TGF-β regulates isoform switching of FGF receptors and epithelial–mesenchymal transition
Both TGF-β and FGF signalling regulate the epithelial–mesenchymal transition. Here, TGF-β is found to promote myofibroblast differentiation, while concomitant FGF pathway activation instead drives cells towards an invasive mesenchymal fate.
Keywords: EMT, FGF-2, FGF receptor, TGF-β, δEF1
Abstract
The epithelial–mesenchymal transition (EMT) is a crucial event in wound healing, tissue repair, and cancer progression in adult tissues. Here, we demonstrate that transforming growth factor (TGF)-β induced EMT and that long-term exposure to TGF-β elicited the epithelial–myofibroblastic transition (EMyoT) by inactivating the MEK-Erk pathway. During the EMT process, TGF-β induced isoform switching of fibroblast growth factor (FGF) receptors, causing the cells to become sensitive to FGF-2. Addition of FGF-2 to TGF-β-treated cells perturbed EMyoT by reactivating the MEK-Erk pathway and subsequently enhanced EMT through the formation of MEK-Erk-dependent complexes of the transcription factor δEF1/ZEB1 with the transcriptional corepressor CtBP1. Consequently, normal epithelial cells that have undergone EMT as a result of combined TGF-β and FGF-2 stimulation promoted the invasion of cancer cells. Thus, TGF-β and FGF-2 may cooperate with each other and may regulate EMT of various kinds of cells in cancer microenvironment during cancer progression.
Introduction
The epithelial–mesenchymal transition (EMT) is a differentiation switch that directs polarized epithelial cells to differentiate into mesenchymal cells. Recent studies have proposed that EMT can be classified into three subtypes based on the biological context (Thiery and Sleeman, 2006; Kalluri and Weinberg, 2009; Kalluri, 2009; Zeisberg and Neilson, 2009). Type 1 EMT involves the transition of primitive epithelial cells to motile mesenchymal cells during gastrulation and the generation of migrating-neural crest cells from primitive neuroepithelial cells. This process can generate mesenchymal cells that have the potential to subsequently undergo the mesenchymal–epithelial transition, leading to the generation of secondary epithelia. Type 2 EMT involves the transition of secondary epithelial or endothelial cells to resident tissue fibroblasts and is associated with wound healing, tissue regeneration, and tissue fibrosis. Type 2 EMT begins as a part of a repair-associated event that normally generates fibroblasts for tissue reconstruction after trauma and inflammatory injury. Type 3 EMT occurs in epithelial carcinoma cells in primary nodules that are transitioning to metastatic tumour cells. This process can affect both oncogenes and anti-oncogenes, and carcinoma cells that undergo type 3 EMT can invade and metastasize, resulting in cancer progression.
Thus far, nearly all cases of EMT in adult tissues have been shown to be regulated by extracellular matrix components and soluble growth factors or cytokines, including epidermal growth factor, hepatocyte growth factor, fibroblast growth factors (FGFs), and transforming growth factor (TGF)-βs (Thiery and Sleeman, 2006; Acloque et al, 2009). Among these factors, TGF-β and FGF are considered key mediators of EMT and are frequently and abundantly expressed in various tumours. TGF-β signalling not only contributes to EMT during embryonic development, but also induces EMT in cancer cells during cancer progression (Moustakas and Heldin, 2007). TGF-β activates Smad proteins and activated Smads transcriptionally regulate several genes including δEF1, Snail, Twist, HMGA2, and Ids, which lead to EMT particularly through the transcriptional repression of E-cadherin (Zavadil and Bottinger, 2005; Horiguchi et al, 2009).
The E-cadherin repressors that are regulated by TGF-β appear to function in a cell context-dependent manner, and δEF1 is one of the most well-characterized factors that is involved in TGF-β-induced EMT in mouse epithelial cells (Shirakihara et al, 2007). δEF1 (also known as ZEB1 or TCF-8) and SIP1 (known as ZEB2) are members of the δEF1 family. δEF1 can bind directly to the E-cadherin promoter and repress the E-cadherin expression. In addition, δEF1 also regulates the TGF-β-mediated epithelial–myofibroblastic transition (EMyoT). δEF1 seems to positively regulate the transcription of α-smooth muscle actin (α-SMA), a representative myofibroblast marker, by binding to its promoter in vascular smooth muscle cells (Nishimura et al, 2006).
Among various mammalian FGFs, FGF-2 and FGF-4 are key regulators of EMT during development and cancer progression (Strutz et al, 2002; Strutz and Neilson, 2003). FGFs execute diverse functions by binding and activating members of the FGF receptor (FGFR) family. There are four FGFR genes (FGFR1–FGFR4) that encode functional receptors that consist of three extracellular immunoglobulin domains (Ig-I, Ig-II, and Ig-III), a single-transmembrane domain, and a cytoplasmic tyrosine kinase domain (Eswarakumar et al, 2005). FGFRs have several isoforms as exon skipping removes the Ig-I domain. In addition, alternative splicing in the second half of the Ig-III domain in FGFR1–FGFR3 produces the IIIb (FGFR1IIIb–FGFR3IIIb) and IIIc (FGFR1IIIc–FGFR3IIIc) isoforms that have distinct FGF-binding specificities and are predominantly expressed in epithelial and mesenchymal cells, respectively. FGF-2 binds preferentially to the IIIc isoforms, whereas FGF-7 and FGF-10 bind exclusively to the IIIb isoforms. FGF-2 is highly expressed in wounds and tumour tissues. However, it has not been determined how FGF-2 transmits signals to induce EMT and promote cancer progression because epithelial cells typically express the FGFRIIIb isoforms, which do not bind to FGF-2. Recently, it was reported that advanced cancer cells overexpress FGFR1IIIc (Foster et al, 1999; Acevedo et al, 2007), but it is still unknown how epithelial cells and cancer cells upregulate the expression of the FGFRIIIc isoforms.
In the present study, we investigated the properties of EMT induced by TGF-β in cooperation with FGFs. TGF-β induced EMT, and prolonged treatment with TGF-β induced EMyoT. During TGF-β-mediated EMT, TGF-β altered the sensitivities of cells from FGF-7 to FGF-2 through FGFR isoform switching. In addition, FGF-2 prevented TGF-β-mediated EMyoT and enhanced EMT with more aggressive characteristics that resemble those of activated fibroblasts. Moreover, the cells generated through EMT mediated by FGF-2 and TGF-β facilitated cancer cell invasion, when the cells undergoing EMT were mixed with cancer cells. Therefore, TGF-β and FGF-2 cooperate with each other and may regulate the EMT of various kinds of cells in cancer microenvironment.
Results
FGFR isoform switching during TGF-β-induced EMT
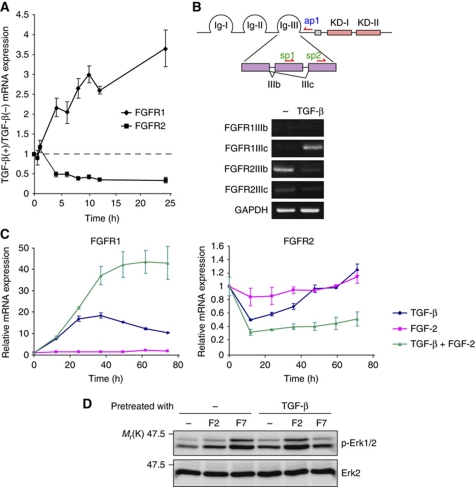
Although the FGFRIIIc isoforms are not predominantly expressed in epithelial cells, their expression and FGF-2 expression have been correlated with cancer progression. We hypothesized that TGF-β primes FGFR isoform switching in cells undergoing EMT and subsequently alters the sensitivity of cells to FGF ligands. To verify this possibility, we first examined the mRNA profiles of FGFR variants in response to TGF-β by quantitative RT–PCR analyses. FGFR1 mRNA gradually increased to ∼3.5-fold in mouse normal mammary epithelial NMuMG cells 24 h after TGF-β stimulation, whereas FGFR2 mRNA levels decreased over this time period (Figure 1A). Conventional RT–PCR analyses using isoform-specific primers revealed that FGFR1 upregulated by TGF-β was FGFR1IIIc (the mesenchymal isoform), whereas the FGFR2 downregulated by TGF-β was FGFR2IIIb (the epithelial isoform) (Figure 1B). FGFR1IIIb, FGFR2IIIc, FGFR3, and FGFR4 were not clearly detected in NMuMG cells. The isoform switching of FGFRs was also detected in mouse mammary gland epithelial Eph4 cells that underwent EMT by TGF-β. Interestingly, the addition of FGF-2 to TGF-β-treated cells sustained expression of FGFR1 and FGFR2 at higher and lower levels, respectively, for 72 h (Figure 1C). Consistent with the changes in the mRNA expression profiles of the FGFR isoforms, Erk phosphorylation was observed in NMuMG cells in response to FGF-7, but not FGF-2, whereas Erk phosphorylation was observed in TGF-β-pretreated cells in response to FGF-2, but not FGF-7 (Figure 1D). These findings indicate that TGF-β primes isoform switching of FGFRs during EMT and thereby changes the sensitivities of cells from FGF-7 to FGF-2.
Figure 1.
Isoform switching of FGFRs during TGF-β-induced EMT. (A) The kinetics of FGFR1 and FGFR2 expression were examined in NMuMG cells treated with 1 ng/ml TGF-β for the indicated periods by quantitative RT–PCR analysis. The ratio of the mRNA levels in TGF-β-treated cells compared with non-treated cells is shown. Each value represents the mean±s.d. of duplicate determinations from a representative experiment. Similar results were obtained in at least three independent experiments. (B) Expression of the alternatively spliced forms of FGFR1 and FGFR2 was examined by RT–PCR using RNA samples from NMuMG cells treated with TGF-β for 36 h. A schematic illustration of the primers is shown at the top. The primers for the IIIb and IIIc isoforms were the sp1–ap1 pair and sp2–ap1 pair, respectively. Ig, extracellular immunoglobulin-like domain; KD, kinase domain. FGFR3 and FGFR4 were not detected in NMuMG cells by quantitative RT–PCR. (C) Regulation of FGFR1 expression (left) and FGFR2 expression (right) by TGF-β and/or FGF-2 were examined by quantitative RT–PCR analysis in NMuMG cells. Values were normalized to the housekeeping gene TBP and are indicated as fold differences compared with the non-treated cells. Each value represents the mean±s.d. of duplicate determinations from a representative experiment. Similar results were obtained in three independent experiments. (D) Erk1/2 phosphorylation (p-Erk1/2) by FGF-2 or FGF-7 was examined by immunoblot analysis. NMuMG cells were preincubated with or without TGF-β for 2 days and then stimulated with 30 ng/ml FGF-2 or 30 ng/ml FGF-7 for 15 min. F2, FGF-2; F7, FGF-7.
Activated phenotype of cells generated by FGF-2 and TGF-β treatment
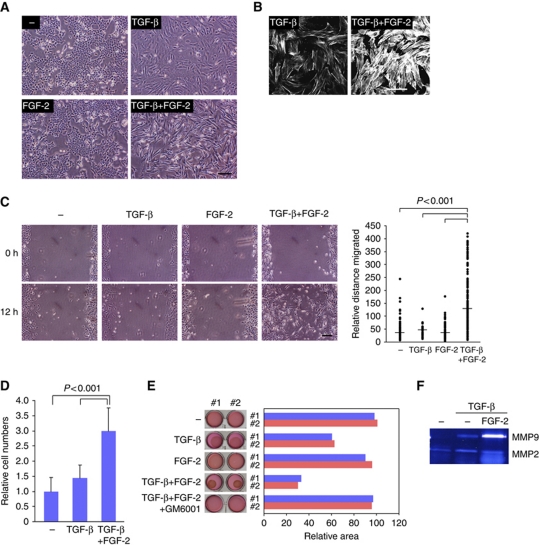
Next, we determined whether FGF-2 enhanced TGF-β-induced EMT after TGF-β-induced FGFR isoform switching, because previous studies have shown that TGF-β-induced EMT can be enhanced with oncogenic signals or growth factors (Horiguchi et al, 2009). To examine this possibility, NMuMG cells were continuously exposed to TGF-β, FGF-2, or both for several days. As we previously reported (Shirakihara et al, 2007), the morphology of NMuMG cells clearly changed from a cobblestone-like shape to a fibroblastic spindle shape upon prolonged TGF-β treatment, whereas cells treated with FGF-2 alone did not exhibit morphological changes (Figure 2A). Compared with cells treated with TGF-β alone, the addition of FGF-2 to TGF-β-treated cells led to drastic changes in cell morphology and actin fibre formation that are typical of fibroblastic differentiation (Figure 2A and B).
Figure 2.
Activated phenotype of cells generated by treating with FGF-2 and TGF-β. (A) NMuMG cells were incubated for 4 days in the absence or presence of 1 ng/ml TGF-β alone, 30 ng/ml FGF-2 alone, or both TGF-β and FGF-2, and visualized by phase-contrast microscopy. Scale bar indicates 100 μm. (B) Actin cytoskeleton reorganization was visualized by TRITC-phalloidin staining 4 days after TGF-β or TGF-β+FGF-2 treatment. Scale bar indicates 100 μm. (C) NMuMG cells were cultured for 4 days in the absence or presence of TGF-β, FGF-2, or both, and then the monolayers were wounded. After 12 h, the migratory behaviours of the cells were determined and quantified (right). Bars represent the median for each category from a representative experiment. Similar results were obtained in two independent experiments. The median migration distance of the cells was significantly different for non-treated versus TGF-β+FGF-2, TGF-β versus TGF-β+FGF-2, and FGF-2 versus TGF-β+FGF-2 (P<0.001; non-parametric Mann–Whitney U test). Scale bar indicates 100 μm. (D) NMuMG cells were cultured for 4 days in the absence or presence of TGF-β or both TGF-β and FGF-2, and then seeded into cell culture inserts. After 12 h, the cells that had migrated to the opposite side were stained and quantified. Each value represents the mean±s.d. of triplicate determinations from a representative experiment. Similar results were obtained in two independent experiments. (E) NMuMG cells were preincubated with or without the indicated combinations of TGF-β, FGF-2, and 10 μM GM6001 for 5 days, and then mixed in collagen matrices. After polymerization, the mixtures were released from the culture dishes and incubated for an additional 2 days. Three independent experiments were performed in duplicate, and representative results are shown (#1 in blue and #2 in red). (F) The conditioned media from NMuMG cells treated with TGF-β or both TGF-β and FGF-2 were collected and applied to 10% polyacrylamide gel impregnated with 1 mg/ml gelatin. After removing SDS, the gels were incubated overnight and stained with CBB.
Next, cell motility was examined using a wound closure assay with NMuMG cells. After treating with TGF-β, FGF-2, or both for 4 days, the cells were wounded by scratching and were then analysed after 12 h. Figure 2C shows that NMuMG cells treated with TGF-β alone had slightly enhanced cell motility, compared with non-treated cells or FGF-2-treated cells. On the other hand, treating with TGF-β and FGF-2 strongly promoted the motility of NMuMG cells within only 12 h. Similar to their ability to enhance migration, combined TGF-β and FGF-2 treatment remarkably promoted the invasion of NMuMG cells in an in vitro invasion assay (Figure 2D). As one of the most characteristic features of fibroblasts is their ability to degrade extracellular matrix, this property was also determined by a collagen gel degradation/contraction assay (Mikko et al, 2008). Cells were pretreated with TGF-β, FGF-2, or both and then suspended in a collagen type I gel. After the collagen had solidified, the gel was detached from the sides and bottoms of the dishes and floated in media containing ligands for 48 h. There was no significant degradation of the collagen gel in either the control or FGF-2-treated cells, but the volume of the collagen gel was reduced by ∼60% in cells treated with TGF-β (Figure 2E). More importantly, the cells treated with TGF-β and FGF-2 drastically decreased the volume of collagen gel by ∼30%, and these changes were inhibited by the matrix metalloprotease inhibitor, GM6001. Gelatin zymography further showed that MMP9 activity was enhanced by TGF-β and FGF-2, compared with that by TGF-β alone (Figure 2F). These findings suggest that NMuMG cells treated with TGF-β alone reveal incomplete EMT, and that the cells exposed to TGF-β and FGF-2 exhibit EMT and more aggressive characteristics that resemble those of activated fibroblasts.
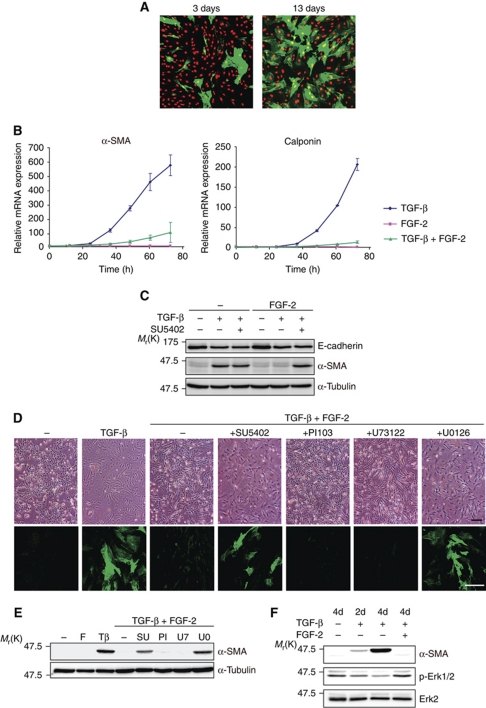
Myofibroblastic differentiation by prolonged TGF-β treatment
To biochemically discriminate between cells that were continuously exposed to TGF-β alone and those exposed to TGF-β and FGF-2, we examined the expression of representative TGF-β-target genes based on our previous study (Kondo et al, 2004). The phosphorylation levels of Smad2 or Smad1/5/8 were not different, and expression levels of some TGF-β-target genes, including Smad7 and fibronectin, were not affected by the addition of FGF-2, indicating that FGF-2 did not affect general TGF-β-Smad signalling (Supplementary Figure S1A and B). Interestingly, the mRNA expression levels of well-known myofibroblast markers, α-SMA and calponin, were increased in TGF-β-treated cells, and α-SMA expression was confirmed in the cells by immunohistochemical analyses (Figure 3A and B), suggesting that TGF-β induced EMyoT. Conversely, the addition of FGF-2 to TGF-β-treated cells markedly decreased the expression of α-SMA and calponin (Figure 3B and C), although the levels of the representative EMT marker, E-cadherin, were repressed by TGF-β and unaffected by addition of FGF-2 (Figure 3C; Supplementary Figure S1C). Inhibition of FGF-2 signalling by SU5402 or short interfering RNA (siRNA)-mediated knockdown of FGFR1IIIc by its specific siRNAs did not obviously alter the expression of E-cadherin regulated by TGF-β (Figure 3C; Supplementary Figure S1D), suggesting that autonomously secreted ligands to FGFR1IIIc exhibit only limited effects on TGF-β-induced EMT. In addition, these findings were not restricted to NMuMG cells, and were also observed in another epithelial cell line α-TN4 (Supplementary Figure S2A and B). These findings indicate that prolonged TGF-β treatment induces the differentiation of epithelial cells into myofibroblastic cells, and that FGF-2 inhibits the TGF-β-mediated EMyoT.
Figure 3.
Prevention of EMyoT by FGF-2. (A) Immunohistochemical analysis was performed with anti-α-SMA antibody (green) and propidium iodide to detect nuclei (red) in cells treated with 1 ng/ml TGF-β for 3 days and 13 days. (B) Cells were treated with either 1 ng/ml TGF-β or 30 ng/ml FGF-2 alone, or both and then examined for the induction of α-SMA (left) and calponin (right) at the indicated time points by quantitative RT–PCR analysis. Each value represents the mean±s.d. of duplicate determinations from a representative experiment. Similar results were obtained at least three independent experiments. (C) Protein levels of endogenous E-cadherin and α-SMA were analysed by immunoblotting 4 days after treating with combinations of TGF-β, FGF-2, and 10 μM SU5402. α-Tubulin levels were monitored as a loading control for the whole-cell extracts. (D) Phase-contrast micrographs (upper panels) and confocal images after anti-α-SMA antibody staining (green in lower panels) of NMuMG cells. The ligand and inhibitor concentrations were as follows: 1 ng/ml TGF-β, 30 ng/ml FGF-2, 10 μM SU5402, 1 μM PI-103, 1 μM U73122, and 10 μM U0126. Scale bar indicates 100 μm. (E, F) Immunoblot analyses were performed with lysates from NMuMG cells treated with the indicated combinations of ligands and inhibitors. α-Tubulin was used as a loading control (E). The ligands and inhibitors were incubated in culture media containing 10% FBS for 4 days (E), and 2 days (2d in F) or 4 days (4d in F). The same reagent concentrations were used as described in (D). F, FGF-2; Tβ, TGF-β; SU, SU5402; PI, PI-103; U7, U73122; U0, U0126.
FGF stimulation leads to tyrosine phosphorylation of FGFR, the recruitment of multiple complexes with adaptor proteins, and activation of the MEK-Erk, phosphoinositide 3 kinase (PI3K) and phosphatidylinositol-specific phospholipase C (PLC) γ pathways. To determine how FGF-2 inhibits TGF-β-mediated EMyoT, we used several inhibitors, including SU5402 for FGFR1, U0126 for MEK 1 and 2, PI-103 for PI3K, and U73122 for PLC. Among these inhibitors, SU5402 and U0126 clearly inhibited the effects of FGF-2 and restored TGF-β-induced α-SMA expression and cell morphology, while neither PI-103 nor U73122 significantly affected these cells (Figure 3D and E). More importantly, TGF-β reduced the levels of Erk phosphorylation, which was accompanied by a reciprocal induction of α-SMA, whereas the addition of FGF-2 returned the Erk phosphorylation levels to baseline (Figure 3F). The cells expressing α-SMA had weaker Erk phosphorylation compared with α-SMA-negative cells, as determined by immunohistochemical analyses (Supplementary Figure S2C). Thus, these findings suggest that Erk inactivation by TGF-β results in α-SMA induction and that restored Erk activation by FGF-2 is required to prevent the induction of α-SMA by TGF-β.
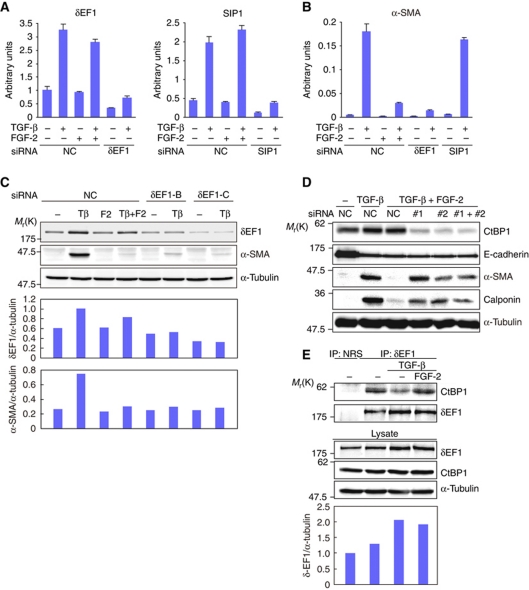
Prevention of α-SMA induction by the CtBP1–δEF1 interaction
We further examined how FGF-2-induced Erk activation suppressed the induction of α-SMA by TGF-β. Based on a previous report and our previous study (Nishimura et al, 2006; Shirakihara et al, 2007), we focused on the δEF1 family proteins, δEF1/ZEB1 and SIP1/ZEB2. After confirming that specific siRNAs successfully knocked down δEF1 and SIP1 at both the mRNA (Figure 4A) and protein levels (Shirakihara et al, 2007), we found that SIP1 siRNA did not affect the induction of α-SMA (Figure 4B). However, in cells transfected with δEF1 siRNAs, TGF-β-mediated α-SMA upregulation was significantly reduced to a level similar to that obtained with the addition of FGF-2 (Figure 4B and C). More interestingly, FGF-2 did not affect the expression of δEF1 at either the mRNA or protein level (Figure 4A and lower panels in Figure 4E), nor did it affect the expression of microRNA-200b that directly targets δEF1 (Supplementary Figure S2D) (Gregory et al, 2008). These findings indicate that δEF1 is required for the TGF-β-mediated induction of α-SMA and that FGF-2 reduces α-SMA expression without affecting the levels of δEF1 that are upregulated by TGF-β. Thus, the function of δEF1 is regulated by mechanisms other than those involved in regulating transcription or protein stability.
Figure 4.
Interaction of CtBP1 with δEF1 to prevent α-SMA induction. (A, B) NMuMG cells transfected with either δEF1 siRNA or SIP1 siRNA were stimulated with or without 1 ng/ml TGF-β, 30 ng/ml FGF-2, or both for 24 h and then examined by quantitative RT–PCR analysis for the expression levels of δEF1 (A, left), SIP1 (A, right), and α-SMA (B). NC, control siRNA. Each value represents the mean±s.d. of duplicate determinations from a representative experiment. Similar results were obtained at least three independent experiments. (C) After transfection with different types of siRNAs against δEF1 (δEF1-B and δEF1-C), α-SMA induction by TGF-β was evaluated by immunoblot analysis. α-Tubulin was used as a loading control. The ratio of δEF1 or α-SMA to α-tubulin was validated by densitometric analysis and shown at the lower panels. NC, control siRNA; Tβ, TGF-β; F2, FGF-2. (D) NMuMG cells transfected with control siRNA (NC) or two different types of CtBP1 siRNAs (#1 and #2) were stimulated with TGF-β, FGF-2, or both for 60 h as indicated. After measuring the protein concentrations, the cell lysates were examined by immunoblot analysis. α-Tubulin was used as a loading control. (E) After NMuMG cells were incubated in culture media containing 10% FBS for 4 days with the indicated ligands, cell lysates from ∼1 × 106 cells were examined for protein concentrations and then subjected to immunoprecipitation with normal rabbit IgG (NRS) or an anti-δEF1 antibody. Copurified CtBP1 was detected by immunoblotting with an anti-CtBP1 antibody (top panel). The expression levels of δEF1, CtBP1, and α-tubulin in the same lysates were verified by immunoblotting (lower three panels). The ratio of δEF1 to α-tubulin was validated by densitometric analysis and shown at the bottom.
The transcriptional function of δEF1 is regulated through interactions with CtBP1, C-terminal binding protein 1 (Chinnadurai, 2009; Vandewalle et al, 2009). We also found that the inhibitory effect of FGF-2 on TGF-β-induced α-SMA expression was abolished by knocking down CtBP1, resulting in restored α-SMA expression, whereas E-cadherin repression by TGF-β was not affected (Figure 4D). Moreover, endogenous CtBP1 coimmunoprecipitated with endogenous δEF1 (Figure 4E). TGF-β treatment reduced the interaction between CtBP1 and δEF1, while the addition of FGF-2 restored this interaction to a level similar to that in untreated cells (Figure 4E). Inhibition of the MEK-Erk pathway with U0126 and PD98059 decreased this interaction compared with that in the cells treated with TGF-β and FGF-2 (Supplementary Figure S3A). To further define this interaction, we transiently transfected HEK-293T cells with expression plasmids encoding Myc-tagged δEF1 and FLAG-tagged CtBP1. The interaction between δEF1 and CtBP1 was confirmed in overexpressing HEK-293T cells (Supplementary Figure S3B). A CtBP1 mutant that harbours mutations in conserved sites putatively phosphorylated by MAP kinases also interacted with δEF1. This interaction was enhanced by FGF-2 stimulation and repressed by U0126 (Supplementary Figure S3B). In addition, FLAG-CtBP1 that was immunoprecipitated from FGF-2-treated cells exhibited an enhanced interaction with Myc-δEF1 compared with that from non-treated cells in an in vitro binding assay (Supplementary Figure S3C). To examine whether the association between CtBP1 and δEF1 was regulated by Erk kinase-mediated phosphorylation of CtBP1, we used alkaline phosphatase, which is widely used in in vitro dephosphorylation assays. Dephosphorylation of immunoprecipitated FLAG-CtBP1 by alkaline phosphatase slightly decreased its ability to interact with Myc-δEF1 (Supplementary Figure S3C). Thus, these findings indicate that the interaction between CtBP1 and δEF1 is highly dependent on MEK-Erk activation and that CtBP1 phosphorylation slightly enhances its ability to interact with δEF1. Therefore, TGF-β upregulates the expression of δEF1 and inactivates the MEK-Erk pathway to facilitate the dissociation of δEF1 from CtBP1, which induces α-SMA expression. Conversely, the addition of FGF-2 to TGF-β-treated cells activates the MEK-Erk pathway to enhance the interaction between δEF1 and CtBP1, which represses α-SMA induction.
Differentiation of epithelial cells to myofibroblastic cells from fibroblastic cells through TGF-β-induced EMyoT was highly dependent on δEF1 (Figure 4). To elucidate the molecular mechanism by which fibroblastic cells differentiate to activated fibroblastic cells by combination of TGF-β with FGF-2, we performed DNA microarray analysis and knocked down δEF1 and CtBP1 using their specific siRNAs. During this process, FGF-2 altered the broad transcriptional program mediated by TGF-β. Among them, Twist, a representative of EMT regulators, was included in the program and remarkably upregulated by combination of TGF-β and FGF-2 (Supplementary Figure S4A). In addition, siRNA-mediated knockdown of δEF1 did not affect the migratory behaviour of cells (Supplementary Figure S4B), indicating that δEF1 is dispensable for differentiation into activated fibroblastic cells. Conversely, silencing CtBP1 in cells treated with FGF-2 and TGF-β affected some properties, including the induction of α-SMA and calponin, cell migration and cell invasion (see Figure 4D and Supplementary Figure S4C and D), while MMP9 activity and E-cadherin expression were not affected by CtBP1 siRNA (see Figure 4D and Supplementary Figure S4E). These findings thus suggest that CtBP1 mediates some properties of EMT induced by TGF-β and FGF-2, and that other EMT regulators following TGF-β-mediated upregulation of δEF1 may be important to differentiate into activated fibroblastic cells.
Effects of cells undergoing EMT by TGF-β and FGF-2 on the invasion of cancer cells
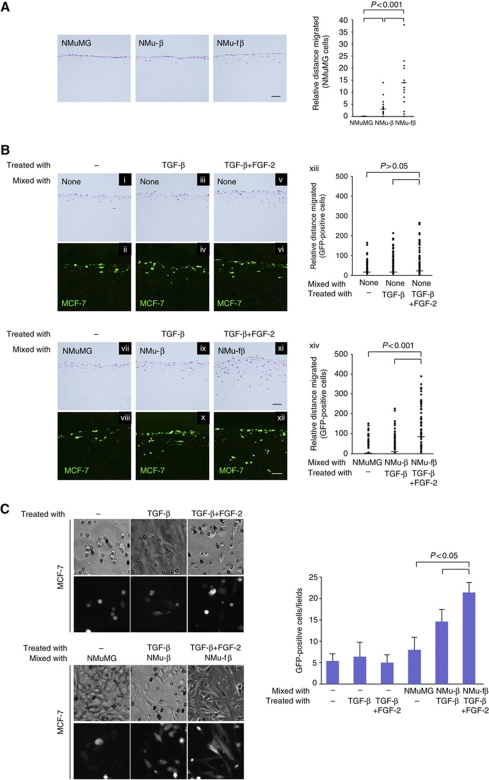
TGF-β induced differentiation of epithelial NMuMG cells into two types of mesenchymal cells: one was α-SMA-positive myofibroblastic cells (thereafter denoted NMu-β cells) generated through EMyoT by TGF-β alone, and the other was activated fibroblastic cells (NMu-fβ cells) generated through EMT by both TGF-β and FGF-2. To determine the biological functions of these cells, the cells were mixed with human breast cancer MCF-7 cells expressing green fluorescent protein (MCF-7-GFP) and seeded on collagen gels to examine the invasive properties of the cancer cells (Margulis et al, 2005; Higashikawa et al, 2007; Ikebe et al, 2007; Horiguchi et al, 2009). In cultures containing NMuMG cells alone, TGF-β slightly enhanced the invasive properties of the cells compared with non-treated cells due to changes in the NMu-β cells, and the addition of FGF-2 and TGF-β further promoted the invasion of the cells due to changes in NMu-fβ cells (Figure 5A). On the other hand, MCF-7-GFP cells seeded on collagen gels with or without NMuMG cells spontaneously invaded into the collagen (Figure 5B-i, ii, vii, and viii). This invasion was slightly, but not significantly, enhanced in MCF-7-GFP cells treated with TGF-β and TGF-β+FGF-2 (Figures 5B-iii, iv, v, and vi). When MCF-7-GFP cells were mixed with NMu-β cells and treated with TGF-β, their invasion was slightly enhanced compared with those mixed with NMuMG cells (Figure 5B-ix and x). More interestingly, MCF-7-GFP cells that were mixed with NMu-fβ cells and treated with TGF-β+FGF-2 invaded more deeply into the collagen gels (Figure 5B-xi and xii). Under these conditions, proliferation of MCF-7 was suppressed in the presence of TGF-β, as determined by immunostaining with anti-Ki67 antibody (Supplementary Figure S5), suggesting that enhanced invasive behaviour does not result from cell proliferation of MCF-7 cells. When MCF-7 cells were cocultured with NMu-fβ cells and treated with TGF-β and FGF-2, the cells exhibited spindle shapes with enhanced invasive behaviour, which was repressed by CtBP1 siRNA (Figure 5C; Supplementary Figure S4F). Therefore, the cells that have undergone EMT by TGF-β and FGF-2 promoted invasion of MCF-7 cells more potently than the α-SMA-positive cells that were generated by TGF-β-induced EMyoT.
Figure 5.
Effects of TGF-β and FGF signalling on cancer cell invasion. (A) NMuMG cells were pretreated with 1 ng/ml TGF-β (NMu-β) or both 1 ng/ml TGF-β and 30 ng/ml FGF-2 (NMu-fβ) for 2 days and then seeded on collagen gels. After 1 week, the gels were fixed, embedded in paraffin, and stained with HE. Bars represent the median for each category from a representative experiment. Similar results were obtained in two independent experiments. Scale bar indicates 100 μm. (B) MCF-7-GFP cells were seeded on a collagen gel and treated with 1 ng/ml TGF-β or 30 ng/ml FGF-2+TGF-β (i–vi). NMuMG cells were pretreated with 1 ng/ml TGF-β (NMu-β cells) or 30 ng/ml FGF-2 together with TGF-β (NMu-fβ cells) for 2 days, and mixed with MCF-7-GFP cells in the presence or absence of TGF-β and TGF-β+FGF-2 (vii–xii). After 7 days, the gel was fixed and stained with HE (i, iii, v, vii, ix, xi). GFP was detected by confocal laser scanning microscopy (ii, iv, vi, viii, x, xii). Scale bars indicate 100 μm. (xiii, xiv) Statistical analyses were performed using a non-parametric Mann–Whitney U test. The median migration distance of cells was significantly different at P>0.05 between the two groups of MCF-7 cells (non-treatment versus TGF-β+FGF-2 treatment, and TGF-β treatment versus TGF-β+FGF-2 treatment) and at P<0.001 between the two groups of MCF-7 cells mixed with the indicated cells (NMuMG cells versus NMu-fβ cells and NMu-β cells versus NMu-fβ cells). Bars represent the median for each category from a representative experiment. (C) NMuMG cells were cultured for 4 days in the absence or presence of TGF-β or both TGF-β and FGF-2, and then seeded into collagen gel-loaded culture inserts with MCF-7-GFP cells, which had been cocultured in cell culture inserts with NMuMG cells. The GFP-positive cells that had invaded to the opposite side were fixed and photographed (left), and typical morphology of the cells were counted and quantified (right).
Discussion
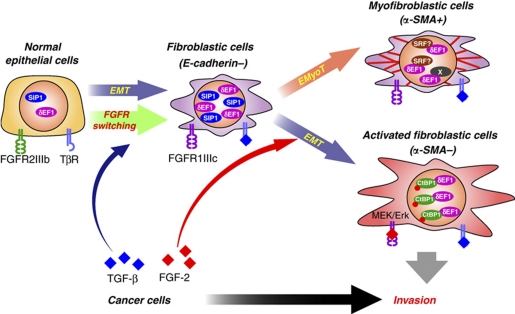
In the present study, we found that TGF-β primes FGFR isoform switching and generates fibroblastic cells by EMT, and that the fibroblastic cells have a potential to differentiate to myofibroblastic cells through EMyoT in the presence of TGF-β and to activated fibroblastic cells through EMT in the presence of FGF-2 and TGF-β (Figure 6). During these processes, δEF1 has critical roles in determining the direction of the differentiation. First, transcriptional upregulation of δEF1 and SIP1 by TGF-β is essential to generate ‘fibroblastic cells' through EMT that is incomplete, as we previously reported (Shirakihara et al, 2007). Second, increased levels of δEF1 are required to induce α-SMA during the differentiation into ‘myofibroblastic cells' through EMyoT. Third, FGF-2 enhances TGF-β-induced EMT and generates ‘activated fibroblastic cells' by promoting the formation of complexes between δEF1 and CtBP1 phosphorylated through the FGF-2-activated MEK-Erk pathway (Figure 6). The activated fibroblastic cells generated by FGF-2 in combination with TGF-β promote the invasion of cancer cells. Thus, TGF-β and FGF-2, which are abundantly present in tumours, may regulate not only the EMT of cancer cells as they acquire metastatic properties but also that of normal epithelial cells as they promote the invasion of cancer cells.
Figure 6.
Schematic illustration of EMT induction by TGF-β and FGF-2. ‘Epithelial cells' differentiate into ‘fibroblastic cells' through EMT induced by TGF-β, and further differentiate into α-SMA-positive ‘myofibroblastic cells' through EMyoT. When FGF-2 is present in this process, FGF-2 induces the differentiation of (myo)fibroblastic cells to ‘activated fibroblastic cells'. The red dot in CtBP1 indicates phosphorylation.
Isoform switching of FGFRs during TGF-β-induced EMT
In the present study, we found that NMuMG cells were sensitive to FGF-7 because they expressed the cognate receptor FGFR2IIIb, whereas TGF-β-pretreated NMuMG cells became sensitive to FGF-2 as FGFR2IIIb was replaced to FGFR1IIIc (Figure 1). The molecular mechanism by which TGF-β controls isoform switching has not been elucidated. Recently, it was reported that RNA-binding proteins, ESRP1 and ESRP2, coordinate to promote a cell-type specific splicing program, which results in isoform switching from FGFR2IIIb to FGFR2IIIc (Warzecha et al, 2009). However, during TGF-β-induced EMT, FGFR2IIIb changed to FGFR1IIIc but not to FGFR2IIIc. Thus, in addition to ESRPs, other transcriptional factors may be involved in the process of TGF-β-mediated isoform switching. Moreover, the addition of FGF-2 to TGF-β-treated cells resulted in sustained expression of FGFR1IIIc at higher levels (Figure 1C). Previous studies reported that FGF-2 induced EMT in some cells through the expression of Snail (de Frutos et al, 2007), and that FGFR1IIIc expression was associated with EMT and correlated with the progression of several advanced cancers (Acevedo et al, 2007). As TGF-β is frequently overexpressed in cancer cells, FGFR1IIIc expression and the effects of FGF-2 during cancer progression may be regulated by TGF-β that is autonomously secreted from cancer cells. Therefore, inhibition of TGF-β signalling may ameliorate cancer progression through FGF-mediated EMT, and thus may be a useful therapeutic target.
Formation of δEF1–CtBP1 complexes by the MEK-Erk pathway
It has been reported that CtBP1 is a corepressor for δEF1 (Chinnadurai, 2009). In the present study, we found that the interaction between δEF1 and CtBP1 was enhanced by FGF-2 and decreased by inhibiting the MEK-Erk pathway (Figure 4E; Supplementary Figure S3B). CtBP1 is localized in the nucleus of the cells. In addition, in vitro dephosphorylation of immunoprecipitated CtBP1 by alkaline phosphatase treatment slightly decreased its interaction with Myc-δEF1 (Supplementary Figure S3C), suggesting that phosphorylation of CtBP1 is partially involved in the interaction with δEF1. It has been reported that PKA and JNK phosphorylate CtBP1 at threonine 144 and serine 422, respectively (Wang et al, 2006; Dammer and Sewer, 2008). Moreover, we identified another consensus sequence (S/T-P site) at threonine 311 in human CtBP1 that is putatively phosphorylated by Erk kinase, and this site is also conserved in mouse CtBP1 and in the corresponding site of CtBP2. When these putative phosphorylation sites in CtBP1 were mutated to alanine residues, the mutations did not affect the interaction between CtBP1 and δEF1 (Supplementary Figure S3B). Recently, novel phosphorylation sites were identified by mass spectrometry (Merrill et al, 2010). Thus, in addition to the phosphorylation sites of 311 and 422, we mutated these phosphorylation sites and have performed coimmunoprecipitation assay. However, FGF-2-dependent complex formation between δEF1 and CtBP1 mutants with alteration of these phosphorylation sites (100, 144, 158, and 176) was not considerably altered, compared with that between δEF1 and CtBP1 (Supplementary Figure S3D). These findings suggest that phosphorylation of CtBP1 at unidentified site(s) by the MEK-Erk pathway regulates this interaction. Alternatively, other MEK-Erk-dependent post-translational modifications of CtBP1, such as methylation, oxidation, and acetylation, which are inhibited by U0126, may regulate the association with δEF1.
δEF1 has multiple consensus sites for phosphorylation by Erk kinases (Costantino et al, 2002), and the bands corresponding to δEF1 by SDS–PAGE migrated faster after in vitro dephosphorylation by alkaline phosphatase (see top right panel in Supplementary Figure S3E). In addition, δEF1 was acetylated in the CtBP1 interaction domain to regulate CtBP1 binding (Postigo et al, 2003). Thus, although in vitro dephosphorylation of δEF1 did not affect its ability to bind to CtBP1 (Supplementary Figure S3E), the possibility that post-translational modification of δEF1 regulates the interaction with CtBP1 could not be completely excluded.
Although the formation of δEF1–CtBP1 complexes inhibited the induction of α-SMA, the molecular mechanism by which TGF-β induces α-SMA was not fully determined in the present study. It was previously reported that δEF1 has important roles in the TGF-β-mediated induction of α-SMA in vascular smooth muscle cells. δEF1 physically interacted with SRF, serum response factor, and these complexes synergistically activated α-SMA expression by directly binding to the promoter of the α-SMA gene (Nishimura et al, 2006). We, however, found that δEF1 did not form a complex with SRF, and TGF-β repressed the protein levels of SRF in NMuMG cells (Supplementary Figure S3F). It appears likely that CtBP1 constitutively binds to δEF1 and suppresses the transcriptional regulatory function of δEF1 under unstimulated conditions, whereas TGF-β upregulates δEF1 expression levels and reduces CtBP1 modifications by inactivating the MEK-Erk pathway, leading to the dissociation of CtBP1 from δEF1. Subsequently, δEF1 can regulate α-SMA induction in association with SRF or unidentified protein(s) (Figure 6). The α-SMA-promoter construct responded to TGF-β stimulation as well as δEF1 overexpression in reporter assays (Supplementary Figure S3G). CtBP1 partially inhibited this α-SMA-reporter activity induced by δEF1, while CtBP1 failed to inhibit the activity induced by δEF1 mutant that does not bind to CtBP1 (Furusawa et al, 1999). Thus, δEF1 acts as an activator or derepressor, and is indispensable for α-SMA induction by TGF-β in NMuMG cells. However, it should be determined in the future whether δEF1 acts as an activator or derepressor of α-SMA transcription.
Differentiation of normal epithelial cells by cancer cells in a cocultivation system
In cancer microenvironments, both TGF-β and FGF-2 are frequently secreted from advanced cancer cells and abundantly expressed in tumour tissues (Memarzadeh et al, 2007; Massague, 2008; Padua and Massague, 2009). We found that cocultivating normal epithelial cells with mouse breast cancer JygMC(A) cells, which secrete TGF-β, led to EMyoT of epithelial cells, accompanied by the induction of α-SMA (Supplementary Figure S6A and B), while cocultures with mouse breast cancer 4T1 cells, which express higher levels of FGF-2 mRNA than JygMC(A) cells (Supplementary Figure S6C), failed to yield α-SMA-positive NMuMG cells (Supplementary Figure S6B). Both breast cancer cells autonomously secreted TGF-β ligands. Therefore, these findings indicate that TGF-β secreted from cancer cells induces the differentiation of normal epithelial cells into α-SMA-expressing cells through EMyoT, a change that is suppressed by FGFs secreted from cancer cells.
Cells that are located in or around tumour tissues and express myofibroblast markers, including α-SMA, calponin, and desmin, are known as cancer-associated fibroblasts (CAFs) and have an important role in cancer progression (Mueller and Fusenig, 2004; Orimo et al, 2005; Ostman and Augsten, 2009). Thus, EMyoT mediated by cancer cells may help generate CAFs. More interestingly, NMu-fβ cells did not express myofibroblast markers but promoted the invasion of MCF-7 cells much more potently than NMu-β cells expressing myofibroblast markers. Expression of MMP9, but not that of MMP2, in MCF-7 cells was upregulated in the cells cocultured with NMu-fβ cells, compared with that in MCF-7 cells alone (Supplementary Figure S6D). However, the increase in MMP9 expression may not be sufficient to explain the phenomena. Thus, the mechanism responsible for this process has not been determined, and specific protein markers of NMu-fβ cells remain to be identified. Cancer cell invasion was recently classified into two categories, so-called ‘collective cancer cell invasion' and ‘fibroblast-led collective invasion' (Gaggioli et al, 2007). The former is a phenomenon in which cancer cells invade by themselves, while the latter highlights the importance of certain fibroblasts that initiate a collective invasion chain of cancer cells. Although the specific fibroblasts associated with ‘fibroblast-led collective invasion' have not been identified, NMu-fβ cells may function as these types of fibroblasts. Therefore, fibroblastic and myofibroblastic cells generated from epithelial cells adjacent to tumours (cancer-associated epithelial cells) by EMT and EMyoT, respectively, might have a unique role in cancer progression that is distinct from those of ordinary fibroblasts.
Materials and methods
Cell culture
All cells used in the present study were from the American Type Culture Collection (Manassas, VA). The cell media and culture conditions were previously described (Shirakihara et al, 2007). NMuMG cells were cultured for several days and passaged every 2 days in medium with or without 1 ng/ml TGF-β1, 30 ng/ml FGF-2, and 100 μg/ml heparin. All cells were grown in a 5% CO2 atmosphere at 37°C.
Reagents and antibodies
Recombinant human TGF-β1, FGF basic (FGF-2), and FGF-7 were purchased from R&D Systems (Minneapolis, MN). SU5402, PI-103, U73122, and GM6001 were purchased from Calbiochem (Darmstadt, Germany). U0126 was obtained from Promega (Madison, WI). The mouse monoclonal anti-α-tubulin and anti-α-SMA antibodies were purchased from Sigma-Aldrich. The mouse monoclonal anti-Erk2 antibody was from Millipore (Bedford, MA). The rabbit polyclonal anti-phospho-Erk1/2 antibody was obtained from Cell Signaling (Beverly, MA). The rabbit polyclonal anti-δEF1 antibody was from Santa Cruz Biotechnology (Lot No. E1408, Santa Cruz, CA), and the mouse monoclonal anti-CtBP1 and anti-E-cadherin antibodies were from BD Transduction Laboratory (Lexington, KY). Rabbit monoclonal anti-calponin antibody was from Abcam (Cambridge, UK). Tetramethylrhodamine isothiocyanate (TRITC)-conjugated phalloidin and propidium iodide (PI) were from Sigma-Aldrich (Saint Louis, MO) and Molecular Probes (Eugene, OR), respectively.
Immunoprecipitation, immunoblotting, and immunofluorescence labelling
The procedures used for immunoprecipitation, immunoblotting, and immunofluorescence were previously described (Shirakihara et al, 2007). Immunodetection was performed with the ECL blotting system (Amersham Bioscience, Piscataway, NJ) and Luminescent Image Analyzer (LAS4000, Fujifilm, Tokyo, Japan). Fluorescence was examined using a confocal laser scanning microscope (Carl Zeiss, Thornwood, NY).
RNA interference
siRNAs were transfected into cells according to the protocol recommended for HiPerFect reagent (Qiagen, Valencia, CA). NMuMG cells were transiently transfected with siRNAs against mouse δEF1 (Stealth RNAi; Invitrogen, Carlsbad, CA), mouse SIP1 (GGAAAAACGUGGUGAACUA; B-Bridge, Sunnyvale, CA), or mouse CtBP1 (Stealth RNAi; Invitrogen). The final concentrations of the siRNAs were 5 nM, except for SIP1 siRNA, which had a final concentration of 10 nM.
RNA extraction and RT–PCR
Total RNA was extracted and analysed by quantitative RT–PCR analysis as previously described (Shirakihara et al, 2007). The specificity of the detected signals was confirmed by a dissociation curve, which consisted of a single peak. Values were normalized to the TATA-binding protein (TBP). Conventional RT–PCR analyses were performed using the PC-708 Program Temp Control System (ASTEC, Fukuoka, Japan). The RT–PCR primer sequences are listed in Supplementary Table S1.
Cell motility assay
NMuMG cells were treated with TGF-β, FGF-2, or both for 4 days and seeded in six-well tissue culture plates. The cell monolayers were wounded by scratching with sterile plastic 200 μl micropipette tips and photographed using phase-contrast microscopy immediately and 12 h after wounding. The assays were independently performed in triplicate. The migration distance of each cell was measured after the photographs were converted to Photoshop files (Adobe, San Jose, CA).
Invasion assay
NMuMG cells were treated with TGF-β or both TGF-β and FGF-2 for 4 days and seeded onto Cell Culture Inserts (8 μm pore size; BD Falcon, Franklin Lakes, NJ) coated with Type IV collagen (BD Biosciences, San Jose, CA). After 24 h, the cells that had not invaded into the lower surface of the filters were removed from the upper face of the filters using cotton swabs. The cells that had invaded into the lower surface of the filters were fixed in methanol and stained with Giemsa. Invasion was quantitated by visually counting the photographed cells.
Collagen gel contraction assay
Type I collagen gel was prepared using an 8:1:1 ratio of cold collagen solution (Cellmatrix I-P; Nitta Gelatin, Osaka, Japan), 10 × concentrated MEM (Invitrogen), and collagen dilution buffer containing 0.05 N NaOH, 2.2% NaHCO3, and 200 mM HEPES, pH 7.4. NMuMG cell suspensions (2.0 × 105 cells/200 μl) were mixed in 800 μl of the collagen gel solution. A measure of 1 ml of the mixture was added to each well of 12-well culture plates and allowed to solidify at 37°C for 30 min. After solidification, 1 ml of DMEM containing 4.5 g/l glucose, 10% FBS, 10 μg/ml insulin, 50 U/ml penicillin, and 50 μg/ml streptomycin was overlaid to float the gel. The floating gels were incubated at 37°C in 5% CO2 for 2 days. The gel surface area was quantified based on pixel number using ImageJ (US National Institutes of Health, Bethesda, MD). The relative changes in the surface area are reported as the percentage of the original surface area.
Gelatin zymography
NMuMG cells were treated with TGF-β, FGF-2, or both for 4 days. Then, the cells were cultured with serum-free media for 24 h, and the conditioned media were collected. Equal amounts of samples were mixed with 3 × non-reducing SDS–PAGE sample buffer. The samples were applied to a 10% polyacrylamide gel impregnated with 1 mg/ml gelatin (Sigma). After running, SDS was removed from the gel by washing three times with 2.5% Triton X-100 solution for 20 min. Then, the gels were incubated overnight at 37°C in buffer containing 50 mM Tris–HCl, pH 7.6, 5 mM CaCl2, and 200 mM NaCl. The gel was stained with 0.5% Coomassie blue R250 (CBB) in 50% methanol and 5% acetic acid for 2 h, and subsequently destained with a solution containing 40% methanol and 10% acetic acid.
In vitro three-dimensional invasion assay
A mixture of collagen I (Koken, Tokyo, Japan) and DMEM containing 10% FBS, 10 mM HEPES, and 0.1% sodium bicarbonate were added to 12-well culture plates. The collagen gel was allowed to solidify at 37°C for 1 h. The final concentration of collagen gel was 2.4 mg/ml. MCF-7 cells stably expressing Ha-ras G12V were infected with lentivirus encoding green fluorescent protein (MCF-7-GFP). NMuMG cells were pretreated with TGF-β or TGF-β and FGF-2 for 2 days and then mixed with MCF-7-GFP cells and seeded on collagen gels. The culture media containing each of the noted ligands was changed every 2 days. After 1 week, the gels were fixed with Mildform (Wako, Osaka, Japan), embedded in paraffin, and stained with haematoxylin–eosin (HE). The same sections were used to detect GFP by confocal laser scanning microscopy.
Online supplemental material
Supplementary Figure S1 shows the effects of FGF-2 on TGF-β signalling. Supplementary Figure S2 shows that FGF-2 prevents EMyoT of α-TN4 cells, and Erk phosphorylation and miR-200b expression in NMuMG cells. Supplementary Figure S3 shows the interaction between CtBP1 and δEF1. Supplementary Figure S4 shows the role of δEF1 in EMT. Supplementary Figure S5 shows cell proliferation of MCF-7 cocultured with NMuMG cells in collagen gel. Supplementary Figure S6 shows that TGF-β and FGF-2 are produced by cancer cells. Supplementary Table S1 lists the primers used in this study.
Supplementary Material
Acknowledgments
We are grateful to M Fujiwara, A Hanyu, and Y Morishita for technical assistance. We thank Drs K Higashikawa, K Tobiume, K Sakamoto, C Iwata, K Nakahama, H Ichijo, K Takeda, N Takemori, and T Imamura for their advice and helpful discussions. We also thank Dr M Nakamura and all the members of the Human Gene Sciences Center of Tokyo Medical and Dental University for consistently supporting this study. This work was supported by KAKENHI (Grants-in-Aid for Scientific Research) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Author contributions: TS performed the experiments, analysed the data, and wrote the paper. KH and SE performed the experiments. KM, TS, and IM wrote the paper. KM and MS designed the research and wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Acevedo VD, Gangula RD, Freeman KW, Li R, Zhang Y, Wang F, Ayala GE, Peterson LE, Ittmann M, Spencer DM (2007) Inducible FGFR-1 activation leads to irreversible prostate adenocarcinoma and an epithelial-to-mesenchymal transition. Cancer Cell 12: 559–571 [DOI] [PubMed] [Google Scholar]
- Acloque H, Adams MS, Fishwick K, Bronner-Fraser M, Nieto MA (2009) Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. J Clin Invest 119: 1438–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnadurai G (2009) The transcriptional corepressor CtBP: a foe of multiple tumor suppressors. Cancer Res 69: 731–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantino ME, Stearman RP, Smith GE, Darling DS (2002) Cell-specific phosphorylation of Zfhep transcription factor. Biochem Biophys Res Commun 296: 368–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammer EB, Sewer MB (2008) Phosphorylation of CtBP1 by cAMP-dependent protein kinase modulates induction of CYP17 by stimulating partnering of CtBP1 and 2. J Biol Chem 283: 6925–6934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Frutos CA, Vega S, Manzanares M, Flores JM, Huertas H, Martinez-Frias ML, Nieto MA (2007) Snail1 is a transcriptional effector of FGFR3 signaling during chondrogenesis and achondroplasias. Dev Cell 13: 872–883 [DOI] [PubMed] [Google Scholar]
- Eswarakumar VP, Lax I, Schlessinger J (2005) Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev 16: 139–149 [DOI] [PubMed] [Google Scholar]
- Foster BA, Kaplan PJ, Greenberg NM (1999) Characterization of the FGF axis and identification of a novel FGFR1IIIc isoform during prostate cancer progression in the TRAMP model. Prostate Cancer Prostatic Dis 2: 76–82 [DOI] [PubMed] [Google Scholar]
- Furusawa T, Moribe H, Kondoh H, Higashi T (1999) Identigication of CtBP1 and CtBP2 as corepressors of zinc finger-homeodomain factor δEF1. Mol Cell Biol 19: 8581–8590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaggioli C, Hooper S, Hidalgo-Carcedo C, Grosse R, Marshall JF, Harrington K, Sahai E (2007) Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat Cell Biol 9: 1392–1400 [DOI] [PubMed] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ (2008) The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 10: 593–601 [DOI] [PubMed] [Google Scholar]
- Higashikawa K, Yoneda S, Tobiume K, Taki M, Shigeishi H, Kamata N (2007) Snail-induced down-regulation of ΔNp63α acquires invasive phenotype of human squamous cell carcinoma. Cancer Res 67: 9207–9213 [DOI] [PubMed] [Google Scholar]
- Horiguchi K, Shirakihara T, Nakano A, Imamura T, Miyazono K, Saitoh M (2009) Role of Ras signaling in the induction of snail by transforming growth factor-β. J Biol Chem 284: 245–253 [DOI] [PubMed] [Google Scholar]
- Ikebe D, Wang B, Suzuki K, Kato M (2007) Suppression of keratinocyte stratification by a dominant negative JunB mutant without blocking cell proliferation. Gens Cells 12: 197–207 [DOI] [PubMed] [Google Scholar]
- Kalluri R (2009) EMT: when epithelial cells decide to become mesenchymal-like cells. J Clin Invest 119: 1417–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R, Weinberg RA (2009) The basics of epithelial-mesenchymal transition. J Clin Invest 119: 1420–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M, Cubillo E, Tobiume K, Shirakihara T, Fukuda N, Suzuki H, Shimizu K, Takehara K, Cano A, Saitoh M, Miyazono K (2004) A role for Id in the regulation of TGF-β-induced epithelial-mesenchymal transdifferentiation. Cell Death Differ 11: 1092–1101 [DOI] [PubMed] [Google Scholar]
- Margulis A, Margulis A, Zhang W, Alt-Holland A, Crawford HC, Fusenig NE, Garlick JA (2005) E-cadherin suppression accelerates squamous cell carcinoma progression in three-dimensional, human tissue constructs. Cancer Res 65: 1783–1791 [DOI] [PubMed] [Google Scholar]
- Massague J (2008) TGF-β in cancer. Cell 134: 215–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memarzadeh S, Xin L, Mulholland DJ, Mansukhani A, Wu H, Teitell MA, Witte ON (2007) Enhanced paracrine FGF10 expression promotes formation of multifocal prostate adenocarcinoma and an increase in epithelial androgen receptor. Cancer Cell 12: 572–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill JC, Kagey MH, Melhuish TA, Powers SE, Zerlanko BJ, Wotton D (2010) Inhibition of CtBP1 activity by Akt-mediated phosphorylation. J Mol Biol 21: 657–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikko M, Fredriksson K, Wahlstrom J, Eriksson P, Grunewald J, Skold CM (2008) Human T cells stimulate fibroblast-mediated degradation of extracellular matrix in vitro. Clin Exp Immunol 151: 317–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustakas A, Heldin CH (2007) Signaling networks guiding epithelial-mesenchymal transitions during embryogenesis and cancer progression. Cancer Sci 98: 1512–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller MM, Fusenig NE (2004) Friends or foes—bipolar effects of the tumour stroma in cancer. Nat Rev Cancer 4: 839–849 [DOI] [PubMed] [Google Scholar]
- Nishimura G, Manabe I, Tsushima K, Fujiu K, Oishi Y, Imai Y, Maemura K, Miyagishi M, Higashi Y, Kondoh H, Nagai R (2006) δEF1 mediates TGF-β signaling in vascular smooth muscle cell differentiation. Dev Cell 11: 93–104 [DOI] [PubMed] [Google Scholar]
- Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, Naeem R, Carey VJ, Richardson AL, Weinberg RA (2005) Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell 121: 335–348 [DOI] [PubMed] [Google Scholar]
- Ostman A, Augsten M (2009) Cancer-associated fibroblasts and tumor growth--bystanders turning into key players. Curr Opin Genet Dev 19: 67–73 [DOI] [PubMed] [Google Scholar]
- Padua D, Massague J (2009) Roles of TGFβ in metastasis. Cell Res 19: 89–102 [DOI] [PubMed] [Google Scholar]
- Postigo AA, Depp JL, Taylor JJ, Kroll KL (2003) Regulation of Smad signaling through a differential recruitment of coactivators and corepressors by ZEB proteins. EMBO J 22: 2453–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirakihara T, Saitoh M, Miyazono K (2007) Differential regulation of epithelial and mesenchymal markers by δEF1 proteins in epithelial mesenchymal transition induced by TGF-β. Mol Biol Cell 18: 3533–3544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutz F, Neilson EG (2003) New insights into mechanisms of fibrosis in immune renal injury. Springer Semin Immunopathol 24: 459–476 [DOI] [PubMed] [Google Scholar]
- Strutz F, Zeisberg M, Ziyadeh FN, Yang CQ, Kalluri R, Muller GA, Neilson EG (2002) Role of basic fibroblast growth factor-2 in epithelial-mesenchymal transformation. Kidney Int 61: 1714–1728 [DOI] [PubMed] [Google Scholar]
- Thiery JP, Sleeman JP (2006) Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol 7: 131–142 [DOI] [PubMed] [Google Scholar]
- Vandewalle C, Van Roy F, Berx G (2009) The role of the ZEB family of transcription factors in development and disease. Cell Mol Life Sci 66: 773–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SY, Iordanov M, Zhang Q (2006) c-Jun NH2-terminal kinase promotes apoptosis by down-regulating the transcriptional co-repressor CtBP. J Biol Chem 281: 34810–34815 [DOI] [PubMed] [Google Scholar]
- Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP (2009) ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell 33: 591–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavadil J, Bottinger EP (2005) TGF-β and epithelial-to-mesenchymal transitions. Oncogene 24: 5764–5774 [DOI] [PubMed] [Google Scholar]
- Zeisberg M, Neilson EG (2009) Biomarkers for epithelial-mesenchymal transitions. J Clin Invest 119: 1429–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.