further investigated whether it would suppress wing vein defects caused by Atg1. We found that co-expression of Sqa-T279A mutant markedly suppressed the Atg1-induced wing phenotypes and the activation of myosin II (Supplementary Figure S5F and G), indicating that Atg1-mediated phosphorylation of Thr279 is required for Sqa activation, which subsequently stimulates the activation of myosin II downstream.
Myosin II activation is required for starvation-induced autophagy
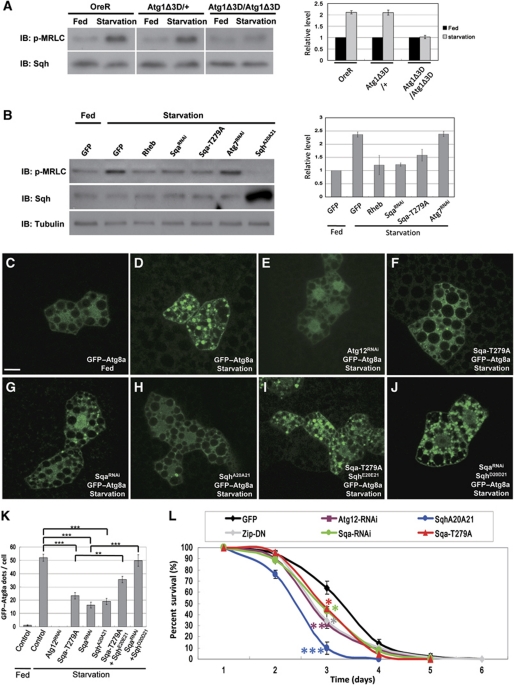
It has been reported that a robust autophagic response is induced in Drosophila larval fat body in response to nutrient deprivation (Scott et al, 2004). This process depends on the downregulation of TOR signalling and activation of Atg1 kinase. To investigate the physiological role of myosin II in autophagy, we first tested whether the myosin II activity of larval fat body was altered during starvation conditions, and found a robust increase in MRLC phosphorylation (Figure 4A). Importantly, starvation-induced myosin II activation was markedly abolished in Atg1 null but not in Atg1 heterozygous animals (Figure 4A), suggesting that the observed MRLC phosphorylation occurred in an Atg1-dependent manner. As the Atg1-induced autophagy could be modulated by the TOR pathway, we tested whether increased TOR activity affected starvation-induced activation of myosin II. As shown in Figure 4B, overexpression of TOR activator, the Rheb GTPase, in the starved larval fat body strongly suppressed myosin activation, compared with GFP controls. Moreover, we found that expression of either Sqa-RNAi or Sqa-T279A in larval fat body significantly blocked the upregulation of myosin activity under starvation conditions (Figure 4B). However, neither ablation of Atg7 nor Atg12 expression affected MRLC phosphorylation (Figure 4B, data not shown). These results suggest that the starvation-induced myosin II activation is regulated by TOR activity and through the Atg1–Sqa-mediated pathway.
Figure 4.
Atg1–spaghetti-squash activator (Sqa)-mediated myosin II activation is required for starvation-induced autophagy. (A, B) Activation of myosin II on nutrient deprivation. The larval fat body of denoted genotypes under fed or starved conditions were dissected, lysed, and subjected to western blot analysis using antibodies specific for phospho-myosin regulatory light chain (MRLC) and total MRLC. The Rheb, SqaRNAi, Sqa-T279A, spaghetti-squash (SqhA20A21), and Atg7RNAi transgenes were expressed under the control of hs–GAL4 driver (B). For quantification, the relative phosphorylation levels of MRLC were quantified as in Figure 3D. Data are represented as mean±s.e. of triplicates. (C–J) Starvation-induced autophagosome formation was compromised by inhibition of myosin II activation. Compared with the fed condition (C), starvation induced a robust formation of GFP–Atg8a puncta in larval fat-body cells (D). Clonal expression of Atg12RNAi (E), Sqa-T279A (F), SqaRNAi (G), or SqhA20A21 (H) markedly suppressed GFP–Atg8a puncta formation during starvation. The autophagic defects caused by Sqa-T279A and SqaRNAi were rescued by co-expression of the constitutively active SqhE20E21 (I) and SqhD20D21 (J), respectively. Bar, 20 μm. (K) The average number of GFP–Atg8a marked autophagosomes per cell is shown (data are represented as mean±s.e. of 20 fat-body samples imaged per genotype; ***P<0.001). (L) Inhibition of myosin II activity increased sensitivity to starvation. Flies carrying transgenes under the control of fat body-specific Cg–GAL4 driver were used for further analysis. Histogram illustrating the survival curve of adult female flies of denoted genotypes when placed under starved conditions. Data are mean±s.e. from triplicate experiments (n=100 flies/genotype/treatment). *P<0.05, **P<0.01, ***P<0.001. See Supplementary data for genotypes.
As we found that the activation of myosin II had a critical role in the Atg1-induced formation of autophagosomes (Supplementary Figure S2H), we investigated whether depletion of Sqa or inhibition of myosin II activation would affect the formation of autophagic vacuoles induced by starvation. For these experiments, we analysed the GFP–Atg8a distribution in fat-body clones generated by the flip-out system. GFP–Atg8a was redistributed from a uniformly diffuse cytoplasmic localization under nutrient-rich conditions to punctate structures in response to starvation (compare Figure 4C and D). The formation of these GTP-Atg8a punctae was blocked by depletion of Atg12 (Figure 4E), suggesting that these punctate structures are autophagy-related vesicles. Co-expression of SqhA20A21 with GFP–Atg8a strongly inhibited the starvation-induced GFP–Atg8a punctae (Figure 4H and K). Consistent with this finding, co-expression of Sqa-T279A or Sqa-RNAi with GFP–Atg8a also resulted in a significant decrease in size and number of GFP–Atg8a punctae (Figure 4F, G and K) in response to starvation. Moreover, we found that the constitutively active Sqh (SqhE20E21 and SqhD20D21) (Dorsten et al, 2007; Mitonaka et al, 2007) restored the autophagic defects caused by Sqa-T279A and Sqa-RNAi (Figure 4I–K). Together, these findings strongly support the hypothesis that Sqa-mediated Sqh activation has an essential role in starvation-induced autophagy.
As autophagy-mediated degradation and recycling of cellular components are known to have an important role in organism survival under unfavourable conditions (Juhasz et al, 2007), we examined the effects of myosin II activity on adult fly viability under starvation conditions. Compared with control flies, those in which myosin II activity was inhibited by expressing the non-phosphorylatable SqhA20A21 or dominant-negative form of myosin heavy chain zipper (Zip-DN) had a significantly shortened life span under starvation conditions (Figure 4L). Similarly, reduced expression of Sqa and overexpression of Sqa-T279A mutant resulted in increased death rate (Figure 4L). Taken together, these observations suggest that Sqa and myosin II activation are required for autophagy and are also crucial for animal survival under starvation conditions.
Myosin II is involved in the regulation of autophagy in mammalian cells
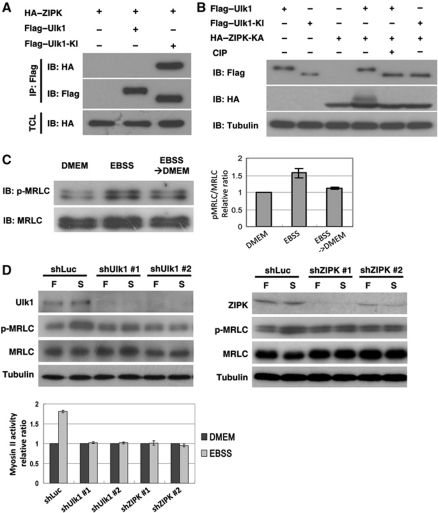
Autophagy has been shown to be a highly conserved process from yeast to mammals (Suzuki and Ohsumi, 2007; Yang and Klionsky, 2010). To determine whether Atg1–Sqa-mediated myosin II activation and autophagosome formation is conserved in mammalian cells, we first performed transient transfection experiments to test whether Ulk1 interacted with the Sqa mammalian homolog ZIPK. Co-immunoprecipitation assays showed that, similar to Atg1–Sqa interaction, ZIPK specifically interacted with the kinase inactive form of Ulk1, Ulk1-KI, but not with the wild-type Ulk1 (Figure 5A). Moreover, we found that the kinase-inactive ZIPK-KA showed a mobility shift in cells co-transected with wild-type Ulk1, but not with Ulk1-KI (Figure 5B). The mobility shift of ZIPK-KA was abolished by the CIP phosphatase treatment. These results together suggest that ZIPK may be a substrate of Ulk1. Next, we investigated whether the myosin II activity would be affected in cells deprived of nutrients. Consistent with our observation in Drosophila, we found a marked increase of phospho-MRLC in MCF7 cells during amino acid and serum starvation (Figure 5C). The myosin II activity was decreased after the medium was replaced with a nutrient-rich medium. Kinetic assays revealed that the activation of MRLC occurred within 30 min after nutrient deprivation, and the phosphorylation of MRLC coincided with the autophagic flux (Supplementary Figure S6). We further used Ulk1- and ZIPK shRNAs to downregulate Ulk1 and ZIPK expression in MCF7 cells. We found that the starvation-induced activation of myosin II was reduced by Ulk1 and ZIPK depletion (Figure 5D), indicating a role of Ulk1 and ZIPK in the regulation of myosin II activation on nutrient deprivation.
Figure 5.
UNC-51-like kinase (Ulk1) and zipper-interacting protein kinase (ZIPK) are essential for starvation-induced myosin II activation. (A) Ulk1 interacted with ZIPK. 293T cells transfected with HA-tagged ZIPK together with Flag-tagged Ulk1 or Ulk1-KI were subjected to immunoprecipitations with anti-Flag antibody. The immunoprecipitated proteins and the total cell lysates were analysed by immunoblotting with antibodies as indicated. (B) HA-tagged kinase-inactive ZIPK-KA (K42A) was expressed in 293T cells with Flag-tagged Ulk1 or Ulk1-KI. Cell lysates were incubated with or without alkaline phosphatase (CIP) and analysed by immunoblotting with antibodies as indicated. (C) Myosin II was activated upon nutrient deprivation in MCF7 cells. MCF7 cells were cultured in serum containing DMEM medium (nutrient-rich condition) or in Earle's balanced salt solution (EBSS; starved condition) for 2 h. The myosin II activity was significantly downregulated after EBSS-starved cells were replenished with fresh DMEM medium containing 10% FBS for 2 h. The myosin II activity was quantified and expressed as a fold changes compared with the DMEM controls. Each value represents mean±s.e. of triplicates. (D) MCF7 cells stably infected with lentivirus expressing control (shLuc), Ulk1 or ZIPK shRNA were cultured in nutrient-rich DMEM medium (F) or EBSS (S) for 2 h. Effects of Ulk1 and ZIPK knockdown on starvation-induced myosin II activation were analysed by immunoblotting with antibodies as indicated. Data are mean±s.e. of triplicates.
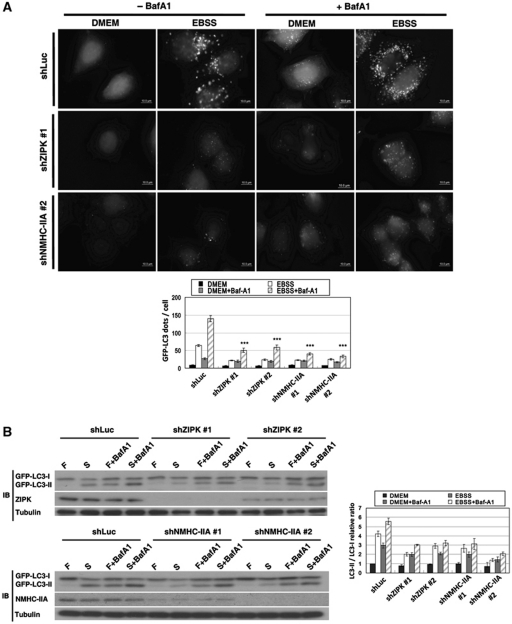
To further test the function of myosin II in autophagy, we used three independent methods to specifically inhibit myosin II function. MCF7/GFP–LC3 cells were transfected with non-muscle myosin heavy chain-IIA (NMHC-IIA) shRNA or treated with ML-7, a highly specific inhibitor of MLCK, or blebbistatin, a myosin II inhibitor. The cells were incubated in a starvation medium (EBSS) in the presence or absence of the lysosomal inhibitor bafilomycin A1 (BafA1). We found that the depletion or inactivation of myosin II significantly reduced the size and number of GFP–LC3 and Atg16 punctae when the cells were deprived of nutrients (Figure 6A and Supplementary Figure S7). Moreover, LC3 conversion assay revealed that myosin II inhibition strongly suppressed the conversion of cytosolic LC3 (LC3-I) to the lipidated form of LC3 (LC3-II) (Figure 6B and Supplementary Figure S7B). Notably, although treatment with BafA1 led to a marked increase of GFP–LC3 signal and LC3II/LC3I ratio in control cells, the fact that a reduction of GFP–LC3 puncta formation and GFP–LC3 lipidation was unchanged when myosin II was inhibited indicates that the decrease in autophagic flux was not due to an increase in autophagosome turnover. It has been shown that expression of the constitutively active form of ZIPK leads to the induction of autophagy (Shani et al, 2004). To investigate the role of ZIPK in starvation-induced autophagy, control and ZIPK knockdown MCF7/GFP–LC3 cells were fed and starved for 2 h with and without BafA1. ZIPK depletion led to a significant decrease in the number and size of GFP–LC3 and Atg16 punctae and in LCII/I ratio, compared with control cells (Figure 6 and Supplementary Figure S7). Taken together, our results demonstrate the importance of ZIPK and myosin II to the starvation-induced autophagy in mammalian cells.
Figure 6.
Involvement of zipper-interacting protein kinase (ZIPK) and myosin II in starvation-induced autophagy. (A) MCF7/GFP–LC3 cells stably infected with lentivirus expressing control (shLuc), ZIPK, or non-muscle myosin heavy chain-IIA (NMHC-IIA) shRNA were cultured in nutrient-rich medium (DMEM) or starvation medium (Earle's balanced salt solution; EBSS) in the presence or absence of lysosomal inhibitor bafilomycin A1 (BafA1) for 2 h. Depletion of ZIPK and NMHC-IIA markedly inhibited starvation-induced GFP–LC3 puncta formation. Quantification of the number of GFP–LC3 dots per cell (lower panel) was shown (data are represented as mean±s.e. of 100 cells, ***P<0.001). (B) Cells as in (A) were cultured in nutrient-rich medium (F) or EBSS (S) with or without BafA1 for 2 h. Effects of ZIPK and NMHC-IIA knockdown on starvation-induced GFP–LC3 conversion were assessed by immunoblotting with anti-LC3, anti-ZIPK, anti-NMHC-IIA, and anti-tubulin antibodies. The relative ratio of LC3II/LC3I is shown at the right panel. Data are mean±s.e. of triplicates.
Redistribution of myosin II during starvation
Myosin II has been shown to function in diverse cellular processes, including cell adhesion, migration, and cytokinesis (Sellers, 2000). Recent findings have indicated that myosin II also has a role in membrane trafficking (Duran et al, 2003; DePina et al, 2007). Myosin II undergoes dynamic subcellular translocation during these processes. To determine the distribution of myosin II during starvation-induced autophagy, we first examined the localization of total and activated myosin II in the larval fat-body cells of sqhAX3; sqh–GFP, in which Sqh–GFP is the only source of MRLC (Royou et al, 2002). Under nutrient-rich conditions, Sqh–GFP and phospho-MRLC were predominantly localized on plasma membranes, especially at cell–cell junctions (Figure 7A). Under starvation conditions, Sqh–GFP and phospho-MRLC not only localized to the cell junctions but also enriched in the perinuclear region of the cell (Figure 7A). Next, we assessed whether the cellular distribution of myosin II was affected by starvation in mammalian cells. MCF7/GFP–LC3 cells were incubated in either full medium or starvation medium. Myosin II was localized to peripheral region of MCF7 cells under nutrient-rich conditions (Figure 7B). We found that both MRLC and phospho-MRLC were redistributed to the perinuclear region and were co-localized with GFP–LC3-labelled autophagosome under starvation condition (Figure 7B). We further tested whether starvation would increase the population of myosin II on membrane using subcellular fractionation. Under nutrient-rich condition, we observed that most myosin II existed in the cytosol compared with the membrane pellet (Figure 7C). However, under the starvation condition, both MRLC and phospho-MRLC were significantly decreased in the cytosolic fraction and increased in the membrane pellet (Figure 7C). These results show that nutrient starvation induces myosin II redistribution and that this process is conserved from Drosophila to mammalian cells.
Figure 7.
Redistribution of myosin II during nutrient starvation. (A) The fat body of spaghetti-squash (sqhAX3); sqh–GFP larva under fed or starved conditions were dissected and subjected to immunofluorescence analysis. Sqh–GFP and phospho-myosin regulatory light chain (MRLC) were enriched in cell–cell junction under nutrient-rich (fed) condition. Under starvation conditions, Sqh–GFP and phospho-MRLC were localized to both cell–cell junction and perinuclear region. Actin was stained with TRITC-labelled phalloidin (red) and nucleus was stained with DAPI (blue). Bar, 20 μm. (B) Starvation induced redistribution of myosin II in MCF7/GFP–LC3 cells. Both phospho-MRLC and MRLC were localized in peripheral region of cells in nutrient-rich medium (DMEM), whereas they redistributed to the perinuclear region and co-localized with GFP–LC3 under starvation conditions (Earle's balanced salt solution; EBSS). Bar, 10 μm. (C) MCF7/GFP–LC3 cells cultured in nutrient-rich (DMEM) or starved (EBSS) conditions were homogenized and subjected to centrifugation, and the resulting post-nuclear supernatant (PNS) was fractionated by high-speed centrifugation into membrane pellet and cytosol. Proteins were resolved by SDS–PAGE and immunoblotted with anti-TGN46 antibody as a control for membrane-association proteins, anti-caspase-3 as a control for cytosolic proteins. The levels of phospho-MRLC and MRLC in each fraction were quantified using ImageJ and plotted relative to their amounts in PNS (n=3). Each value represents mean±s.e. of three experiments. *P<0.05, **P<0.01.
The starvation-induced mAtg9 trafficking is regulated by myosin II
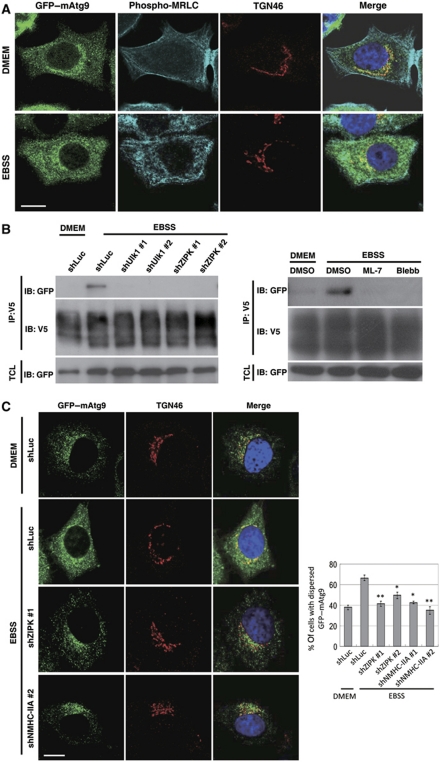
Atg9 is a multispanning membrane protein that cycles between the PAS and peripheral punctate structures (Reggiori et al, 2004). As the cycling of Ag9 is essential for autophagy, it has been proposed that Atg9 may have a pivotal role in supplying lipids to forming autophagosomes (Mari and Reggiori, 2007; Webber et al, 2007). In yeast, Atg1, actin filaments, and a myosin-like protein Atg11 have been shown to regulate the cycling of Atg9 (He et al, 2006); however, the molecular mechanism underlying this process is not clear. In this study, we assessed whether myosin II might act as a motor protein in the regulation of Atg9 trafficking during autophagy. In mammalian cells, mAtg9 has been found to cycle from the TGN to a dispersed peripheral pool upon nutrient deprivation (Young et al, 2006). We generated MCF7 cells that stably expressed GFP–mAtg9 and found that, consistent with previous findings, GFP–mAtg9 is enriched in a juxta-nuclear region and co-localizes with the trans-Golgi marker TGN46 under nutrient-rich conditions (Figure 8A). GFP–mAtg9 also partially co-localizes with the cis-Golgi marker GM130 but not with the marker for lysosomes (LAMP1) (Supplementary Figure S8A and B). During starvation, GFP–mAtg9 was dispersed to a peripheral region and appeared to co-localize with late endosomes and autophagosomes (Supplementary Figure S8C and D) (Young et al, 2006). Intriguingly, we found that NMHC-IIA, MRLC, and phospho-MRLC co-localized with GFP–mAtg9 under starvation conditions (Figure 8A and data no shown). Furthermore, our co-immunoprecipitation experiments revealed that mAtg9 physically interacted with NMHC-IIA on nutrient deprivation (Figure 8B). RNAi-mediated depletion of Ulk1 and ZIPK, or myosin II inhibition disrupted the interaction between mAtg9 and NMHC-IIA (Figure 8B). As Ulk1 is essential for the starvation-induced redistribution of mAtg9 (Young et al, 2006), we tested whether ZIPK and myosin II were also involved in the regulation of mAtg9 trafficking. Confocal microscopic analysis revealed that ZIPK depletion and myosin II inhibition significantly restricted mAtg9 localization to the TGN during nutrient starvation conditions (Figure 8C and Supplementary Figure S9). Taken together, our results demonstrate that Ulk1-ZIPK-mediated myosin II activation is required in starvation-induced mAtg9 trafficking and suggest that myosin II might act as the motor protein involved in regulating the movement of Atg9-mediated complex.
Figure 8.
The starvation-dependent cycling of mammalian Atg9 (mAtg9) is blocked by inhibition of myosin II activity. (A) Immunofluorescence analysis of GFP–mAtg9 and phospho-myosin regulatory light chain (MRLC) in MCF7/GFP–mAtg9 cells. GFP–mAtg9 was enriched in the trans-Golgi network (TGN) labelled with anti-TGN46 antibody and phospho-MRLC localized to cell peripheral in nutrient-rich condition (DMEM). GFP–mAtg9 redistributed and co-localized with phospho-MRLC in starvation condition (Earle's balanced salt solution; EBSS). (B) mAtg9 interacted with non-muscle myosin heavy chain-IIA (NMHC-IIA) under starvation conditions. 293T cells (right panel) or 293T cells stably infected with lentivirus expressing control (shLuc), UNC-51-like kinases (Ulk1) or zipper-interacting protein kinase (ZIPK) shRNA (left panel) were transfected with V5-tagged mAtg9 and GFP–tagged NMHC-IIA. At 48 h after transfection, cells were incubated in either serum-containing medium (DMEM) or EBSS for 2 h with or without the treatment of 50 μM ML-7 and blebbistatin (Blebb). Cell lysates were immunoprecipitated with anti-V5 antibody and immunoblotted with indicated antibodies. (C) MCF7/GFP–mAtg9 cells were stably infected with lentivirus expressing control (shLuc), ZIPK, or NMHC-IIA shRNA. Under starved conditions, mAtg9 was redistributed from the TGN to a dispersed peripheral pool in control cells (shLuc). Starvation-induced GFP–mAtg9 redistribution was blocked in ZIPK and NMHC-IIA knockdown cells. The images were analysed by quantifying mAtg9 localization in cells for either a TGN-enriched or dispersed pattern. Data are represented as mean±s.e. of 70 cells. *P<0.05, **P<0.01. Bar, 10 μm.
Discussion
In this study, we found that Atg1 is involved in the regulation of actomyosin activity that is achieved through the mediation of an Atg1-interacting protein, which we newly identified in the study, Sqa. We demonstrated that Sqa and myosin II activity are essential to the starvation-induced autophagy in Drosophila larval fat-body cells. Moreover, we found that the redistribution of mAtg9 requires myosin activity in mammalian cells, suggesting that myosin II functions as a motor protein in Atg1-dependent cycling of Atg9 during the formation of autophagosomes.
Myosin II is a conventional two-headed myosin composed of two heavy chains, two essential light chains, and two regulatory light chains (Sellers, 2000). Myosin II activation is regulated by the phosphorylation of its regulatory light chain via MLCKs. Rho GTPase and Rho kinase have been implicated in the regulation of myosin activation (Fukata et al, 2001). However, we found that neither RNA-mediated knockdown of dRok nor mutations in Rho1 or dRhoGEF2 could suppress the Atg1-induced wing defects (G-C Chen, unpublished data). Instead, we did find that depletion of Sqa rescued Atg1-induced wing defects. Our epistasis analysis showed that Sqa functioned downstream of Atg1. Moreover, we found that Sqa but not Atg1 could directly phosphorylate Sqh in the in vitro kinase assay, suggesting that Atg1 stimulates myosin activity via Sqa. Importantly, Atg1 phosphorylates and interacts with Sqa, indicating that Atg1–Sqa functions in a kinase cascade to regulate myosin II activation. Moreover, Atg1 has been found to have a critical role in the regulation of autophagy induction under stress conditions in yeast, Drosophila, and mammalian cells (Kamada et al, 2000; Scott et al, 2004; Hara et al, 2008). Our results provide the first evidence that nutrient starvation stimulates myosin II activation in an Atg1–Sqa-dependent manner. Most significantly, we found a dramatic decrease in the size and number of autophagosomes in cells expressing Sqa-T279A, Sqa-RNAi, and SqhA20A21 on nutrient deprivation, indicating that Atg1–Sqa-mediated actomyosin activation has a critical role in autophagy.
The kinase domain of Sqa is also highly homologous to that of the mammalian DAPK family proteins. Recent studies have indicated that DAPK1 regulates autophagy through its association with MAP1B and Beclin1, or by modulating the Tor signalling pathway (Harrison et al, 2008; Stevens et al, 2009; Zalckvar et al, 2009). As DAPK family proteins also regulate myosin II phosphorylation (Bialik et al, 2004), one might speculate that Sqa may be the Drosophila counterpart of DAPK protein. Indeed, although overexpression of Sqa does not induce cell death (H-W Tang, G-C Chen, unpublished data), Sqa shares several characteristics with DAPK3/ZIPK. First, unlike MLCK family proteins, both Sqa and ZIPK contain an amino-terminal kinase domain that has 42% sequence identity and 61% similarity. Moreover, like ZIPK, our recent sequence analysis from FlyBase identified a Sqa isoform (FBpp0289207) that also contains a leucine-zipper domain (G-C Chen, unpublished data). Second, as phosphorylation of Thr-265 in ZIPK is essential for its kinase activity (Sato et al, 2006), we found that Atg1 phosphorylates Sqa at the corresponding Thr-279, and is critical for Sqa activity. Third, just as Sqa specifically associates with kinase-inactive Atg1, our results indicate a similar interaction between ZIPK and Ulk1. Importantly, depletion of Sqa and ZIPK resulted in autophagic defects in response to nutrient deprivation. These findings together suggest that ZIPK may act as a mammalian homolog of Sqa during starvation-induced autophagy. Further investigation is needed to determine whether the mammalian Atg1 (Ulk1) directly phosphorylates ZIPK at Thr-265, and the role of this regulation in autophagy.
In autophagy, the source of the autophagosomal membrane and dynamics of autophagosome formation are fundamental questions. Studies in yeast and mammalian cells have identified several intracellular compartments as potential sources for the PAS (also termed isolation membrane/phagophore) (Tooze and Yoshimori, 2010). Axe et al (2008) reported on the formation of PI(3)P-enriched ER subdomains (omegasomes) during nutrient starvation and autophagy induction, and Hayashi-Nishino et al (2009) observed a direct connection between ER and the phagophore using the 3D electron tomography (Yla-Anttila et al, 2009). In addition, recent studies in yeast cells have suggested Atg9 and the Golgi complex have a role in the formation of autophagosomes (Geng et al, 2010; Ohashi and Munro, 2010; van der Vaart et al, 2010). It has been proposed that the integral membrane protein Atg9 may respond to the induction signal in promoting lipid transport to the forming autophagosomes (Mari and Reggiori, 2007; Webber et al, 2007). The mAtg9 has been found to localize on the TGN and the endosomes in nutrient-rich conditions and translocate to LC3-positive autophagosomes on nutrient deprivation (Young et al, 2006). Although several proteins, including Ulk1, mAtg13, and p38IP, have been found to regulate starvation-induced mAtg9 trafficking (Young et al, 2006; Chan et al, 2009; Webber and Tooze, 2010), the molecular motor that controls the movement of mAtg9 between different subcellular compartments remains unknown.
Our finding that myosin II redistributes from peripheral to the perinuclear region of cells on starvation suggests that myosin II has a role in membrane trafficking (Figure 7). In fact, it has been reported that myosin II is required for the trafficking of major histocompatibility complex (MHC) class II molecules and antigen presentation in B lymphocytes (Vascotto et al, 2007). Myosin II has also been found to be involved in the protein transport between ER and Golgi (Duran et al, 2003). As seen in Figure 8, there is a molecule link between mAtg9 and the actomyosin network, indicating that myosin II may function as a motor protein for mAtg9 trafficking during early autophagosome formation. In conclusion, our work has unravelled a regulatory mechanism between Atg1 activity and the Atg9-mediated formation of autophagosomes (Supplementary Figure S10). Further studies are needed to determine the involvement of this signalling process in other stress-induced or developmentally regulated autophagy.
Materials and methods
Drosophila strains and genetics
Flies were raised at 25°C following standard procedures unless otherwise noted. The following Drosophila strains were used: Atg1Δ3D, UAS-Atg7RNAi, UAS-Atg12RNAi, UAS-GFP-Atg8a, Cg-Gal4 (Scott et al, 2004, 2007), UAS-SqhA20A21, UAS-SqhE20E21 (Dorsten et al, 2007), UAS-SqhD20D21 (Mitonaka et al, 2007), UAS-Rheb (Saucedo et al, 2003), UAS-ZIP-DN (Escudero et al, 2007), and sqhAX3; P(w+ sqh-GFP) (Royou et al, 2002). The gal4 lines, act>CD2>gal4 UAS-GFP, and UAS-p35 were obtained from the Bloomington Stock Center. UAS-Atg1, UAS-Atg1-KR, and UAS-Atg1RNAi were generated in our laboratory as previously described (Chen et al, 2008). Viability assays were performed as described previously (Kim et al, 2008).
Plasmids
Plasmid constructions and the details of various shRNAs are included in the Supplementary data. GFP–NMHC-IIA and RFP-LC3 were obtained from Addgene. GFP–LC3 was a gift from T Yoshimori (Kabeya et al, 2000). ZIPK cDNA was a gift from T Haystead (Graves et al, 2005). Flag–Ulk1 and Flag–Ulk1-KI were gifts from J Chung (Lee et al, 2007). mAtg9 cDNA was a gift from W-Y Yang (Academia Sinica).
Cell culture and transfection
HEK293T and MCF7 cells were cultured at 37°C in DMEM (Invitrogen) medium supplemented with 10% FBS (complete medium). For nutrient starvation, cells were starved for 2 h in serum-free Earle's balanced salt solution (EBSS) medium (Sigma) plus 100 nM BafA1 (Merck) where indicated. Lipofectamine 2000 reagent (Invitrogen) and PolyJet reagent (SignaGen) were used for transfection.
Antibodies and reagents
Antibodies used for the study were: anti-cleaved Caspase-3 (Cell Signaling), anti-Sqh (gift from Tien Hsu, Boston University), anti-pSer19-MRLC (Cell Signaling), anti-MRLC (Abcam), anti-NMHC-IIA (Cell Signaling), anti-LC3 (Novus), anti-Ulk1 (Novus), anti-ZIPK (Cell Signaling), anti-TGN46 (Serotec), anti-Atg16L (MBL), anti-LAMP1 (Abcam), and anti-GM130 (BD Biosciences). F-actin was stained using TRITC-labelled phalloidin (Sigma). DAPI (1 μg/ml) was used to stain nuclei.
Immunofluorescence
GFP-marked overexpression clones in the larval fat body were generated through heat shock-independent induction as previously described (Scott et al, 2007). MCF7 cells stably expressing GFP–LC3 or GFP–mAtg9 were fixed with 4% paraformaldehyde and then permeabilized with 0.1% triton. Cells were then processed for immunostaining. Samples were examined using an epifluorescent microscope (Olympus BX51) equipped with a × 100 objective lens or a confocal laser scanning microscope (LSM510; Carl Zeiss Inc.) equipped with a × 63 Plan-Apochromat (NA1.4) objective lens.
Immunoblotting
Larvae carrying transgenes under the control of hs–GAL4 driver were collected 72–96 h after the eggs were laid, subjected to heat shock for 2 h at 37°C, followed by 24 h recovery at room temperature, and cultured in fresh fly media supplemented with yeast paste (fed) or in vials containing 20% sucrose (starved) for 4 h before dissection. After dissection, the samples were then boiled in SDS sample buffer, run on 15% polyacrylamide gel, and then transferred to Immobilon-P polyvinylidene fluoride (PVDF) membrane (Millipore). The blot was probed with primary antibody, followed by HRP-conjugated secondary antibody, and signal was detected by enhanced chemiluminescence (ECL; Amersham). The signal intensities were analysed using ImageJ software (NIH).
In vitro kinase assay
In vitro kinase assay was performed as previously described (Chen et al, 2008, and detailed protocol in the Supplementary data).
Subcellular fractionation
Membrane association of myosin II was analysed according to a previous reference (Webber and Tooze, 2010).
Supplementary Material
Acknowledgments
We thank T Neufeld, R-H Chen, J Chung, B Edgar, M Freeman, T Haystead, T Hsu, R Karess, Y Nishida, M VanBerkum, T Yoshimori, W-Y Yang, the National RNAi Core Facility and the Bloomington Stock Center for reagents. We are grateful to Drs R-H Chen, H-J Kung and J Settleman for helpful discussions and comments on the manuscript, and J Steed for English editing. Special thanks to C-C Hung for confocal assistance and NRPGM Core Facilities for Proteomics and Glycomics for mass spectrometry analyses. This work was supported by the National Science Council of Taiwan (NSC96-2311-B-001-033-MY3) and Academia Sinica Thematic Research Program.
Author contributions: HT and GC carried out the Drosophila experiments, HT, YW, SW, and MW carried out the mammalian experiments, and SL contributed to the MS work. GC and HT designed the experiments and wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aplin A, Jasionowski T, Tuttle DL, Lenk SE, Dunn WA Jr (1992) Cytoskeletal elements are required for the formation and maturation of autophagic vacuoles. J Cell Physiol 152: 458–466 [DOI] [PubMed] [Google Scholar]
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT (2008) Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 182: 685–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialik S, Bresnick AR, Kimchi A (2004) DAP-kinase-mediated morphological changes are localization dependent and involve myosin-II phosphorylation. Cell Death Differ 11: 631–644 [DOI] [PubMed] [Google Scholar]
- Chan EY, Longatti A, McKnight NC, Tooze SA (2009) Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domain using an Atg13-independent mechanism. Mol Cell Biol 29: 157–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GC, Lee JY, Tang HW, Debnath J, Thomas SM, Settleman J (2008) Genetic interactions between Drosophila melanogaster Atg1 and paxillin reveal a role for paxillin in autophagosome formation. Autophagy 4: 37–45 [DOI] [PubMed] [Google Scholar]
- Deminoff SJ, Howard SC, Hester A, Warner S, Herman PK (2006) Using substrate-binding variants of the cAMP-dependent protein kinase to identify novel targets and a kinase domain important for substrate interactions in Saccharomyces cerevisiae. Genetics 173: 1909–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePina AS, Wollert T, Langford GM (2007) Membrane associated nonmuscle myosin II functions as a motor for actin-based vesicle transport in clam oocyte extracts. Cell Motil Cytoskeleton 64: 739–755 [DOI] [PubMed] [Google Scholar]
- Dorsten JN, Kolodziej PA, VanBerkum MF (2007) Frazzled regulation of myosin II activity in the Drosophila embryonic CNS. Dev Biol 308: 120–132 [DOI] [PubMed] [Google Scholar]
- Duran JM, Valderrama F, Castel S, Magdalena J, Tomas M, Hosoya H, Renau-Piqueras J, Malhotra V, Egea G (2003) Myosin motors and not actin comets are mediators of the actin-based Golgi-to-endoplasmic reticulum protein transport. Mol Biol Cell 14: 445–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escudero LM, Bischoff M, Freeman M (2007) Myosin II regulates complex cellular arrangement and epithelial architecture in Drosophila. Dev Cell 13: 717–729 [DOI] [PubMed] [Google Scholar]
- Findlay GM, Yan L, Procter J, Mieulet V, Lamb RF (2007) A MAP4 kinase related to Ste20 is a nutrient-sensitive regulator of mTOR signalling. Biochem J 403: 13–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukata Y, Amano M, Kaibuchi K (2001) Rho-Rho-kinase pathway in smooth muscle contraction and cytoskeletal reorganization of non-muscle cells. Trends Pharmacol Sci 22: 32–39 [DOI] [PubMed] [Google Scholar]
- Gallagher PJ, Herring BP, Stull JT (1997) Myosin light chain kinases. J Muscle Res Cell Motil 18: 1–16 [DOI] [PubMed] [Google Scholar]
- Geng J, Nair U, Yasumura-Yorimitsu K, Klionsky DJ (2010) Post-golgi sec proteins are required for autophagy in Saccharomyces cerevisiae. Mol Biol Cell 21: 2257–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves PR, Winkfield KM, Haystead TA (2005) Regulation of zipper-interacting protein kinase activity in vitro and in vivo by multisite phosphorylation. J Biol Chem 280: 9363–9374 [DOI] [PubMed] [Google Scholar]
- Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N (2008) FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol 181: 497–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison B, Kraus M, Burch L, Stevens C, Craig A, Gordon-Weeks P, Hupp TR (2008) DAPK-1 binding to a linear peptide motif in MAP1B stimulates autophagy and membrane blebbing. J Biol Chem 283: 9999–10014 [DOI] [PubMed] [Google Scholar]
- Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A (2009) A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol 11: 1433–1437 [DOI] [PubMed] [Google Scholar]
- He C, Song H, Yorimitsu T, Monastyrska I, Yen WL, Legakis JE, Klionsky DJ (2006) Recruitment of Atg9 to the preautophagosomal structure by Atg11 is essential for selective autophagy in budding yeast. J Cell Biol 175: 925–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, Guan JL, Oshiro N, Mizushima N (2009a) Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell 20: 1981–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, Sasaki T, Iemura S, Natsume T, Hara T, Mizushima N (2009b) Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy 5: 973–979 [DOI] [PubMed] [Google Scholar]
- Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D (1997) The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurons and glial cells. Development 124: 761–771 [DOI] [PubMed] [Google Scholar]
- Jordan P, Karess R (1997) Myosin light chain-activating phosphorylation sites are required for oogenesis in Drosophila. J Cell Biol 139: 1805–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhasz G, Erdi B, Sass M, Neufeld TP (2007) Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev 21: 3061–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH (2009) ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell 20: 1992–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Kamada Y, Baba M, Takikawa H, Sasaki M, Ohsumi Y (2005) Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol Biol Cell 16: 2544–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19: 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y (2000) Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol 150: 1507–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata T, Kamada Y, Kabeya Y, Sekito T, Ohsumi Y (2008) Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol Biol Cell 19: 2039–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL (2008) Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol 10: 935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ (2007) Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol 8: 931–937 [DOI] [PubMed] [Google Scholar]
- Kuroyanagi H, Yan J, Seki N, Yamanouchi Y, Suzuki Y, Takano T, Muramatsu M, Shirasawa T (1998) Human ULK1, a novel serine/threonine kinase related to UNC-51 kinase of Caenorhabditis elegans: cDNA cloning, expression, and chromosomal assignment. Genomics 51: 76–85 [DOI] [PubMed] [Google Scholar]
- Lanzetti L (2007) Actin in membrane trafficking. Curr Opin Cell Biol 19: 453–458 [DOI] [PubMed] [Google Scholar]
- Lee SB, Kim S, Lee J, Park J, Lee G, Kim Y, Kim JM, Chung J (2007) ATG1, an autophagy regulator, inhibits cell growth by negatively regulating S6 kinase. EMBO Rep 8: 360–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning BD, Cantley LC (2002) Hitting the target: emerging technologies in the search for kinase substrates. Sci STKE 2002: pe49. [DOI] [PubMed] [Google Scholar]
- Mari M, Reggiori F (2007) Atg9 trafficking in the yeast Saccharomyces cerevisiae. Autophagy 3: 145–148 [DOI] [PubMed] [Google Scholar]
- Matsumura F, Ono S, Yamakita Y, Totsukawa G, Yamashiro S (1998) Specific localization of serine 19 phosphorylated myosin II during cell locomotion and mitosis of cultured cells. J Cell Biol 140: 119–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitonaka T, Muramatsu Y, Sugiyama S, Mizuno T, Nishida Y (2007) Essential roles of myosin phosphatase in the maintenance of epithelial cell integrity of Drosophila imaginal disc cells. Dev Biol 309: 78–86 [DOI] [PubMed] [Google Scholar]
- Mizushima N (2007) Autophagy: process and function. Genes Dev 21: 2861–2873 [DOI] [PubMed] [Google Scholar]
- Monastyrska I, He C, Geng J, Hoppe AD, Li Z, Klionsky DJ (2008) Arp2 links autophagic machinery with the actin cytoskeleton. Mol Biol Cell 19: 1962–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi Y, Munro S (2010) Membrane delivery to the yeast autophagosome from the Golgi-endosomal system. Mol Biol Cell 21: 3998–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Klionsky DJ (2005) Autophagosomes: biogenesis from scratch? Curr Opin Cell Biol 17: 415–422 [DOI] [PubMed] [Google Scholar]
- Reggiori F, Monastyrska I, Shintani T, Klionsky DJ (2005) The actin cytoskeleton is required for selective types of autophagy, but not nonspecific autophagy, in the yeast Saccharomyces cerevisiae. Mol Biol Cell 16: 5843–5856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ (2004) The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell 6: 79–90 [DOI] [PubMed] [Google Scholar]
- Royou A, Sullivan W, Karess R (2002) Cortical recruitment of nonmuscle myosin II∣ in early syncytial Drosophila embryos: its role in nuclear axial expansion and its regulation by Cdc2 activity. J Cell Biol 158: 127–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, Kamada N, Muromoto R, Kawai T, Sugiyama K, Watanabe T, Imoto S, Sekine Y, Ohbayashi N, Ishida M, Akira S, Matsuda T (2006) Phosphorylation of threonine-265 in Zipper-interacting protein kinase plays an important role in its activity and is induced by IL-6 family cytokines. Immunol Lett 103: 127–134 [DOI] [PubMed] [Google Scholar]
- Saucedo LJ, Gao X, Chiarelli DA, Li L, Pan D, Edgar BA (2003) Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat Cell Biol 5: 566–571 [DOI] [PubMed] [Google Scholar]
- Scott RC, Juhasz G, Neufeld TP (2007) Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr Biol 17: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RC, Schuldiner O, Neufeld TP (2004) Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell 7: 167–178 [DOI] [PubMed] [Google Scholar]
- Seglen PO, Berg TO, Blankson H, Fengsrud M, Holen I, Stromhaug PE (1996) Structural aspects of autophagy. Adv Exp Med Biol 389: 103–111 [DOI] [PubMed] [Google Scholar]
- Sellers JR (2000) Myosins: a diverse superfamily. Biochim Biophys Acta 1496: 3–22 [DOI] [PubMed] [Google Scholar]
- Shani G, Marash L, Gozuacik D, Bialik S, Teitelbaum L, Shohat G, Kimchi A (2004) Death-associated protein kinase phosphorylates ZIP kinase, forming a unique kinase hierarchy to activate its cell death functions. Mol Cell Biol 24: 8611–8626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens C, Lin Y, Harrison B, Burch L, Ridgway RA, Sansom O, Hupp T (2009) Peptide combinatorial libraries identify TSC2 as a death-associated protein kinase (DAPK) death domain-binding protein and reveal a stimulatory role for DAPK in mTORC1 signaling. J Biol Chem 284: 334–344 [DOI] [PubMed] [Google Scholar]
- Stull JT, Tansey MG, Tang DC, Word RA, Kamm KE (1993) Phosphorylation of myosin light chain kinase: a cellular mechanism for Ca2+ desensitization. Mol Cell Biochem 127–128: 229–237 [DOI] [PubMed] [Google Scholar]
- Suzuki K, Ohsumi Y (2007) Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett 581: 2156–2161 [DOI] [PubMed] [Google Scholar]
- Tooze SA, Yoshimori T (2010) The origin of the autophagosomal membrane. Nat Cell Biol 12: 831–835 [DOI] [PubMed] [Google Scholar]
- van der Vaart A, Griffith J, Reggiori F (2010) Exit from the golgi is required for the expansion of the autophagosomal phagophore in yeast Saccharomyces cerevisiae. Mol Biol Cell 21: 2270–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vascotto F, Lankar D, Faure-Andre G, Vargas P, Diaz J, Le Roux D, Yuseff MI, Sibarita JB, Boes M, Raposo G, Mougneau E, Glaichenhaus N, Bonnerot C, Manoury B, Lennon-Dumenil AM (2007) The actin-based motor protein myosin II regulates MHC class II trafficking and BCR-driven antigen presentation. J Cell Biol 176: 1007–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR (2009) Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol 10: 778–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber JL, Tooze SA (2010) Coordinated regulation of autophagy by p38alpha MAPK through mAtg9 and p38IP. EMBO J 29: 27–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber JL, Young AR, Tooze SA (2007) Atg9 trafficking in Mammalian cells. Autophagy 3: 54–56 [DOI] [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ (2007) Autophagosome formation: core machinery and adaptations. Nat Cell Biol 9: 1102–1109 [DOI] [PubMed] [Google Scholar]
- Yan J, Kuroyanagi H, Tomemori T, Okazaki N, Asato K, Matsuda Y, Suzuki Y, Ohshima Y, Mitani S, Masuho Y, Shirasawa T, Muramatsu M (1999) Mouse ULK2, a novel member of the UNC-51-like protein kinases: unique features of functional domains. Oncogene 18: 5850–5859 [DOI] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ (2010) Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol 22: 124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yla-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL (2009) 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy 5: 1180–1185 [DOI] [PubMed] [Google Scholar]
- Yorimitsu T, Klionsky DJ (2005) Autophagy: molecular machinery for self-eating. Cell Death Differ 12 (Suppl 2): 1542–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AR, Chan EY, Hu XW, Kochl R, Crawshaw SG, High S, Hailey DW, Lippincott-Schwartz J, Tooze SA (2006) Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci 119: 3888–3900 [DOI] [PubMed] [Google Scholar]
- Zalckvar E, Berissi H, Mizrachy L, Idelchuk Y, Koren I, Eisenstein M, Sabanay H, Pinkas-Kramarski R, Kimchi A (2009) DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep 10: 285–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.