Retinoblastoma protein is essential for early meiotic events in Arabidopsis
The tumour suppressor retinoblastoma, Rb, is a key cell-cycle regulator, but has so far not been implicated in meiosis. The findings show here that Rb is important for meiosis in Arabidopsis, and raise the possibility that this function might be conserved in other multicellular organisms.
Keywords: Arabidopsis, meiosis, meiotic recombination, retinoblastoma
Abstract
We have analysed the role of RBR (retinoblastoma related), the Arabidopsis homologue of the tumour suppressor Retinoblastoma protein (pRb), during meiosis. We characterise the rbr-2 mutation, which causes a loss of RBR in male meiocytes. The rbr-2 plants exhibit strongly reduced fertility, while vegetative growth is generally unaffected. The reduced fertility is due to a meiotic defect that results in reduced chiasma formation and subsequent errors in chromosome disjunction. Immunolocalisation studies in wild-type meiocytes reveal that RBR is recruited as foci to the chromosomes during early prophase I in a DNA double-strand-break-dependent manner. In the absence of RBR, expression of several meiotic genes is reduced. The localisation of the recombinases AtRAD51 and AtDMC1 is normal. However, localisation of the MutS homologue AtMSH4 is compromised. Additionally, polymerisation of the synaptonemal complex protein AtZYP1 is abnormal. Together, these data indicate that loss of RBR during meiosis results in a reduction of crossover formation and an associated failure in chromosome synapsis. Our results indicate that RBR has an important role in meiosis affecting different aspects of this complex process.
Introduction
Meiosis is a specialised cell division in which a single round of DNA replication is followed by two rounds of nuclear division to produce four haploid products. In most species, recombination between homologous pairs of chromosomes is an essential feature of meiosis. Recombination during prophase I of meiosis results in genetic crossover (CO) formation, which establishes physical links between the homologues that are cytologically visible as chiasmata (Jones, 1984; Jones and Franklin, 2006). The chiasmata, in conjunction with sister chromatid cohesion, which is established during or soon after S phase, facilitates accurate chromosome disjunction at the metaphase I-to-anaphase I transition. At this first division, sister chromatid cohesion is lost along the chromosome arms, but is maintained at the centromeric region. This allows segregation of the homologues, but maintains the inter-sister chromatid links until the second meiotic division.
In budding yeast, recombination is initiated by the formation of DNA double-strand breaks (DSBs) catalysed by the topoisomerase-related protein Spo11 (Keeney et al, 1997). The DSBs are then resected to generate 3′-single-stranded tails that interact with the RecA homologues Rad51 and Dmc1 to form nucleoprotein filaments. The filament on one side of the break then invades the homologous duplex DNA of one of the two non-sister chromatids to form a stable single-end invasion intermediate. The displaced DNA strand forms a D-loop that extends as the invading strand is extended. This enables the capture of the 3′-end on the other side of the break. Subsequent ligation of the broken DNA strands results in the formation of the double Holliday junction (dHj) recombination intermediate that is then resolved to form a CO (reviewed in Neale and Keeney, 2006). Studies suggest that meiotic recombination in Arabidopsis shares many features in common with that of budding yeast and mammals (Hamant et al, 2006), although some differences have emerged during evolution. For instance, DSB formation in Arabidopsis involves two Spo11 paralogues, AtSPO11-1 and AtSPO11-2, rather than a single Spo11 protein in yeast (Grelon et al, 2001; Stacey et al, 2006).
A prominent feature of meiosis is that the chromosomes undergo spatial and structural reorganisation that are dependent on the recombination machinery. At leptotene, the sister chromatids form looped chromatin arrays that are associated by a closely apposed linear protein axis that is elaborated along the base of each looped array. Recombination complexes assemble at Spo11-dependent DSBs that form at sites within chromatin loops that have become spatially associated with the chromosome axes (reviewed in Zickler and Kleckner, 1999). In most organisms, this enables alignment of the homologous chromosomes and requires the activity of the strand-exchange proteins Rad51 and Dmc1 that are recruited to the break site following strand resection. The mechanism of chromosome alignment is likely the same in Arabidopsis, as loss of AtDMC1 results in asynapsis, such that only univalent chromosomes are observed at metaphase I (Doutriaux et al, 1998; Sanchez-Moran et al, 2007). During zygotene, assembly of the synaptonemal complex (SC) occurs. This is a tripartite proteinaceous structure that intimately links the homologous chromosomes along their entire length (Page and Hawley, 2004). Completion of SC formation denotes the onset of pachytene, during which recombination is completed. At the end of pachytene, the SC breaks down and the chromosomes condense during diplotene/diakinesis to reach a maximum condensation at metaphase I before the first meiotic division. This is followed by the second meiotic division, leading to the formation of a tetrad containing four haploid spores. Each spore undergoes two mitotic divisions, leading to the development of mature pollen.
Although meiosis is not a cell cycle, coordination and progression through meiosis requires many of the components of the mitotic cell cycle (Marston and Amon, 2004). Cyclin-dependent kinases (CDKs) have been extensively studied in the context of the mitotic cell cycle in yeast, mammals and plants. They are also required during meiosis (Kishimoto, 2003; McKim and Hawley, 1995). Studies in budding yeast have shown that the CDK, Cdc28, forms a complex with B-type cyclins to promote chromosome replication before the onset of meiosis and has a role in the formation of Spo11-dependent DSBs (Stuart and Wittenberg, 1998; Henderson et al, 2006). Cdc28 has also been shown to be required for normal SC formation, indicating that it has an important role in meiotic chromosome morphogenesis (Zhu et al, 2010). A cdk2 knockout mouse was found to be defective in SC formation, indicating that CDKs are also important during meiosis in multicellular organisms (Ortega et al, 2003). Relatively little is known about the meiotic role of cell-cycle regulators in plants. SOLO DANCERS (SDS) is thought to encode a plant-specific cyclin that is required for chromosome synapsis and recombination during prophase I in Arabidopsis (Wang et al, 2004). A second cyclin, CYCLIN A1;2, encoded by TARDY ASYNCHRONOUS MEIOSIS (TAM) is required for timely and synchronous meiotic progression (Magnard et al, 2001). A weak allele of the CDKA (CDKA;1), which is the Arabidopsis homologue of Cdk2/Cdc28, was shown to affect meiosis (Dissmeyer et al, 2007). Plants carrying the cdka;1 allele appeared to initiate meiosis normally, but cytokinesis occurred after the first meiotic division rather than after the second division. SMG7, an Arabidopsis protein that is thought to have a role in nonsense-mediated RNA decay, is proposed to promote exit from meiosis by regulating CDK activity (Riehs et al, 2008).
The human tumour suppressor gene retinoblastoma (Rb) is an important target for CDK phosphorylation during the cell cycle, wherein it regulates the G1/S transition by acting as a repressor of E2F transcription factors (E2Fs; Friend et al, 1986). Activated CDK–cyclin complexes phosphorylate Retinoblastoma protein (pRb) at a later stage of G1 phase causing the protein to lose its binding affinity for E2Fs (Weinberg, 1995). E2Fs then regulate the expression of a variety of genes required for cell-cycle progression and cell differentiation (Dimova and Dyson, 2005). The Rb-E2F pathway is also conserved in plants (Shen, 2002).
In addition to its key cell-cycle regulatory function, pRb also recruits chromatin remodelling factors that exert a broad range of cellular functions distinct from cell-cycle control (Brehm and Kouzarides, 1999), including cell fate regulation (Macaluso et al, 2006), senescence (Funayama and Ishikawa, 2007) and apoptosis (Harbour and Dean, 2000). pRb also interacts directly with structural chromatin components such as condensins (Longworth et al, 2008) and origin of replication proteins (Ahlander et al, 2008). In mice, knockout of Rb function causes excessive proliferation and abnormal placental development, leading to embryonic lethality (Clarke et al, 1992; Jacks et al, 1992; Lee et al, 1992; Wu et al, 2003).
The Arabidopsis genome contains a single Rb-related gene (RBR; Ebel et al., 2004). The loss of function of RBR prevents proper gametogenesis, leading to extremely reduced transmission of rbr alleles. RBR is also expressed in adult cells; however, analysis of this role has been restricted because of the profound effect that its loss has on embryogenesis. RNA interference using a root-specific promoter allowed downregulation of RBR in Arabidopsis roots, revealing that the protein regulates root stem-cell maintenance (Wildwater et al, 2005). Ectopic expression and repression of RBR transcription in somatic tissues have enabled further progress towards gaining a fuller understanding of the role of the protein during post-embryonic development (Park et al, 2005; Desvoyes et al, 2006; Wyrzykowska et al, 2006; Borghi et al, 2010).
Thus far, a role for pRb or RBR in meiosis has not been established. The animal germ line is difficult to manipulate. However, establishment of the germ line in plants takes place during post-embryonic development and is, therefore, amenable to genetic dissection (Berger et al, 2008). The diploid vegetative phase dominates the life cycle until flowers are produced. Flowers bear sexual organs in which meiosis takes place, and produces haploid structures in which gametes differentiate (Berger et al, 2008; Drews and Yadegari, 2002). Thus, despite the fact that loss of RBR is gametophyte lethal, it has been possible to investigate the role of RBR during both male and female gametogenesis by taking advantage of the fact that half the gametes produced by a heterozygous rbr mutant possess a wild-type copy of the gene and the remainder carry a defective allele (Ingouff et al, 2006; Johnston et al, 2008, Chen et al, 2009). As a result, it has been established that the primary defect in both male and female gametogenesis is a failure in cell-cycle regulation, leading to overproliferation of the cells within the developing gametophyte. Consequently, genetic transmission of mutant rbr alleles is completely prevented on the female side and strongly reduced on the male side.
In this study, we have been able to take advantage of this reproductive programme to determine the potential role of RBR in meiosis. We provide evidence that clearly indicates a crucial role for RBR during prophase I of meiosis. Our studies reveal that loss of RBR results in reduced CO formation and aberrant chromosome synapsis, leading to chromosome mis-segregation and gamete lethality.
Results
The rbr-2 allele results in aberrant RBR mRNA splicing and is associated with reduced fertility
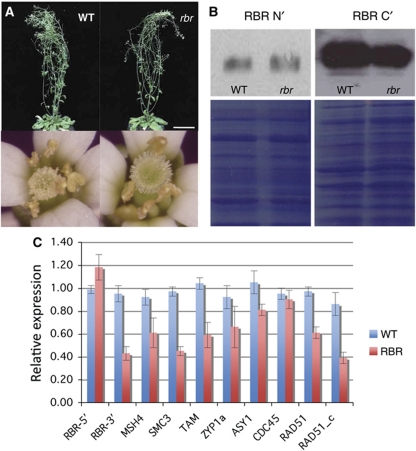
As loss of RBR results in cellular proliferation in the male gametophyte, it was postulated that it would be possible to rescue paternal transmission of an rbr mutant in a heterozygous background by downregulation of CDKA;1, as this would be expected to restore cellular proliferation in the rbr pollen to the wild-type level (Chen et al, 2009). When this hypothesis was tested in a rbr-2/+; cdka;1/+ background, the proportion of viable pollen increased in line with expectation (Chen et al, 2009). Subsequently, we found that maternal transmission of rbr-2 was also rescued by cdka;1 (Table I). However, an unanticipated outcome to these experiments was the finding that a quarter of the progeny from rbr-2/+; cdka;1/+ self-fertilised plants were rbr-2 homozygous plants. This indicated that rbr-2 homozygous seeds survived, irrespective of the presence of CDKA;1 (Figure 1A). This rescue was not observed with the allele rbr-3 (Table I), which suggested special properties associated with the rbr-2 allele. Although, the vegetative growth of rbr-2 homozygote plants revealed some differences to that of wild-type plants, such as an increase in hypocotyl length and a slight elevation in ploidy level, no dramatic effects were apparent (Figure 1A, Supplementary Figure S1A and B). This was surprising because earlier reports had shown that partial loss of RBR causes hyperproliferation and alters differentiation in various vegetative cell types (Ingouff et al, 2006; Park et al, 2005; Rossi et al, 2003; Sabelli et al, 2005; Wildwater et al, 2005; Wyrzykowska et al, 2006). However, the rbr-2 plants exhibited a large reduction in fertility (Figure 1A), producing approximately 1–5 seeds per silique, compared with wild-type plants, which produce between 50 and 60 seeds per silique.
Table 1. Genetic rescue of rbr-2 transmission by cdka;1.
| Female × male | F1 progeny genotype (%) | n | ||
|---|---|---|---|---|
| Wild type | rbr-2/+ | rbr-2/rbr-2 | ||
| rbr-2/+ × Col | 100 | 0 | NA | 268 |
| rbr-2/+; cdka;1/+ × Col | 51.2 | 48.8 | NA | 342 |
| Col × rbr-2/+ | 92.0 | 8.0 | NA | 637 |
| Col × rbr-2/+; cdka;1/+ | 53.0 | 47.0 | NA | 430 |
| rbr-2/+ × rbr-2/+ | 79.1 | 20.9 | 0 | 1212 |
| rbr-2/+; cdka;1/+ × rbr-2/+; cdka;1/+ | 29.2 | 48.6 | 22.2 | 370 |
| Wild type | rbr-3/+ | rbr-3/rbr-3 | ||
| rbr-3/+ × Col | 100 | 0 | NA | 190 |
| rbr-3/+; cdka;1/+ × Col | 100 | 0 | NA | 187 |
| Col × rbr-3/+ | 94.3 | 5.7 | NA | 280 |
| Col × rbr-3/+; cdka;1/+ | 93.9 | 6.1 | NA | 278 |
| rbr-3/+ × rbr-3/+ | 93.7 | 6.3 | 0 | 380 |
| rbr-3/+; cdka;1/+ × rbr-3/+; cdka;1/+ | 93.9 | 6.1 | 0 | 374 |
Figure 1.
RBR expression in vegetative tissues and in meiocytes of rbr-2 plants. (A) Morphological features of wild-type and homozygous rbr-2 plants. The vegetative development is similar, but rbr-2 plant produce flowers that bear reduced amount of pollen. As a consequence, very few pollen grains are deposited on the stigma causing sterility. (B) Western blots for detection of RBR proteins in leaves produced by wild-type and rbr-2 plants using anti-RBR-C and anti-RBR-N antibodies. The loading controls are shown under the western blots. (C) Expression analysis in buds containing meiocytes of genes encoding meiotic proteins and RBR transcripts representing the wild-type unspliced (RBR-3′) and both the wild-type and spliced transcripts (RBR-5′) produced by rbr-2. Scale bar, 8 cm and 1 mm (inset).
Molecular analysis of rbr-2 revealed the presence of two RBR transcripts in vegetative tissues (Supplementary Figure S2A). Expression of the full-length RBR product was nearly normal (Supplementary Figure S1C). However, it was reduced in bud tissues, which comprised both vegetative somatic tissues and meiocytes (Figure 1C). DNA sequencing indicated that the novel shorter form resulted from an aberrant splicing event caused by the presence of the T-DNA insert in an intron in the rbr-2 allele (Supplementary Figure S2B). The T-DNA insert could also be spliced out with intron 12 to generate a transcript identical to the wild-type RBR transcript. The alternate form removed exon 12 and the remaining five base pairs from intron 13 (5′-ATAAG-3′), resulting in a truncated RBR transcript. The shorter transcript was predicted to produce a truncated RBR protein (RBR_m) devoid of the B-box (Supplementary Figure S2C), which is essential for RBR function (Brehm et al, 1998).
Expression of RBR protein in wild-type and rbr-2 meiocytes during prophase I
The reduced fertility observed in rbr-2 was consistent with an effect on meiosis. We investigated further the intracellular distribution of RBR during meiosis. To selectively detect the full-length RBR protein, we raised an anti-RBR peptide antibody (Ab), against a sequence within the RBR C-terminal region (RBR-C; see Materials and methods) that was predicted to be absent in the product of the shorter transcript. We also used an Ab (designated as anti-RBR-N) raised against the amino-terminal region (amino acids 1–374; Borghi et al, 2010) to detect the expression of the predicted truncated RBR protein together with the full-length RBR protein. We detected a single band of ∼125 kDa when protein blots from leaf tissues were probed with anti-RBR-N and anti-RBR-C antibodies (Figure 1B). Although low amounts of mutant-spliced RBR transcripts were detected in leaf tissues (supplementary Figure 2A), the anti-RBR-N Ab did not detect a band corresponding to the truncated RBR protein with the expected lower molecular weight. This suggests that either the amount of truncated protein in leaf tissue is below the detection threshold or that this truncated protein is degraded. It would appear that wild-type RBR and the truncated product are both expressed in vegetative cells with no major development effects (Supplementary Figure S1). In addition, when the truncated transcript was expressed under the RBR promoter in transgenic wild-type lines, their vegetative phenotype was indistinguishable from that of the rbr-2 mutant. In contrast to rbr-2, the transgenic plants expressing RBR_m appeared fully fertile (three transgenic lines observed and n=10 siliques per line). This suggested that the truncated RBR_m protein encoded by the rbr-2 allele did not negatively compete with RBR when co-expressed in a wild-type background. In summary, vegetative tissues of rbr-2 plants expressed levels of wild-type RBR that did not affect development.
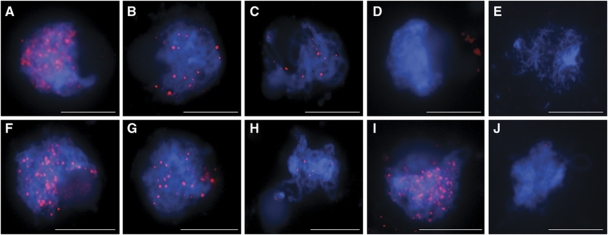
In buds containing meiocytes from rbr-2 plants (400–800 μm in length), we detected a lower amount of unspliced wild-type RBR transcript than in wild-type buds (Figure 1C). However, most cells within a developing bud are vegetative in nature and only a small proportion are reproductive. Thus, any differences in RBR expression in reproductive cells would be potentially masked. It is also possible that RBR expression levels in wild-type meiocytes is relatively much higher than in surrounding wild-type somatic cells, which would explain the unexpected high impact of rbr-2 on the level of wild-type RBR transcript. To assess and compare the distribution of RBR protein during meiosis in wild type and rbr-2, immunolocalisation studies were conducted on chromosome-spread preparations from pollen mother cells (PMCs) during prophase I using anti-RBR-C antibodies (Figure 2A–E). In wild-type meiocytes, the RBR protein was found to localise along the chromosome axes forming numerous foci (96±7, n=5) during late G2/early leptotene (Figure 2A). As prophase I progressed, the number of RBR foci decreased, with 38 (±2, n=5) foci detectable at zygotene, which was reduced to 18 (±2, n=5) foci at pachytene (Figure 2B and C). The RBR protein was not detected at later stages (data not shown). When chromosome-spread preparations from rbr-2 meiocytes were analysed, we could not detect RBR protein (Figure 2D). In contrast, immunolocalisation of RBR on chromosome-spread preparations from wild-type meiocytes using the anti-RBR-N Ab was indistinguishable from that using the RBR-C Ab (Figure 2F–H). When the anti-RBR-N Ab was applied to chromosome spreads from rbr-2 meiocytes, numerous chromosome-associated foci were detected at early leptotene, and their number reduced as prophase I progressed (Figure 2I). As these foci were not detected with the RBR-C Ab, it seems likely that they correspond to the predicted truncated RBR_m protein encoded by the spliced RBR transcript. In conclusion, in rbr-2 meiocytes, the truncated RBR_m is by far the major protein encoded by RBR, suggesting that rbr-2 meiocytes essentially produce the spliced RBR transcript. As reported above, the expression of truncated RBR_m under the control of pRBR promoter does not cause any phenotype change, suggesting that the absence of wild-type RBR protein is the basis for the reduced fertility in rbr-2 plants.
Figure 2.
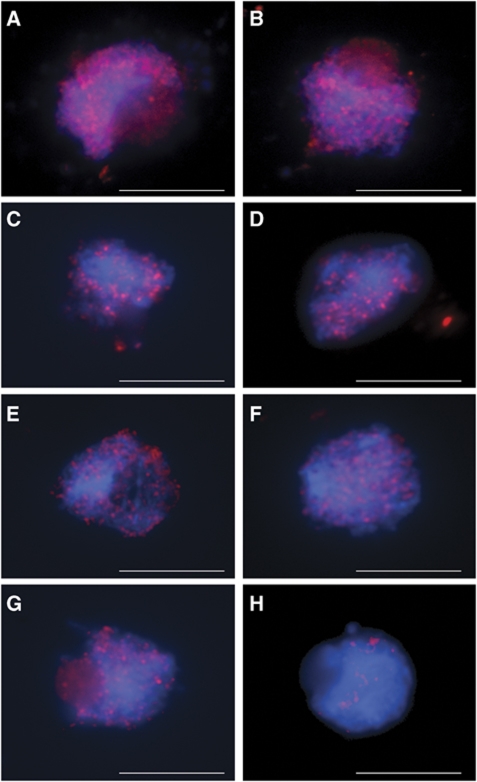
Immunolocalisation of RBR and RBR_m in prophase I nuclei. (A–C) Immunolocalisation of RBR in wild-type nuclei with anti-RBR C-terminal Ab (RBR-C). (F–H) Immunolocalisation of RBR in wild-type nuclei with anti-RBR N-terminal Ab (RBR-N). (A, F) Leptotene, (B, G) zygotene, (C, H) pachytene; (D) RBR is not detected in rbr-2 plants by anti-RBR-C Ab; (I) detection of RBR_m in rbr-2 plants using anti-RBR-N Ab; (E, J) RBR foci are absent in Atspo11-1-4 meiocytes, as determined using both anti-RBR-C Ab (E) and anti-RBR-N Ab (J). Scale bar, 10 μm.
Loss of RBR reduces meiotic gene expression
RBR controls transcription, at least in part, by inhibiting E2Fs (Macaluso et al, 2006). A large number of Arabidopsis genes involved in meiosis contain the consensus E2F-binding sites and might be E2F regulated (Supplementary Table S1). Hence, given the lack of wild-type RBR expression in rbr-2 meiocytes and reduced fertility, we investigated whether loss of protein affected meiotic gene expression in buds (approximately 400–800 μm in length). In rbr-2 buds containing meiocytes, the expression of genes important for meiotic events during prophase I, including AtMSH4 (Higgins et al, 2004), STRUCTURAL MAINTENANCE OF CHROMOSOMES 3 (SMC3; Liu et al, 2002), TAM (Magnard et al, 2001) and AtZYP1a (Higgins et al, 2005) was reduced markedly (Figure 1C). Although the presence of predicted E2F-binding sites in the promoters of several of these genes (Supplementary Table S1) suggested that RBR may act directly on E2F to regulate the expression of key meiotic proteins, it was also possible that the absence of RBR in meiocytes had indirect effects on transcription of meiotic genes. This was suggested by the fact that the loss of RBR does not affect the expression of CDC45 (Stevens et al, 2004), although its promoter contains a putative E2F-binding site, but does affect the expression of meiotic genes AtMSH4, AtRAD51 and AtRAD51C, the promoter of which does not contain an E2F-binding site (Figure 1C and Supplementary Table S1).
rbr-2 is defective in CO formation and synapsis
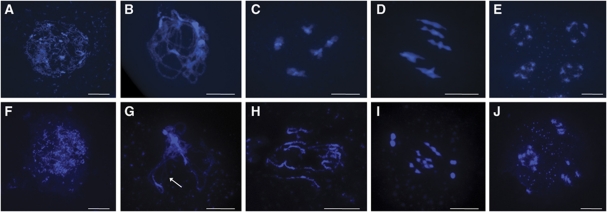
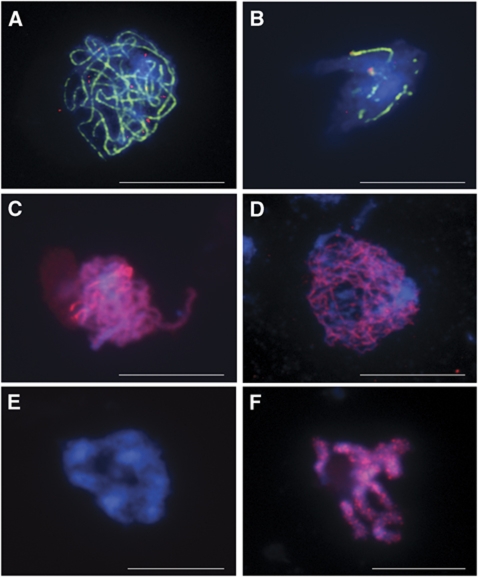
To confirm that the reduced fertility observed in rbr-2 was due to an effect on meiosis and to establish its nature, a cytological analysis of chromosome-spread preparations from PMCs of mutant and wild-type plants was carried out (Figure 3). All meiotic stages were observed in rbr-2, indicating that loss of RBR does not prevent initiation and progression of the meiotic programme. Nevertheless, a number of meiotic defects were clearly apparent, and it was notable that the size range of the buds containing meiocytes in prophase I was much greater for rbr-2 than for wild type. In both cases, buds of ∼400 μm length contained meiocytes at leptotene. In wild type, buds of ∼600 μm had completed meiosis and contained only tetrads, but at this stage, rbr-2 buds still contained cells at zygotene/diplotene. Tetrads were not observed in rbr-2 buds shorter than 800–900 μm. In addition, several different stages were often observed in meiocytes from anthers within the same bud, whereas wild-type anthers from the same bud contain meiocytes at more or less the same meiotic stage (Armstrong and Jones, 2003). This suggested that loss of RBR may affect the rate of meiotic progression. During early prophase I (leptotene), rbr-2 nuclei were indistinguishable from that of wild type (Figure 3A and F). However, in contrast to wild type, which progressed through zygotene to achieve full chromosome synapsis at pachytene, the majority of rbr-2 chromosomes appeared to achieve only partial synapsis, with unpaired chromosome axes clearly visible (Figure 3B and G). This was confirmed by immunolocalisation of the SC transverse filament protein AtZYP1 that marks synapsed chromosomes and AtASY1, a component of the chromosome axes (Higgins et al, 2005; Armstrong et al, 2002; Sanchez-Moran et al, 2007). In wild-type meiocytes, AtZYP1 polymerisation commenced at zygotene, forming a linear signal between each homologous chromosome pair. By pachytene, the AtZYP1 signal was continuous, denoting formation of the mature SC (Figure 4A). In contrast, the polymerisation of AtZYP1 in the rbr-2 nuclei was incomplete or occasionally absent (Figure 4B). In the wild-type meiocytes, AtASY1 dissociates from the axes during pachytene/early diplotene, as the SC breaks down (Figure 4C and E). In rbr-2 meiocytes, AtASY1 localisation appeared normal during much of prophase I, but it did not dissociate in a timely manner and persisted through to diakinesis (Figure 4D and F).
Figure 3.
Meiotic stages from wild-type and rbr-2 pollen mother cells. (A–E) Wild type; (F–J) rbr-2; (A, F) leptotene, (B, G) pachytene, note the unsynapsed axis in the rbr-2 mutant (arrows), (C, H) diakinesis, (D, I) metaphase I, (E, J) tetrad stage. Nuclei are stained with 4′,6-diamidino-2-phenylindole (DAPI). Scale bar, 10 μm.
Figure 4.
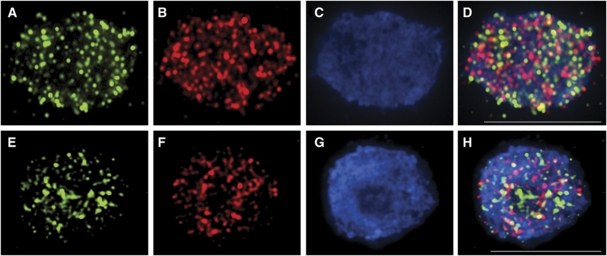
Immunolocalisation of meiotic proteins in wild-type and rbr-2 prophase I nuclei. (A, C, E) Wild-type; (B, D, F) rbr-2 prophase I nuclei. (A, B) AtZYP1 (green), AtMLH1 (red), DAPI (blue); (C, D) AtASY1 (red), DAPI (blue); (E, F) AtASY1 (red), DAPI (blue) at diakinesis. Scale bar, 10 μm.
Wild-type diakinesis is marked by closely apposed homologous chromosome pairs (bivalents) linked by chiasmata, the cytological manifestation of genetic COs (Figure 3C). In contrast, the corresponding rbr-2 chromosomes appeared to be longer than wild type (mean length 60.06 versus 50.17 μm, respectively;P<0.05) and most lacked chiasmata (Figure 3H). At metaphase I, the wild-type nuclei contained five bivalents (Figure 3D), which following the first and second meiotic divisions formed normal tetrads (Figure 3E). However, at metaphase I, most of the chromosomes in the rbr-2 nuclei were present as univalents (Figure 3I) that mis-segregated, leading to the formation of four meiotic products, some containing fewer and others more than the wild-type complement of five chromosomes (Figure 3E and J). FISH analysis of metaphase I chromosomes confirmed the reduction of the number of chiasmata in rbr-2 nuclei to an average of 1.7 (n=76), compared with 9–10 chiasmata typically detected in wild-type nuclei (Supplementary Figure S3A and B; Higgins et al, 2004). In addition, close inspection of the rbr-2 nuclei suggested a tendency for precocious separation of the sister chromatids (Figure 3I). Although, there was no suggestion of a problem with sister chromatid cohesion at diakinesis, this observation could suggest some underlying problem, particularly as the transcription analysis suggested that expression of AtSMC3, which encodes one of the subunits of the cohesin complex (Lam et al, 2005), was significantly reduced (Figure 1C). However, immunolocalisation on chromosome-spread preparations from rbr-2 meiocytes using an anti-SMC3 Ab suggested that there was no obvious deficiency of the protein (Supplementary Figure 3E and F). Moreover, we have observed a tendency for sister chromatid separation at metaphase I in other Arabidopsis mutants, such as Atmsh4 and Atmsh5, that have low levels of chiasma frequency, yet presumably normal expression of the cohesion complex (JD Higgins and FCH Franklin, unpublished observation; Supplementary Figure 3C and D). Although, a subtle effect could not be excluded, it would appear there are no major defects in sister chromatid cohesion in rbr-2.
Loss of RBR affects recruitment of meiotic proteins
Immunolocalisation studies using a panel of antibodies were conducted to gain further insight into the basis of the meiotic defects identified in rbr-2. In Arabidopsis, initiation of meiotic recombination is dependent on the formation of DNA DSBs by AtSPO11-1 and AtSPO11-2 (Grelon et al, 2001; Stacey et al, 2006). DSBs may be detected using an Ab to monitor phosphorylation of histone H2AX (γH2AX) at the break site (Mahadevaiah et al, 2001). In rbr-2, we detected γH2AX foci at wild-type levels (>100) in early prophase I (Figure 5A and B). This indicated that RBR was not required for DSB formation. However, the formation of RBR foci observed in wild-type meiocytes (Figure 2A–C and F–H) was absent in the Atspo11-1–4 mutant, which does not form DSBs (Sanchez-Moran et al, 2007; Figure 2E and J). This suggests that formation of RBR foci is dependent on the formation of DSBs.
Figure 5.
DNA double-strand break formation and localisation of early recombination proteins in wild-type and rbr-2 nuclei. (A, C, E, G) Wild-type; (B, D, F, H) rbr-2 nuclei. (A, B) Immunolocalisation of γH2AX at prophase I indicates that DNA double-strand break formation is normal in rbr-2; (C, D) localisation of AtRAD51 is normal during early prophase I; (E, F) localisation of AtDMC1 is normal during early prophase I. (G, H) Localisation of AtMSH4 reveals a drastic reduction of foci in rbr-2. Scale bar, 10 μm.
During crossing over in budding yeast, the meiotic-specific recombinase Dmc1, either in conjunction with or independently of the general recombinase Rad51, catalyses invasion of homologous DNA sequences on one of the non-sister chromatids by the single-stranded DNA from one side of the DSB (Neale and Keeney, 2006). Although, it has not been demonstrated biochemically, analysis of the corresponding Arabidopsis proteins AtDMC1 and AtRAD51 would suggest that this process is conserved in plants (Couteau et al, 1999; Doutriaux et al, 1998; Sanchez-Moran et al, 2007). Immunolocalisation of AtDMC1 and AtRAD51 in chromosome spreads from wild-type and rbr-2 meiocytes at leptotene were indistinguishable revealing, in both cases, the accumulation of >100 axis-associated foci (Figure 5C–F).
In Arabidopsis, wild-type levels of chiasma formation are dependent on the combined activity of the MutS homologues AtMSH4 and AtMSH5 (Higgins et al, 2004; Higgins et al, 2008b). Immunolocalisation of AtMSH4 on chromosome-spread preparations from wild type revealed numerous (>100) foci that were first detectable at late leptotene (Figure 5G). These then gradually reduced in number throughout zygotene, with only a few foci (<10) persisting until mid-pachytene (Higgins et al, 2004). The number of AtMSH4 foci detected in rbr-2 meiocytes at late leptotene was drastically reduced. In the majority of nuclei (12/22) AtMSH4 foci were entirely absent, with fewer than 50 detected in each of the remaining nuclei inspected (Figure 5H).
In wild-type Arabidopsis, it appears that the sites of COs are marked by the MutL homologue, AtMLH1, which is believed to promote CO formation (Wang et al, 1999). Dual immunolocalisation in wild-type meiocytes of AtMLH1 together with AtZYP1 reveals 9–11 AtMLH1 foci associated with a fully synapsed SC at pachytene (Figure 4A). In rbr-2, the number of AtMLH1 foci was typically in the range 1–3 (Figure 4B). These co-localised to the short stretches of linear AtZYP1 that were present in the mutant. Thus, the reduction in AtMLH1 in rbr-2 corresponded with the reduction in chiasmata observed at metaphase I in the mutant.
The number and turnover of RBR foci observed in wild-type meiocytes during prophase I was similar to that previously observed for AtDMC1 and AtMSH4 (Higgins et al, 2004; Sanchez-Moran et al, 2007). It was of interest to determine whether there was evidence of co-localisation between RBR and the recombination pathway proteins. Dual immunolocalisation of RBR and AtDMC1 in wild-type meiocytes at leptotene showed numerous foci corresponding to each protein associated with the chromosomes at this stage. In many cases, the foci appeared to co-localise (co-localisation 44%±3, n=10) or show a close association, but this was incomplete (Figure 6A–D). Dual labelling of RBR and AtMSH4 revealed a similar overlap of foci at leptotene (co-localisation 43%±2, n=10), but this reduced to only 19% (±4.6%, n=5) at zygotene (Figure 6E–H). Co-localisation controls suggested that ∼40% of these could be random associations.
Figure 6.
Dual immunolocalisation of RBR with recombination with AtDMC1 and AtMSH4 in wild-type prophase I nuclei. (A–D) RBR and AtDMC1 (leptotene); (E–H) RBR and AtMSH4 (zygotene); (A) AtDMC1, (B) RBR, (C) DAPI, (D) merge, (E) AtMSH4, (F) RBR (G), DAPI, (H) merge. Scale bar, 10 μm.
Loss of RBR compromises both Class I and Class II COs
Previous studies have shown that CO formation in Arabidopsis, as in budding yeast, occurs via at least two pathways. Class I COs, which account for ∼85% of the total, are dependent on AtMSH4 and exhibit CO interference, the phenomenon whereby COs do not occur in close proximity to each other (Higgins et al, 2004; Jones and Franklin, 2006). The remaining Class II COs, some of which are dependent on AtMUS81, do not exhibit CO interference (Higgins et al, 2008a). Although, CO formation is dramatically reduced in rbr-2, it is not absent. Indeed, the mean CO frequency at 1.7 is similar to that observed in Atmsh4 (Higgins et al, 2004, 2008a, 2008b). This could imply that RBR is primarily involved in the formation of Class I COs. To clarify whether RBR is implicated in Class I COs or both Class I and II, we constructed an Atmsh4/rbr-2 double-knockout line. Chiasma counts on metaphase I chromosome-spread preparations from homozygous mutants revealed that the mean chiasma frequency was significantly reduced compared with the Atmsh4 single mutant (0.45 versus 1.29, n=115; P<0.001). This indicates that RBR is important for normal levels of both Class I and Class II COs.
Discussion
The isolation of a homozygous rbr-2 line has enabled us to investigate the role of RBR during meiosis. Our studies have revealed that loss of RBR results in a reduction in the formation of meiotic COs and a failure of chromosome synapsis, leading to a dramatic reduction in fertility.
RBR_m is specifically expressed in rbr-2 meiocytes
Although the wild-type RBR protein was expressed in vegetative cells in the rbr-2 line, only the truncated RBR_m form was detected in meiocytes. DNA sequence analysis suggested that this was due to an aberrant splicing event. Cell-type-specific splicing is common in vertebrates (Matlin et al, 2005), but has rarely been observed in plants. There is extensive evidence of meiosis-specific splicing in other species such as budding yeast, fission yeast and mouse (Engebrecht et al, 1991; Juneau et al, 2007; Averbeck et al, 2005; Bellani et al, 2010). As yet, there is no direct evidence that alternative splicing might have a role during plant meiosis. However, in Arabidopsis, mutations in genes encoding essential components of the spliceosome affect gametogenesis, which supports the existence of splicing activity unique to gametes in plants (Gross-Hardt et al, 2007; Moll et al, 2008). Although the expression of RBR_m arises from an aberrant splicing event, it does perhaps highlight some differences in the activity of the splicing machinery in meiocytes compared with vegetative cells.
RBR is essential for inter-homologue recombination and synapsis
It would appear that early events in the meiotic pathway occur normally in rbr-2. As RBR is a negative regulator of the G1/S transition in mitotic cells (Friend et al., 1986), and hence most probably in meiocytes, the loss of RBR activity is not expected to prevent meiotic S phase. DSB formation occurs at or near wild-type levels, and there is no evidence of DNA fragmentation in rbr-2. This suggests that despite reduced CO formation, the DSBs are efficiently repaired, either through non-CO recombination or via repair using a sister chromatid as the repair template. Our studies indicate that loss of RBR does not prevent meiotic progression, but it may delay progression through prophase I. In many organisms, mutations in recombination proteins lead to the activation of checkpoints that result in meiotic arrest or apoptosis (Roeder and Bailis, 2000). Whether similar checkpoints existed in Arabidopsis was initially a matter of debate, as many recombination mutants complete meiosis. However, mutants lacking AtTOP3α and AtRMI1/BLAP75, which are implicated in the dissolution of dHjs, undergo arrest at telophase I (Chelysheva et al, 2008; Hartung et al, 2008). Mutants defective in recombination pathway proteins such as AtMSH4 and AtMLH3 show a significant delay in prophase I progression (Higgins et al, 2004; Jackson et al, 2006). Despite a reduction in CO formation in these mutants, there is no evidence of chromosome fragmentation, indicating that DSBs are eventually repaired. Extrapolating from studies in budding yeast, it has been proposed that an intra-prophase I surveillance system that detects recombination defects may delay prophase I progression in Arabidopsis until DSB repair is complete (Boerner et al, 2004; Jackson et al, 2006). Thus, it seems likely that the delayed prophase I progression in rbr-2 is due to a recombination defect rather than a direct consequence of the loss of RBR.
In Arabidopsis, inter-homologue recombination requires the activity of the meiosis-specific recombinase AtDMC1. Loss of the protein results in a complete absence of chiasmata. No chromosome fragmentation is observed at metaphase I. This suggests that the AtSPO11-induced DSBs are efficiently repaired using the sister chromatid as the repair template (Couteau et al, 1999; Doutriaux et al, 1998; Sanchez-Moran et al, 2007). Following strand-exchange and stable single-end invasion, subsequent CO formation requires the activity of AtMSH4 (Higgins et al, 2004). In rbr-2 meiocytes, AtDMC1 localisation appears normal, while AtMSH4 foci are reduced or absent. Formation of the SC is also compromised with little polymerisation of the transverse filament protein AtZYP1. Loss of AtDMC1 results in a complete failure of SC formation (Couteau et al, 1999; Doutriaux et al, 1998; Sanchez-Moran et al, 2007). In contrast, Atmsh4 forms an intact SC, although with some delay (Higgins et al, 2004). Thus, in this respect, rbr-2 resembles Atdmc1 rather than Atmsh4. Together, these observations suggest that RBR has a critical role at some early stage in the recombination pathway following DSB formation and AtDMC1 loading, but before subsequent AtMSH4 loading. This is supported by our finding that RBR is essential for normal levels of both Class I and Class II COs.
The effect of rbr on meiotic gene transcription is probably not the basis of the meiotic defect in rbr-2
In contrast to mice (Satyanarayana and Kaldis, 2009; Susiarjo et al, 2009), very few plant cell-cycle genes have been found to have roles in meiosis, probably due to the overlapping function of these genes in mitosis and meiosis (Mercier and Grelon, 2008). Although RBR affect the expression of meiotic genes under the control of a promoter containing a putative E2F-binding factor, RBR appears to activate their transcription. However, a direct control of gene expression by the E2F pathway is usually repressive in somatic tissues (Park et al, 2005; Desvoyes et al, 2006) and in reproductive tissues (Jullien et al, 2008). In addition, the meiotic genes that are controlled by RBR and E2F are not overexpressed in vegetative tissues that overexpress E2F (Vandepoele et al, 2005), which suggests that additional transcriptional controls specific to meiotic tissues are required. The fact that rbr-2 also affects the expression of meiotic genes that do not possess a predicted E2F-binding site in their promoter, further supports the idea that the effect of rbr-2 on meiotic gene expression is probably independent from a direct control by E2F.
Although the expression of AtMSH4 and AtZYP1 is reduced in rbr-2, they are nevertheless transcribed at over 60% of the wild-type level. Heterozygous lines carrying a null allele of a wide range of Arabidopsis meiotic genes are fully fertile, suggesting that the pathway is resilient to reductions in gene expression. This is supported by a study of AtSPO11–1 that found that the gene was expressed at ∼60% of wild-type level in a heterozygous Atspo11-1-4−/+ mutant, yet this had no apparent reduction in DSB formation or subsequent chiasma frequency (Roberts, 2010). Of the meiotic genes included in the analysis of transcription, the cohesin AtSMC3 was most reduced in rbr-2. However, immunolocalisation of AtSMC3 appeared normal; while this is not quantitative, it does suggest that the protein is not substantially depleted.
While there was a tendency for precocious separation of sister chromatids at metaphase I, there was no obvious loss of chromosome cohesion in rbr-2 meiocytes at diakinesis. Precocious sister chromatid separation has been observed in the maize mutant desynaptic (Maguire, 1978). It is thought that, in this mutant COs are formed at normal levels, but there is a failure to maintain chiasmata because of an underlying cohesion problem. However, there are obvious defects in both recombination and synapsis in Arabidopsis rbr-2, which were not observed in the maize mutant desynaptic. This suggests that the basis of the reduced chiasma frequency in these mutants is different. Furthermore, precocious sister chromatid separation is also observed in recombination pathway mutants such as Atmsh4 and Atmsh5 (Supplementary Figure 3C and D), in which expression of the cohesin complex is presumably normal. In summary, the level of reduction in meiotic gene expression reported here may be in itself not sufficient to account for the rbr-2 phenotype. Nevertheless, it is potentially a contributing factor, particularly as the expression of several meiotic genes is reduced in parallel.
A link between RBR and recombination
The fact that foci corresponding to RBR and recombination pathway proteins are localised to the chromosomes during early prophase I could suggest that RBR foci form at or in the vicinity of recombination complexes, particularly as they form about the same number of foci at leptotene following and dependent on AtSPO11-1-mediated DSB formation. A study in budding yeast has shown that the CDK, Cdc28, forms foci during early meiotic prophase I, coinciding with the formation of Rad51 foci (Zhu et al, 2010). However, the level of co-localisation was relatively low at 34%. Determining the degree to which there might be a direct spatial relationship between RBR and recombination complexes containing AtDMC1 and AtMSH4 was subject to the limited resolution afforded by immunofluorescence microscopy using fixed material combined with high numbers of foci present on the chromosomes during early prophase I. Any co-localisation of the RBR and AtMSH4 at zygotene appeared limited, which would support the immunolocalisation studies in rbr-2. Although this suggests that the role of RBR precedes AtMSH4 function in relation to CO formation, further analysis will be required to resolve this issue.
Current evidence suggests that the key elements of the core recombination machinery are conserved between plants and budding yeast (Hamant et al, 2006). As the latter does not possess an Rb homologue, a direct biochemical interaction between RBR and the recombination machinery is perhaps unlikely, regardless of any spatial relationship. However, studies in budding yeast show that meiotic recombination occurs in the context of chromatin-associated complexes that are juxtaposed to the chromosome axes. Perturbation of the structural organisation of the chromosomes or their underlying axes can adversely influence stable inter-homologue exchange (Kleckner, 2006; Storlazzi et al, 2010). In Arabidopsis, loss of the chromosome axis protein AtASY1 has been shown to result in the destabilisation of AtDMC1 foci, resulting in a dramatic reduction in CO formation (Sanchez-Moran et al, 2007). That loss of pRb could directly affect chromosome organisation is supported by the finding that the Drosophila Rb family member RBF1 interacts with the condensin II complex and is essential for proper segregation of mitotic chromosomes (Longworth et al, 2008). It is therefore of potential significance that in the rbr-2 mutant, the chromosomes at diakinesis are significantly longer than those in wild-type meiocytes at the same stage. In addition, while initial localisation of the chromosome axis protein AtASY1 appears normal in the rbr-2 meiocytes, its depletion from the axes is delayed until diakinesis. Thus, RBR may influence chromosome structure, possibly in the vicinity of recombination sites, to promote the formation of COs. The fact that a few COs can still occur may reflect that, in some cases, DSBs occur at sites where localised modification to the chromosome organisation is not essential. It may be significant that the C-terminus deletion in RBR corresponds to a region that, in other members of the pRb family, interacts with proteins that are important for chromatin remodelling/modification, such as SWI/SNF, histone deacetylases and histone methyltransferases (Longworth and Dyson, 2010).
A link between chromosome organisation and recombination progression has been proposed (Kleckner et al, 2004). In this model, meiotic (and mitotic) chromosome conformation undergoes cyclic periods of expansion and contraction, which relate to the status of the chromatin and the underlying axes. In the case of meiosis, these stress/relief transitions are coupled to distinct stages in CO formation, including single-end invasion, to form stable strand-exchange products. The defects observed in rbr-2 are consistent with the mechanisms proposed by such a model. DSB formation is not affected, as RBR does not appear to participate in this event. However, subsequent stress-dependent CO recombination may be compromised because of defects in chromosome/axis organisation arising from loss of RBR, which interferes with programmed stress/relief transitions of the meiotic chromosomes. Interestingly, it has been demonstrated that the transcriptional co-activator, mediator and cohesin interact to provide a link between the gene transcription machinery and chromatin architecture (Kagey et al, 2010). Furthermore, in Drosophila, RBF1 interacts with the origin recognition complex and chromatin to directly influence DNA replication (Ahlander et al, 2008). Hence, it is conceivable that RBR may function in an analogous manner to couple meiotic recombination with the cell cycle.
Conclusions
Our study indicates that RBR is important for normal levels of meiotic COs and SC formation in Arabidopsis. Whether the meiotic defects in rbr-2 are attributable to one or more of these will require further investigation. At present there is little evidence as to whether an Rb function in meiosis is widely conserved. E2F-binding elements in genes controlling meiosis have been identified in mice (Kehoe et al, 2008). In addition, the loss of Rb function in the nematode Caenorhabditis elegans causes sterility of unknown origin (Cui et al, 2004). Despite the paucity of experimental evidence, given the similarity in meiotic control shared between plants and animals, it seems likely that this role of pRb is conserved in other multicellular organisms.
Materials and methods
Experimental materials
The wild-type ecotype Columbia (Col-0) was provided by the Nottingham Arabidopsis Stock Centre. The Arabidopsis thaliana mutant alleles (Columbia accession) used in this study were rbr-2 (SALK_002946; SALK collection) and cdka-1 (SALK_106809; SALK collection). After 3 days at 4°C in dark, seeds were germinated and grown on soil. Plants were cultured in a growth chamber under short days (8 h of light at 20°C/16 h of dark at 16°C; 60–70% relative humidity) until rosettes were formed. Plants were transferred to long days at 20°C (16 h of light/8 h of dark) to induce flowering and grown until seeds were harvested.
RNA extraction and RT–PCR
Sample tissues were collected from Arabidopsis plants and frozen in liquid nitrogen immediately. Tissues were ground by Tissue-lyser machine (Qiagen) and total RNA was prepared using the RNeasy mini kit (Qiagen). DNase treatment was done on 2 mg of total RNA using the Dnase-free kit (Ambion). For reverse transcription, 500 ng of total RNA was incubated at 42°C for 1 h with 200 U of murine leukemia virus reverse transcriptase (New England Biolabs) in a 20 μl reaction mixture containing 4 mM oligo (dT) primer, RT reaction buffer, 1 mM deoxynucleotide triphosphate and 40 U of recombinant RNasin ribonuclease inhibitor (Promega). The reaction was terminated by incubation at 90°C for 10 min. (Primers used are listed in Supplementary Table S2.)
Quantitative real-time RT–PCR
Real-time PCR assays were performed using Power SYBR Green PCR Master Mix (Applied Biosystems). A volume of 0.25 μl of the reverse transcript product was used to perform each PCR reaction. Amplification reaction was carried out using specific primers at a concentration of 0.2 mM in a 10 μl reaction mixture. The specificity of the amplification was determined by running a step of dissociation curve analysis. Thermal cycling parameters were 2 min at 50°C, 10 min at 95°C and 50 cycles of 15 s at 95°C and 60 s at 60°C. Three technical replicates were carried out for each sample. The PCR reaction and quantitative measurements were achieved with 7900HT Fast Real-Time PCR System SDS2.3 and RQ manager 1.2 software (Applied Biosystems). The ΔCt was calculated using ACT11 as endogenous control. Relative quantisation (RQ) values were calculated by the 2−ΔCt method (RQ=2−ΔCt). RQ values represent the average of three biological replicates. (Primers used are listed in Supplementary Table S2.)
Western blotting
RBR polyclonal Ab recognising RBR-C was produced from rabbit immunised with synthesised peptide of the amino acid sequence: CNRLNNSSSNRKRTL. The cysteine residue was employed for single point, site-directed conjugation to KLH. RBR was quantified by means of SDS–PAGE followed by western blotting. The extracted total protein content of the cleared supernatant was measured (protein assay kit; Bio-Rad) and equal amounts of total protein were loaded. Proteins were separated on a 10% polyacrylamide gel and transferred to PVDF membrane. Polyclonal Ab diluted in the ratio 1:5000 was applied followed by an anti-mouse/rabbit IgG–POD coupled to HRP and visualised by BM Chemiluminescence Western Blotting Kit (Roche).
Cytological procedures
The cytological methods were carried out as previously described (Higgins et al, 2004). The following antibodies were used in this study: anti-RBR-C terminal (rabbit, 1/200 dilution), anti-RBR N-terminal (Borghi et al, 2010), anti-AtASY1 (rabbit/rat, 1/500 dilution), anti-AtMSH4 (rabbit, 1/500 dilution), anti-AtZYP1 (rabbit/rat, 1/500 dilution), anti-AtDMC1 (rabbit 1/500 dilution), anti-AtRAD51 (rabbit, 1/500 dilution), anti-AtMLH1 (rabbit/rat, 1/200 dilution) and anti-γH2AX (ser 139, Upstate Biotechnology, catalogue no. 07-164; rabbit, 1/100 dilution; Higgins et al, 2004; Higgins et al, 2005; Mercier et al, 2003). To confirm the specificity of anti-RBR-C Ab, comparative immunolocalisation was conducted on wild type and rbr-2. This confirmed that the signal present in wild type was absent in the mutant (Figure 2A–D). FISH on metaphase I chromosomes was carried out using the 45S rDNA and 5S rDNA probes, as previously described (Higgins et al, 2004). Microscopy was conducted using a Nikon 90i Fluorescence Microscope (Tokyo, Japan). Image capture and analysis was carried out using NIS Elements F software (Nikon, Tokyo, Japan).
Statistical procedures
The statistical procedures were carried out as described previously (Higgins et al, 2004).
Supplementary Material
Acknowledgments
We are most grateful to Wilhelm Gruissem (ETH, Zurich) for providing the N-terminal RBR antibody. We thank Philipp Kaldis and Stephen Cohen for critical reading of the manuscript and the reviewers for their constructive comments. FB and ZC were funded by the Temasek Life Sciences Laboratory. ZC was funded by the Singapore Millenium Foundation. The laboratory of FCHF is funded by the Biological and Biotechnology Research Council, UK. ZC contributed all genetic and molecular analysis of the rbr-2 homologous mutant line. JDH contributed the cytological and immunological analysis of meiotic defects. JTLH performed the cell sorting analysis. JL contributed the Q-PCR. FB and FCHF conceived the project, participated in data analyses and wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ahlander J, Chen XB, Bosco G (2008) The N-terminal domain of the Drosophila retinoblastoma protein Rbf1 interacts with ORC and associates with chromatin in an E2F independent manner. PLoS One 3: e2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong SJ, Caryl AP, Jones GH, Franklin FC (2002) Asy1, a protein required for meiotic chromosome synapsis, localises to axis-associated chromatin in Arabidopsis and Brassica. J Cell Sci 115: 3645–3655 [DOI] [PubMed] [Google Scholar]
- Armstrong SJ, Jones GH (2003) Meiotic cytology and chromosome behaviour in wild-type Arabidopsis thaliana. J Exp Bot 54: 1–10 [DOI] [PubMed] [Google Scholar]
- Averbeck N, Sunder S, Sample N, Wise JA, Leatherwood J (2005) Negative control contributes to an extensive program of meiotic splicing in fission yeast. Mol Cell 18: 491–498 [DOI] [PubMed] [Google Scholar]
- Bellani MA, Boateng KA, McLeod D, Camerini-Otero RD (2010) The Expression profile of the major mouse SPO11 isoforms indicates that SPO11 beta introduces double strand breaks and suggests that SPO11 alpha has an additional role in prophase in both spermatocytes and oocytes. Mol Cell Biol 30: 4391–4403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger F, Hamamura Y, Ingouff M, Higashiyama T (2008) Double fertilization—caught in the act. Trends Plant Sci 13: 437–443 [DOI] [PubMed] [Google Scholar]
- Boerner GV, Kleckner N, Hunter N (2004) Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117: 29–45 [DOI] [PubMed] [Google Scholar]
- Borghi L, Gutzat R, Futterer J, Laizet Y, Hennig L, Gruissem W (2010) Arabidopsis RETINOBLASTOMA-RELATED is required for stem cell maintenance, cell differentiation, and lateral organ production. Plant Cell 22: 1792–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm A, Kouzarides T (1999) Retinoblastoma protein meets chromatin. Trends Biochem Sci 24: 142–145 [DOI] [PubMed] [Google Scholar]
- Brehm A, Miska EA, McCance DJ, Reid JL, Bannister AJ, Kouzarides T (1998) Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 391: 597–601 [DOI] [PubMed] [Google Scholar]
- Chelysheva L, Vezon D, Belcram K, Gendrot G, Grelon M (2008) The Arabidopsis BLAP75/Rmi1 homologue plays crucial roles in meiotic double-strand break repair. PLoS Genet 4: e1000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Hafidh S, Poh SH, Twell D, Berger F (2009) Proliferation and cell fate establishment during Arabidopsis male gametogenesis depends on the Retinoblastoma protein. Proc Natl Acad Sci USA 106: 7257–7262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke AR, Maandag ER, van Roon M, van der Lugt NM, van der Valk M, Hooper ML, Berns A, te Riele H (1992) Requirement for a functional Rb-1 gene in murine development. Nature 359: 328–330 [DOI] [PubMed] [Google Scholar]
- Couteau F, Belzile F, Horlow C, Grandjean O, Vezon D, Doutriaux MP (1999) Random chromosome segregation without meiotic arrest in both male and female meiocytes of a dmc1 mutant of Arabidopsis. Plant Cell 11: 1623–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M, Fay DS, Han M (2004) lin-35/Rb cooperates with the SWI/SNF complex to control Caenorhabditis elegans larval development. Genetics 167: 1177–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desvoyes B, Ramirez-Parra E, Xie Q, Chua NH, Gutierrez C (2006) Cell type-specific role of the retinoblastoma/E2F pathway during Arabidopsis leaf development. Plant Physiol 140: 67–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimova DK, Dyson NJ (2005) The E2F transcriptional network: old acquaintances with new faces. Oncogene 24: 2810–2826 [DOI] [PubMed] [Google Scholar]
- Dissmeyer N, Nowack MK, Pusch S, Stals H, Inze D, Grini PE, Schnittger A (2007) T-loop phosphorylation of Arabidopsis CDKA; 1 is required for its function and can be partially substituted by an aspartate residue. Plant Cell 19: 972–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doutriaux MP, Couteau F, Bergounioux C, White C (1998) Isolation and characterisation of the RAD51 and DMC1 homologs from Arabidopsis thaliana. Mol Gen Genet 257: 283–291 [DOI] [PubMed] [Google Scholar]
- Drews GN, Yadegari R (2002) Development and function of the angiosperm female gametophyte. Annu Rev Genet 36: 99–124 [DOI] [PubMed] [Google Scholar]
- Ebel C, Mariconti L, Gruissem W (2004) Plant retinoblastoma homologues control nuclear proliferation in the female gametophyte. Nature 429: 776–780 [DOI] [PubMed] [Google Scholar]
- Engebrecht J, Voelkelmeiman K, Roeder GS (1991) Meiosis-specific RNA splicing in yeast. Cell 66: 1257–1268 [DOI] [PubMed] [Google Scholar]
- Friend SH, Bernards R, Rogelj S, Weinberg RA, Rapaport JM, Albert DM, Dryja TP (1986) A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature 323: 643–646 [DOI] [PubMed] [Google Scholar]
- Funayama R, Ishikawa F (2007) Cellular senescence and chromatin structure. Chromosoma 116: 431–440 [DOI] [PubMed] [Google Scholar]
- Grelon M, Vezon D, Gendrot G, Pelletier G (2001) AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J 20: 589–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross-Hardt R, Kagi C, Baumann N, Moore JM, Baskar R, Gagliano WB, Jurgens G, Grossniklaus U (2007) LACHESIS restricts gametic cell fate in the female gametophyte of Arabidopsis. PLoS Biol 5: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamant O, Ma H, Cande WZ (2006) Genetics of meiotic prophase I in plants. Annu Rev Plant Biol 57: 267–302 [DOI] [PubMed] [Google Scholar]
- Harbour JW, Dean DC (2000) Rb function in cell-cycle regulation and apoptosis. Nat Cell Biol 2: E65–E67 [DOI] [PubMed] [Google Scholar]
- Hartung F, Suer S, Knoll A, Wurz-Wildersinn R, Puchta H (2008) Topoisomerase 3alpha and RMI1 suppress somatic crossovers and are essential for resolution of meiotic recombination intermediates in Arabidopsis thaliana. PLoS Genet 4: e1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson KA, Kee K, Maleki S, Santini PA, Keeney S (2006) Cyclin-dependent kinase directly regulates initiation of meiotic recombination. Cell 125: 1321–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JD, Armstrong SJ, Franklin FCH, Jones GH (2004) The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: evidence for two classes of recombination in Arabidopsis. Genes Dev 18: 2557–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JD, Buckling EF, Franklin FCH, Jones GH (2008a) Expression and functional analysis of AtMUS81 in Arabidopsis meiosis reveals a role in the second pathway of crossing-over. Plant J 54: 152–162 [DOI] [PubMed] [Google Scholar]
- Higgins JD, Sanchez-Moran E, Armstrong SJ, Jones GH, Franklin FCH (2005) The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev 19: 2488–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JD, Vignard J, Mercier R, Pugh AG, Franklin FCH, Jones GH (2008b) AtMSH5 partners AtMSH4 in the class I meiotic crossover pathway in Arabidopsis thaliana, but is not required for synapsis. Plant J 55: 28–39 [DOI] [PubMed] [Google Scholar]
- Ingouff M, Jullien PE, Berger F (2006) The female gametophyte and the endosperm control cell proliferation and differentiation of the seed coat in Arabidopsis. Plant Cell 18: 3491–3501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Fazeli A, Schmitt EM, Bronson RT, Goodell MA, Weinberg RA (1992) Effects of an Rb mutation in the mouse. Nature 359: 295–300 [DOI] [PubMed] [Google Scholar]
- Jackson N, Sanchez-Moran E, Buckling E, Armstrong SJ, Jones GH, Franklin FCH (2006) Reduced meiotic crossovers and delayed prophase I progression At-MLH3-deficient Arabidopsis. EMBO J 25: 1315–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston AJ, Matveeva E, Kirioukhova O, Grossniklaus U, Gruissem W (2008) A dynamic reciprocal RBR-PRC2 regulatory circuit controls Arabidopsis gametophyte development. Curr Biol 18: 1680–1686 [DOI] [PubMed] [Google Scholar]
- Jones GH (1984) The control of chiasma distribution. SEB Symp 38: 293–320 [PubMed] [Google Scholar]
- Jones GH, Franklin FC (2006) Meiotic crossing-over: obligation and interference. Cell 126: 246–248 [DOI] [PubMed] [Google Scholar]
- Jullien PE, Mosquna A, Ingouff M, Sakata T, Ohad N, Berger F (2008) Retinoblastoma and its binding partner MSI1 control imprinting in Arabidopsis. PLoS Biol 6: e194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juneau K, Palm C, Miranda M, Davis RW (2007) High-density yeast-tiling array reveals previously undiscovered introns and extensive regulation of meiotic splicing. Proc Natl Acad Sci USA 104: 1522–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JL, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taajes DJ, Dekker J, Young RA (2010) Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, Kleckner N (1997) Meiosis-specific DNA double strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384 [DOI] [PubMed] [Google Scholar]
- Kehoe SM, Oka M, Hankowski KE, Reichert N, Garcia S, McCarrey JR, Gaubatz S, Terada N (2008) A conserved E2F6-binding element in murine meiosis-specific gene promoters. Biol Reprod 79: 921–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto T (2003) Cell-cycle control during meiotic maturation. Curr Opin Cell Biol 15: 654–663 [DOI] [PubMed] [Google Scholar]
- Kleckner N, Zickler D, Jones GH, Dekker J, Padmore R, Henle J, Hutchinson J (2004) A mechanical basis for chromosome function. Proc Natl Acad Sci USA 101: 12592–12597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N (2006) Chiasma formation: chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma 115: 175–194 [DOI] [PubMed] [Google Scholar]
- Lam WS, Yang XH, Makaroff CA (2005) Characterization of Arabidopsis thaliana SMC1 and SMC3: evidence that AtSMC3 may function beyond chromosome cohesion. J Cell Sci 118: 3037–3048 [DOI] [PubMed] [Google Scholar]
- Lee EY, Chang CY, Hu N, Wang YC, Lai CC, Herrup K, Lee WH, Bradley A (1992) Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature 359: 288–294 [DOI] [PubMed] [Google Scholar]
- Liu CM, McElver J, Tzafrir I, Joosen R, Wittich P, Patton D, Van Lammeren AA, Meinke D (2002) Condensin and cohesin knockouts in Arabidopsis exhibit a titan seed phenotype. Plant J 29: 405–415 [DOI] [PubMed] [Google Scholar]
- Longworth MS, Dyson NJ (2010) pRb, a local chromatin organizer with global possibilities. Chromosoma 119: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longworth MS, Herr A, Ji JY, Dyson NJ (2008) RBF1 promotes chromatin condensation through a conserved interaction with the Condensin II protein dCAP-D3. Genes Dev 22: 1011–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaluso M, Montanari M, Giordano A (2006) Rb family proteins as modulators of gene expression and new aspects regarding the interaction with chromatin remodeling enzymes. Oncogene 25: 5263–5267 [DOI] [PubMed] [Google Scholar]
- Magnard JL, Yang M, Chen YC, Leary M, McCormick S (2001) The Arabidopsis gene tardy asynchronous meiosis is required for the normal pace and synchrony of cell division during male meiosis. Plant Physiol 127: 1157–1166 [PMC free article] [PubMed] [Google Scholar]
- Maguire M (1978) Evidence for separate genetic control of crossing over and chiasma maintenance in maize. Chromosoma 65: 173–183 [Google Scholar]
- Mahadevaiah SK, Turner JM, Baudat F, Rogakou EP, de Boer P, Blanco-Rodriguez J, Jasin M, Keeney S, Bonner WM, Burgoyne PS (2001) Recombinational DNA double-strand breaks in mice precede synapsis. Nat Genet 27: 271–276 [DOI] [PubMed] [Google Scholar]
- Marston AL, Amon A (2004) Meiosis: cell-cycle controls shuffle and deal. Nat Rev Mol Cell Biol 5: 983–997 [DOI] [PubMed] [Google Scholar]
- Matlin AJ, Clark F, Smith CW (2005) Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol 6: 386–398 [DOI] [PubMed] [Google Scholar]
- McKim KS, Hawley RS (1995) Chromosomal control of meiotic cell division. Science 270: 1595–1601 [DOI] [PubMed] [Google Scholar]
- Mercier R, Armstrong SJ, Horlow C, Jackson NP, Makaroff CA, Vezon D, Pelletier G, Jones GH, Franklin FCH (2003) The meiotic protein SWI1 is required for axial element formation and recombination initiation in Arabidopsis. Development 130: 3309–3318 [DOI] [PubMed] [Google Scholar]
- Mercier R, Grelon M (2008) Meiosis in plants: ten years of gene discovery. Cytogenet Genome Res 120: 281–290 [DOI] [PubMed] [Google Scholar]
- Moll C, von Lyncker L, Zimmermann S, Kagi C, Baumann N, Twell D, Grossniklaus U, Gross-Hardt R (2008) CLO/GFA1 and ATO are novel regulators of gametic cell fate in plants. Plant J 56: 913–921 [DOI] [PubMed] [Google Scholar]
- Neale MJ, Keeney S (2006) Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature 442: 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega S, Prieto I, Odajima J, Martin A, Dubus P, Sotillo R, Barbero JL, Malumbres M, Barbacid M (2003) Cyclin-dependent kinase 2 is essential for meiosis but not for mitotic cell division in mice. Nat Genet 35: 25–31 [DOI] [PubMed] [Google Scholar]
- Page SL, Hawley RS (2004) The genetics and molecular biology of the synaptonemal complex. Annu Rev Cell Dev Biol 20: 525–558 [DOI] [PubMed] [Google Scholar]
- Park JA, Ahn JW, Kim YK, Kim SJ, Kim JK, Kim WT, Pai HS (2005) Retinoblastoma protein regulates cell proliferation, differentiation, and endoreduplication in plants. Plant J 42: 153–163 [DOI] [PubMed] [Google Scholar]
- Riehs N, Akimcheva S, Puizina J, Bulankova P, Idol RA, Siroky J, Schleiffer A, Schweizer D, Shippen DE, Riha K (2008) Arabidopsis SMG7 protein is required for exit from meiosis. J Cell Sci 121: 2208–2216 [DOI] [PubMed] [Google Scholar]
- Roberts NY (2010) Investigating the control of homologous chromosome pairing and crossover formation in meiosis of Arabidopsis thaliana. PhD Thesis, University of Birmingham, UK
- Roeder GS, Bailis JM (2000) The pachytene checkpoint. Trends Genet 16: 395–403 [DOI] [PubMed] [Google Scholar]
- Rossi V, Locatelli S, Lanzanova C, Boniotti MB, Varotto S, Pipal A, Goralik-Schramel M, Lusser A, Gatz C, Gutierrez C, Motto M (2003) A maize histone deacetylase and retinoblastoma-related protein physically interact and cooperate in repressing gene transcription. Plant Mol Biol 51: 401–413 [DOI] [PubMed] [Google Scholar]
- Sabelli PA, Dante RA, Leiva-Neto JT, Jung R, Gordon-Kamm WJ, Larkins BA (2005) RBR3, a member of the retinoblastoma-related family from maize, is regulated by the RBR/E2F pathway. Proc Natl Acad Sci USA 102: 13005–13012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Moran E, Santos JL, Jones GH, Franklin FC (2007) ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes Dev 21: 2220–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyanarayana A, Kaldis P (2009) Mammalian cell-cycle regulation: several Cdks, numerous cyclins and diverse compensatory mechanisms. Oncogene 28: 2925–2939 [DOI] [PubMed] [Google Scholar]
- Shen WH (2002) The plant E2F-Rb pathway and epigenetic control. Trends Plant Sci 7: 505–511 [DOI] [PubMed] [Google Scholar]
- Stacey NJ, Kuromori T, Azumi Y, Roberts G, Breuer C, Wada T, Maxwell A, Roberts K, Sugimoto-Shirasu K (2006) Arabidopsis SPO11-2 functions with SPO11-1 in meiotic recombination. Plant J 48: 206–216 [DOI] [PubMed] [Google Scholar]
- Stevens R, Grelon M, Vezon D, Oh J, Meyer P, Perennes C, Domenichini S, Bergounioux C (2004) A CDC45 homolog in Arabidopsis is essential for meiosis, as shown by RNA interference-induced gene silencing. Plant Cell 16: 99–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storlazzi A, Gargano S, Ruprich-Robert G, Falque M, David M, Kleckner N, Zickler D (2010) Recombination proteins mediate meiotic spatial chromosome organization and pairing. Cell 141: 94–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart D, Wittenberg C (1998) CLB5 and CLB6 are required for premeiotic DNA replication and activation of the meiotic S/M checkpoint. Genes Dev 12: 2698–2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susiarjo M, Rubio C, Hunt P (2009) Analyzing mammalian female meiosis. Methods Mol Biol 558: 339–354 [DOI] [PubMed] [Google Scholar]
- Vandepoele K, Vlieghe K, Florquin K, Hennig L, Beemster GT, Gruissem W, Van de Peer Y, Inze D, De Veylder L (2005) Genome-wide identification of potential plant E2F target genes. Plant Physiol 139: 316–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TF, Kleckner N, Hunter N (1999) Functional specificity of MutL homologs in yeast: evidence for three Mlh1-based heterocomplexes with distinct roles during meiosis in recombination and mismatch correction. Proc Natl Acad Sci USA 96: 13914–13919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YX, Magnard JL, McCormick S, Yang M (2004) Progression through meiosis I and meiosis II in Arabidopsis anthers is regulated by an A-type cyclin predominately expressed in prophase I. Plant Physiol 136: 4127–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg RA (1995) The retinoblastoma protein and cell cycle control. Cell 81: 323–330 [DOI] [PubMed] [Google Scholar]
- Wildwater M, Campilho A, Perez-Perez JM, Heidstra R, Blilou I, Korthout H, Chatterjee J, Mariconti L, Gruissem W, Scheres B (2005) The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell 123: 1337–1349 [DOI] [PubMed] [Google Scholar]
- Wu L, de Bruin A, Saavedra HI, Starovic M, Trimboli A, Yang Y, Opavska J, Wilson P, Thompson JC, Ostrowski MC, Rosol TJ, Woollett LA, Weinstein M, Cross JC, Robinson ML, Leone G (2003) Extra-embryonic function of Rb is essential for embryonic development and viability. Nature 421: 942–947 [DOI] [PubMed] [Google Scholar]
- Wyrzykowska J, Schorderet M, Pien S, Gruissem W, Fleming AJ (2006) Induction of differentiation in the shoot apical meristem by transient overexpression of a retinoblastoma-related protein. Plant Physiol 141: 1338–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Mori S, Oshiumi H, Matsuzaki K, Shinohara M, Shinohara A (2010) Cyclin-dependent kinase promotes formation of the synaptonemal complex in yeast meiosis. Genes Cells 15: 1036–1050 [DOI] [PubMed] [Google Scholar]
- Zickler D, Kleckner N (1999) Meiotic chromosomes: integrating structure and function. Annu Rev Genet 33: 603–754 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.