Abstract
The phyllosphere, i.e., the aerial parts of the plant, provides one of the most important niches for microbial colonization. This niche supports the survival and, often, proliferation of microbes such as fungi and bacteria with diverse lifestyles including epiphytes, saprophytes, and pathogens. Although most microbes may complete the life cycle on the leaf surface, pathogens must enter the leaf and multiply aggressively in the leaf interior. Natural surface openings, such as stomata, are important entry sites for bacteria. Stomata are known for their vital role in water transpiration and gas exchange between the plant and the environment that is essential for plant growth. Recent studies have shown that stomata can also play an active role in limiting bacterial invasion of both human and plant pathogenic bacteria as part of the plant innate immune system. As counter-defense, plant pathogens such as Pseudomonas syringae pv. tomato (Pst) DC3000 use the virulence factor coronatine to suppress stomata-based defense. A novel and crucial early battleground in host-pathogen interaction in the phyllosphere has been discovered with broad implications in the study of bacterial pathogenesis, host immunity, and molecular ecology of bacterial diseases.
Keywords: Guard cell, Plant innate immunity, Plant hormones, Bacterial pathogenesis, Food safety
Introduction
It is genuinely important that economically useful plants are safeguarded from detrimental factors that can diminish their output. One such a factor is biotic stress caused by pathogens’, insects’, and herbivores’ attack to the plant. It is estimated that biotic stress reduces 31–42% of the yield capacity of crops worldwide, representing US$500 billion of annual loss (FAO, www.fao.org). Loss due to pathogens alone accounts for 14% of yield reduction (US$220 billion annually) (1).
The phyllosphere may seem to be a very harsh environment for the survival of pathogens. The leaf surface in particular is regularly exposed to extreme conditions such as lack of moisture, ultraviolet irradiation, strong winds, and heat. Nonetheless, bacteria, the most abundant organisms on the leaf surface (2), can reach a high population density (106–107 cells/cm2 of leaf) (3). They have evolved mechanisms to either avoid or tolerate these stresses. The leaf surface is, therefore, a dynamic environment where various bacterial and other microbial activities take place.
Plant responses to its associated microbes have been extensively studied with special attention to pathogen contaminations of the plant intracellular space, i.e., apoplast. One of the most studied pathosystems is Arabidopsis-Pseudomonas syringae pv. tomato (Pst) DC3000 since the genomes of both organisms have been sequenced (4,5) and the availability of genetics and genomic resources (http://www.arabidopsis.org and http://www.pseudomonas-syringae.org/home.html) has greatly facilitated research efforts to understand the molecular basis for plant disease development. In this review, we will focus on recent advances towards understanding the close interaction between plants and foliar pathogenic bacteria early in the infection process.
Pseudomonas syringae: a model phyllosphere bacterium
Pseudomonas syringae is a Gram-negative bacterium that produces a broad variety of symptoms in a wide range of plants including blights, cankers, wilting, and leaf spots (6). P. syringae is differentiated into more than 40 different pathogenic variants or pathovars (pvs.) depending on the host-range of the bacterium isolate. For instance, Pst is the causal agent of bacterial speck of tomato and Arabidopsis (7). Each pathovar (pv.) can be further classified into different strains based on the disease reaction that it causes in specific genotypes of the host, exhibiting a very high degree of specificity.
Disease spread in crop fields occurs due to many possible sources of inoculum including infected seeds, crop and plant debris, infested seedlings and weeds, water, soil, agricultural tools, and volunteer plants (1). Additionally, P. syringae is able to survive and overwinter in plant debris. Upon arrival on the surface of a healthy plant, the infection cycle of P. syringae begins with epiphytic (surface) colonization of the plant phyllosphere (resident phase), followed by a subsequent endophytic phase in the apoplast (6). The size of epiphytic populations of P. syringae is strongly correlated with their ability to cause disease in the host plant (2).
Pseudomonas syringae, the causal agent of bacterial speck, is a hemibiotrophic pathogen as it obtains nutrients from living host cells, multiplies in the apoplast, and infects neighboring tissues. Bacterial speck disease is favored by weather and environmental conditions including a high relative humidity and cooler temperatures ranging from 13 to 28°C. Disease outbreaks occur more frequently after adverse weather conditions such as hard rains (6) and conditions that induce leaf wounding and enable bacteria to bypass natural points of entry. In the absence of wounds, however, P. syringae and other foliar pathogens may still invade plant through natural openings to become an endophytic pathogen (8).
There are many natural openings for bacterial penetration into leaves. Bacterial pathogens may be specialized to invade the plant through only one of them (reviewed in Ref. 9); stomata are the main route for Pst DC3000 penetration. Internal leaf tissues infected with Pst show water-soaked patches and form necrotic lesions surrounded by chlorosis (bleaching or yellowing of plant tissues due to degradation of chlorophyll).
Although much has been learned about the mechanisms of Pst DC3000 virulence (10) and the genetics of the Arabidopsis/Pst DC3000 pathogenic interaction (11), how P. syringae (and other foliar bacterial pathogens) makes the transition from epiphytic to endophytic life styles during a successful infection cycle is not well understood (12). This is clearly one of the most outstanding questions in bacterial disease epidemiology, yet we have little understanding of the process.
Does stomatal defense prevent bacterial contamination of plants?
Stomata are formed by a pair of specialized epidermal cells known as guard cells (Figure 1). Movement of guard cells due to changes in turgor pressure regulates the opening and closing of the stomatal pore (13). Several environmental stimuli such as light, relative humidity, and CO2 concentration control stomatal movement (readers are directed to Refs. 13 and 14 for extensive reviews on this subject). Foliar infection of plants by bacteria such as Pst occurs through stomata (12), which serve as critical entry sites and allow bacteria to transition from epiphytic to endophytic lifestyle.
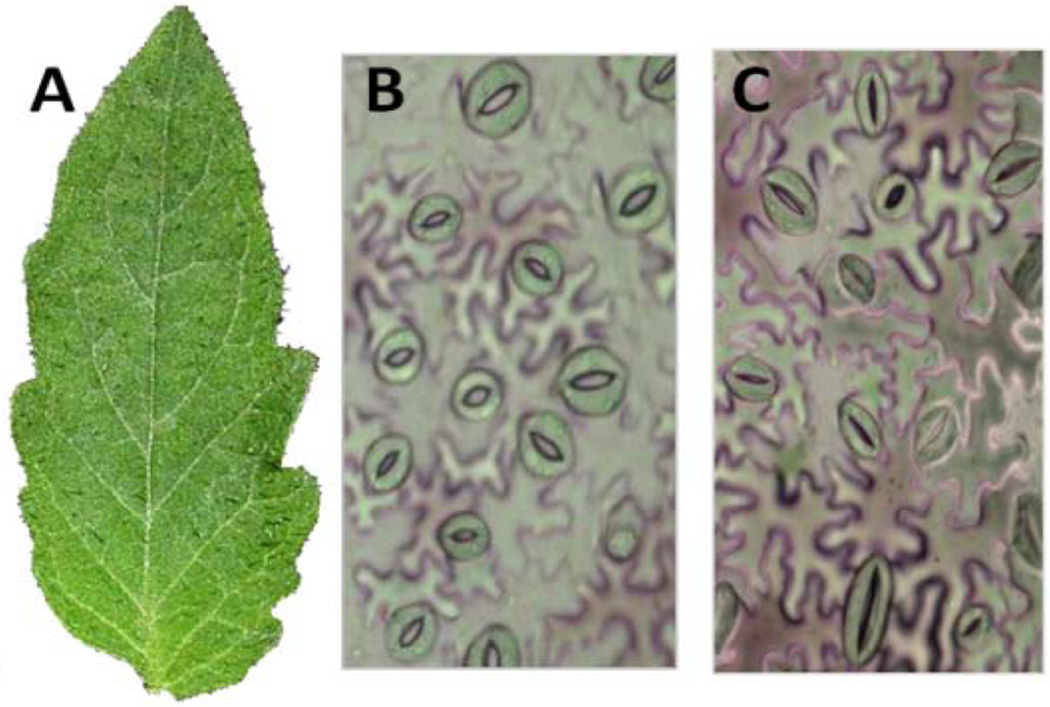
Figure 1.
Light-conditioned tomato leaf (A) and leaf surface under the microscope prior to exposure to Pst DC3000 showing mostly open stomata (B). The same leaf was exposed to Pseudomonas syringae pv. tomato (Pst DC3000) and after 1 h of exposure most stomata were closed (C).
It was previously assumed that the entry of bacteria into leaf tissues through natural openings was a passive process, where the plant lacked mechanisms for preventing bacterial entry, and the bacterium lacked active virulence mechanisms to promote entry (12). Recent studies have shown that entry of bacteria into leaf tissue through stomata is more complex and dynamic than the simple act of swimming into the leaf through passive openings (8,15). Several lines of evidence suggest that stomata actively close in response to plant pathogenic and human pathogenic bacteria or when exposed to conserved molecules found on the surface of bacterial cells known as pathogen/microbe-associated molecular patterns (PAMPs/MAMPs) (8). By definition MAMPs are the molecular motifs of microbes that are recognized by receptors in the host cell called pattern recognition receptors (16). Some examples of MAMPs are lipopolysaccharide, bacterial flagellin, and lipoteichoic acid. Bacterium-induced stomatal closure is part of plant immune defenses and requires the FLS2 receptor, production of nitric oxide, salicylic acid homeostasis, abscisic acid signaling components, such as the guard-cell-specific OST1 kinase (8), K+ channel regulation via heterotrimeric G-protein (17), mitogen-activated protein kinase 3 (MPK3) (15), cAMP, cyclic nucleotide gated channel (CNGC2/DND1), and Ca2+ (18). Thus, stomatal closure is an integral basal plant defense mechanism to restrict the invasion of pathogenic bacteria into plant tissues. In addition, pathogen-derived signals integrate into the dynamic hormonal regulation of guard cell movement (19). Figure 2 depicts the connection between molecular components that have been implicated in pathogen modulation of stomatal responses.
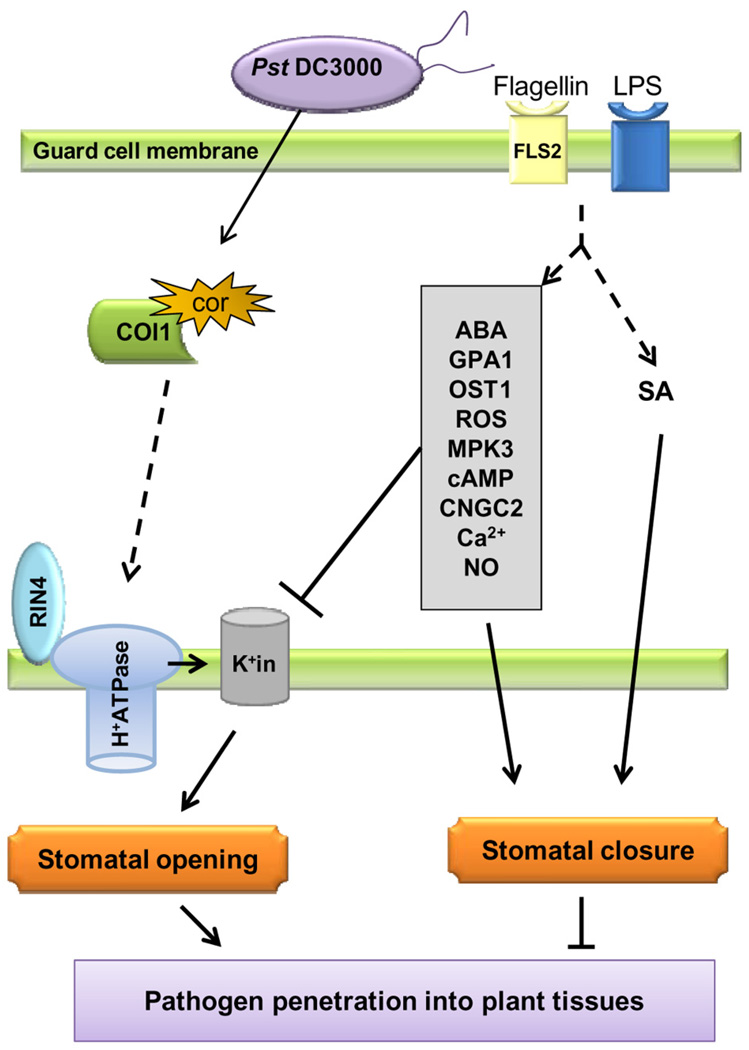
Figure 2.
Model illustrating the signaling components and interactions between molecules that have been experimentally demonstrated to be involved in stomatal defense and bacterial counter defense. In the stomatal guard cell, MAMPs (e.g., flagellin and LPS) are perceived by cognate immune receptors (e.g., flagellin receptor FLS2). Perception of MAMPs triggers stomatal closure, which requires the phytohormones SA and ABA, as well as ABA signaling components listed in the grey rectangle (components are listed in order where the top one is the most upstream. Their localization in the cell is not shown). Flagellin also prevents stomatal opening by inhibiting inwardly rectifying K+ channels (K+in) through ABA signaling components (GPA1 and possibly others). COR-mediated inhibition of MAMP-triggered stomatal closure requires the plant proteins COI1 (a COR receptor) and RIN4. While COI1 physically binds to COR, RIN4 binds and activates the proton pump (H+ATPase) causing membrane hyperpolarization and activation of K+ influx (K+in), a condition that promotes stomatal opening. Dashed arrows indicate possibly indirect pathways. MAMPs = microbe-associated molecular patterns; LPS = lipopolysaccharide; SA = salicylic acid; ABA = abscisic acid; COR = coronatine.
Bacterial counter defense: the virulence factor coronatine promotes entry into leaves
Coronatine (COR) is one of the most well-studied bacterial phytotoxins. COR is a non-host-specific phytotoxin, and its structure consists of two distinct moieties that function as intermediates in the biosynthetic pathway: a) the polyketide coronafacic acid, which is structurally and functionally similar to the jasmonate family of plant signaling molecules induced in response to stress, and b) coronamic acid, an ethylcyclopropyl amino acid that resembles aminocyclopropyl carboxylic acid, a precursor of the plant defense hormone ethylene. Coronamic acid and coronafacic acid are synthesized by separate pathways and joined by an amide bond to form COR (20).
Emerging evidence suggests that COR plays multiple roles in bacterial pathogenesis including promoting entry of bacteria through stomata at the initial stages of infection (8) and suppression of defenses mediated by the plant hormone salicylic acid later in the infection process (21). COR is produced by several pathovars of P. syringae including tomato, maculicola, glycinea, and atropurpurea, where it is known to function as a virulence factor promoting chlorosis in several host plants (22,23). It induces modifications in the plant’s physiology such as anthocyanin production, alkaloid accumulation, ethylene emission, tendril coiling, and root inhibition (20). This toxin acts as a virulence factor and contributes to disease development.
The possibility that COR could suppress early defense responses during Pst DC3000 infection of Arabidopsis and tomato was suggested more than a decade ago (23) and confirmed recently (8). The discovery that COR is required for re-opening stomata by Pst DC3000 represents the first identification of a bacterial virulence factor that suppresses stomatal closure. COR defective mutants have reduced multiplication and symptom production in planta when compared to the wild-type (22,23). Studies in both Pst and P. syringae pv. glycinea (Psg) have suggested that COR may be important for bacterial invasion of plant tissue. COR+ and COR− strains of both Pst and Psg were able to reach similar population densities when infiltrated into host plant tissues (23,24). However, when host plants were inoculated by dipping (23) or spraying (24), the COR− mutants were unable to attain the growth levels of the wild-type COR+ strains. Spray or dip inoculations closely mimic natural infections, whereas infiltration delivers bacteria directly inside the leaves bypassing the penetration step for bacterial infection. The broad use of this artificial inoculation method has masked the functions of COR in the initial steps of the plant-microbe interaction.
The discovery of this virulence mechanism of Pst DC3000 has generated a lot of interest in elucidating the mode of action of coronatine at the molecular level. Interestingly, Mino et al. (25) have shown that 10 µM COR promotes opening of dark-closed stomata of broad bean and Italian ryegrass. The effect of COR on the stomatal aperture was more pronounced on Italian ryegrass. These authors also pointed out that COR activates membrane-bound ATPase activity inducing stomatal opening. More recently, the plant protein RIN4 (RPM1 interacting protein 4) and activation of H+-ATPase have been found to be necessary for COR to re-open stomata (26). How exactly COR functions via RIN4 and activation of H+ATPase is not yet clear. COI1 (coronatine insensitive1) is another plant protein necessary for COR function in the guard cell (Figure 2) (8). In fact, COI1 has been shown to be a receptor for COR (27) and the molecular mechanisms through which COR may operate in the plant cell to promote disease is discussed below.
Molecular action of coronatine in plant cells
Coronatine is a structural and functional mimic of the plant hormone jasmonate (JA) conjugated to the amino acid isoleucine (JA-Ile) (28). Biological concentrations of COR activate the JA signaling pathway in the plant. cDNA microarray analysis indicated the induction of JA-responsive genes in the tomato-Pst DC3000 interaction depends on the bacterial production of COR (29).
JA regulates diverse aspects of plant growth, development, immunity, as well as plant responses to the environment and biotic stresses (30). Identification and characterization of JA-deficient and JA-insensitive mutants have revealed the underlying mechanism of defense responsive genes. The protein COI1 was identified by Arabidopsis mutant screenings and shown to be a key regulator of the JA signaling pathway (31). COI1 is an F-box protein associated with the SCF protein complex; an E3 ubiquitin ligase consisting of SKP1, CULLIN1, and F-box proteins that targets proteins for degradation through the 26S proteasome pathway. It has been shown that the SCFCOI1 ubiquitin complex is required for JA response in Arabidopsis (32), indicating that certain proteins repressing the JA-responsive genes may be targeted for degradation by this complex. Supporting this hypothesis, JAZ proteins have been identified as such repressors and shown to interact with COI1 in a ligand-dependent manner (33).
To identify these JAZ proteins in Arabidopsis, Thines et al. (33) studied the jasmonate synthesis mutant opr3 (12-oxophytodienoic acid reductase 3), which is unable to convert 12-oxophytodienoic acid to JA. Upon treatment of opr3 mutant plants with JA, these scientists observed a significant induction of 32 genes after 30 min of treatment. Among those genes, eight were annotated as encoding proteins of unknown function with very similar sequence structure consisting of two highly conserved domains: the TIFY motif-containing the ZIM domain and the Jas domain at the C-terminus. Bioinformatic analysis of the whole Arabidopsis genome revealed a gene family of 12 genes encoding 19 protein variants. Since these proteins have a 28-amino acid ZIM domain they were named jasmonate ZIM-domain (JAZ) proteins (33,34).
The ZIM domain is involved in mediating homo- and heteromeric interactions between JAZ proteins (35,36). In addition, a protein named novel interactor of JAZ (NINJA) has been found to interact with the TIFY motif of JAZ and act as an adaptor to recruit other co-repressors [Groucho/Tup1-type co-repressor TOPLESS (TPL) and TPL-related proteins] of JA responses in the plant cell (37). The Jas domain of JAZ proteins has been shown to interact with COI1 and the transcription factor MYC2 (27,34,38). While JAZ-COI1 interaction requires COR or JA-Ile, JAZ-MYC2 interaction does not. Experimental evidence also suggests that the region in the Jas domain of JAZ proteins responsible for the interaction with COI1 and MYC2 is not the same (34,39).
Recently, COI1 has been demonstrated to be a receptor for JA-Ile and COR (27). Therefore, a plausible model (Figure 3) can be developed for the entire set of interactions where COR produced by the bacterium binds to COI1 and leads to the degradation of JAZ proteins through the SCFCOI1 complex. In this model, JAZ proteins and other adaptor proteins act as repressors of JA signaling. Degradation of JAZ proteins allows for the expression of JA-responsive genes in the plant cell (33–35,37) blocking plant innate immune responses including stomatal defense.
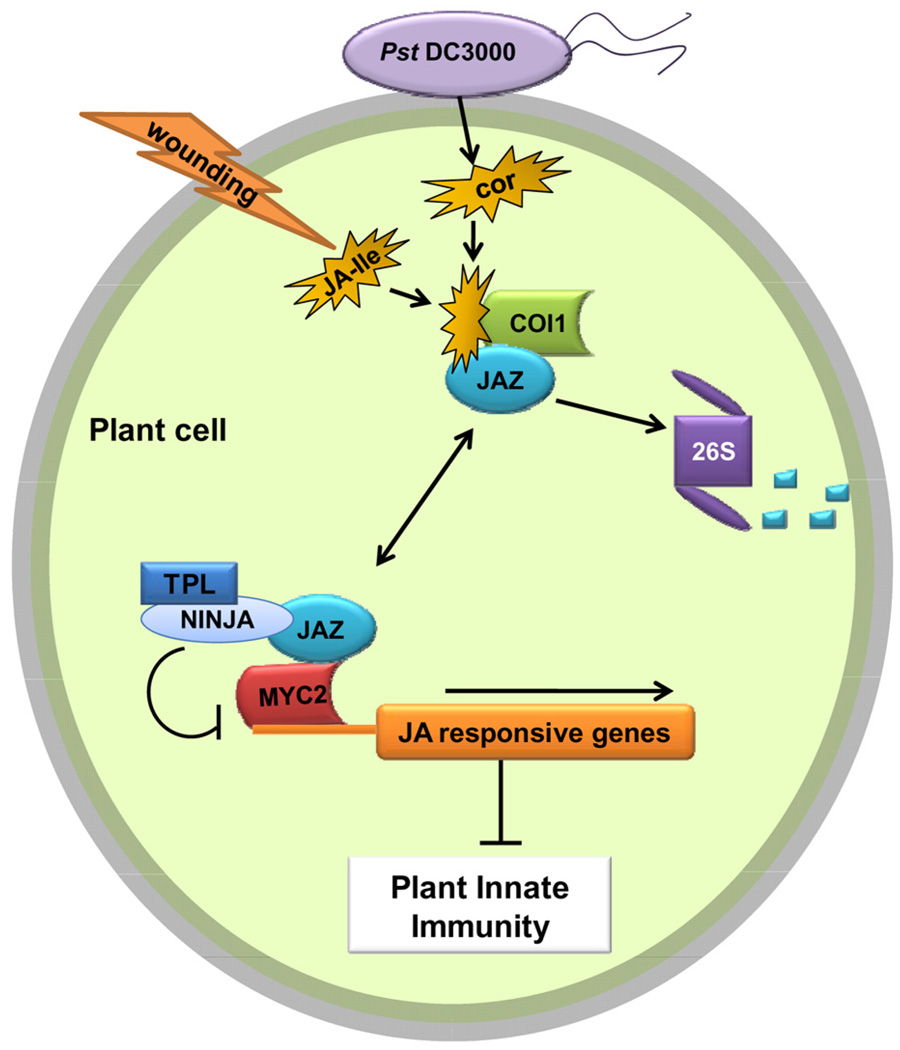
Figure 3.
A model depicting the molecular action of coronatine in plant cells (possibly all cell types). COR is secreted by Pst DC3000 into the plant cell and increases the affinity of the COI1 protein (as part of the SCFCOI1 ubiquitin-ligase complex, not shown here) toward the JAZ repressor. The SCFCOI1 complex catalyzes ubiquitination of JAZ, which is then degraded through the 26S proteasome (denoted as “26S”). JAZ protein is part of a repressor complex that also contains NINJA and TPL, and physically binds to transcriptional activators (such as MYC2) of jasmonate response genes. Upon degradation of JAZ, JA response genes are activated blocking plant innate immune responses including stomatal closure. COR = coronatine; JA = jasmonate; JAZ = jasmonate ZIM-domain; NINJA = novel interactor of JAZ; TPL = TOPLESS.
Virulence strategies to overcome stomatal defense in other pathosystems
The involvement of conserved molecular components and the innate immunity response in stomatal defense against invading bacteria suggest that this form of defense may be widespread across plant species. Therefore, considering that stomatal closure successfully avoids microbial invasion, it is likely that phytopathogens employ distinct virulence factors or lifestyles to overcome or circumvent stomatal closure. New evidence suggests that other bacterial factors are involved in suppressing stomatal closure. For instance, P. syringae pv. tabaci, which does not produce COR, induced initial closure of stomata in tobacco and was able to re-open them at later times, similar to Pst DC3000 (8). The nature of the virulence factor of P. syringae pv. tabaci responsible for overcoming stomatal defense remains to be determined.
The relevance of stomatal innate immunity in the Arabidopsis-Xanthomonas campestris pv. campestris (Xcc) pathosystem has also been studied recently (15). Xcc can penetrate Arabidopsis leaves through both hydathodes and stomata depending on the ecotype and environmental conditions. Live Xcc cells and extracts of its culture supernatant are capable of reversing stomatal closure in Arabidopsis leaves (15). Interestingly, Xcc-triggered stomatal re-opening is dependent on the ability of this bacterium to synthesize or perceive diffusible signals through the rpf/ diffusible signal factor system (15), suggesting that cell-to-cell signaling may regulate virulence factors to overcome stomatal defense. However, the chemical nature of the virulence factor in Xcc responsible for stomatal opening has not been elucidated.
Stomatal responses also differ between incompatible and compatible interactions based on the presence or absence of a resistance gene-avr gene interaction. Specifically, the stomatal responses of wild-type Arabidopsis thaliana ecotype Columbia (Col-0) plants to two bacteria: Pst DC3000 (representing a susceptible interaction) and Pst DC3000/avrRpt2 (representing a resistant interaction) was also tested by Melotto et al. (8). Like Pst DC3000, the avirulent strain Pst DC3000/avrRpt2 caused stomatal closure within 1 h. However, the avirulent strain was less effective in re-opening stomata than the virulent strain at 3 h after incubation. This result suggests that the gene-for-gene resistance mediated by avrRpt2/RPS2 has a positive effect on promoting stomatal closure (8). An independent study has also shown that gene-for-gene resistance through Avr-Rpm1 in Arabidopsis suppresses growth of bacteria, at least in part, by coupling restricted vascular flow to the infection site with stomatal closure (40), further supporting a model in which stomate-based innate immunity also contributes to gene-for-gene resistance.
In addition to plant-bacterial interactions, stomatal regulation has also been observed in some plant-fungal and plant-oomycete interactions. For example, Guimaraes and Stotz (41) made the interesting observation that stomatal pores of Vicia faba leaves infected with an oxalate-deficient mutant of Sclerotinia sclerotiorum were partially closed, whereas the wild-type fungus caused stomatal opening. Furthermore, exogenous application of oxalic acid, a virulence factor of several phytopathogenic fungi including S. sclerotiorum, induces stomatal opening. Open stomata seem to be the exit sites of many fungal hyphae from infected leaves. Plasmopara viticola is also able to prevent dark-and drought-induced stomatal closure in grapevine leaves at the site of infection (42). This oomycete is an obligate biotrophic organism that enters plant tissue through stomata and a localized response would probably be an important adaptation to infect the host while keeping it alive.
Stomata have also been implicated in the response to resistance gene-mediated recognition of the fungal defense elicitor, Avr9. Recognition of the Avr9 protein from the fungus Cladosporium fulvum by transgenic Nicotiana tabacum plants expressing the Cf-9 resistance protein from tomato led to the activation of current through outward-rectifying K+ channels and the inactivation of current through inward-rectifying K+ channels (43). This pattern of regulation of cation channels would be predicted to promote stomatal closure, suggesting that resistant plants also control fungal penetration through the stomata. In another pathosystem, soybean/Phytophthora sojae, stomatal closure was observed within 2 h of contact with the fungus in an incompatible reaction (i.e., plant resistance); whereas during a compatible reaction (i.e., plant susceptibility) between these two organisms, stomata closed slightly initially and remained open as disease progressed (44). It is therefore possible that stomate-based defense and counter defense also occur in some plant-fungal interactions.
The fungal toxin fusicoccin has long been known to promote stomatal opening and to antagonize abscisic acid-induced stomatal closure through activation of a plasma membrane H+ ATPase (45), similar to what has been proposed for COR (25,26). Oligogalacturonic acid, an elicitor derived from the degradation of the plant cell wall by fungal cell wall-degrading enzymes, and chitosan, a component of the fungal cell wall, were both shown to affect stomatal movements in tomato (46). Both oligogalacturonic acid and chitosan elicited H2O2 production in guard cells and inhibited light-induced opening of closed stomata. The biological relevance of the stomatal closure in response to these fungal-derived compounds with respect to plant defense or fungal invasion is not yet clear.
Can stomatal closure prevent plant contamination with human pathogens?
In addition to phytopathogenic bacteria, human pathogens are also capable of occupying the phyllosphere, an aspect of biology of plant-microbe interactions that has major implications for the safety of fresh fruits and vegetables. It is estimated that 76 million cases of food- borne diseases occur yearly in the US (CDC, www.cdc.gov) and over 35 major outbreaks occurred in the last decade (47). The number of serious cases leading to death has been increasing and outbreaks associated with fresh produce have emerged as an important public health concern. In particular, enterohemorrhagic Escherichia coli and Salmonella enterica appear to be two of the most common causal agents of food poisoning associated with the consumption of fresh leafy vegetables (47).
The route of human pathogen internalization into plant tissue has been a subject for extensive discussion (48). Both surface and interior contamination of leaves with human pathogens can be dangerous, but internal contamination can be very difficult, if not impossible, to remove by standard disinfection procedures. Human pathogen internalization through plant stomata has been studied for both E. coli O157:H7 (8) and S. enteric serovar Typhimurium (49). Research suggests that E. coli O157:H7 triggers stomatal closure, but it is not able to overcome this plant immune response when inoculated as pure cultures in laboratory settings (8,50). Interestingly, however, a recent study documented a remarkable ability of S. enteric serovar Typhimurium to migrate toward stomata and enter plant tissues without triggering stomatal immune response (49). This finding raises the possibility that not only plant pathogens, but also some human pathogens have evolved mechanisms to subvert plant stomate-based defense to enter plant tissues. The underlying mechanism of this observation is not understood and is a topic of active research.
Acknowledgments
We thank National Institutes of Health (5R01AI068718) for financial support and the University of Texas at Arlington for startup funds provided to M. Melotto.
References
- 1.Agrios GN. Plant pathology. 5th edn. San Diego: Elsevier Academic Press; 2005. [Google Scholar]
- 2.Lindow SE, Brandl MT. Microbiology of the phyllosphere. Appl Environ Microbiol. 2003;69:1875–1883. doi: 10.1128/AEM.69.4.1875-1883.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrews JH, Harris RF. The ecology and biogeography of microorganisms on plant surfaces. Annu Rev Phytopathol. 2000;38:145–180. doi: 10.1146/annurev.phyto.38.1.145. [DOI] [PubMed] [Google Scholar]
- 4.Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 5.Buell CR, Joardar V, Lindeberg M, Selengut J, Paulsen IT, Gwinn ML, et al. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci U S A. 2003;100:10181–10186. doi: 10.1073/pnas.1731982100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirano SS, Upper CD. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae - a pathogen, ice nucleus, and epiphyte. Microbiol Mol Biol Rev. 2000;64:624–653. doi: 10.1128/mmbr.64.3.624-653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whalen MC, Innes RW, Bent AF, Staskawicz BJ. Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell. 1991;3:49–59. doi: 10.1105/tpc.3.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Melotto M, Underwood W, Koczan J, Nomura K, He SY. Plant stomata function in innate immunity against bacterial invasion. Cell. 2006;126:969–980. doi: 10.1016/j.cell.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 9.Melotto M, Underwood W, He SY. Role of stomata in plant innate immunity and foliar bacterial diseases. Annu Rev Phytopathol. 2008;46:101–122. doi: 10.1146/annurev.phyto.121107.104959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nomura K, Melotto M, He SY. Suppression of host defense in compatible plant-Pseudomonas syringae interactions. Curr Opin Plant Biol. 2005;8:361–368. doi: 10.1016/j.pbi.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 11.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 12.Underwood W, Melotto M, He SY. Role of plant stomata in bacterial invasion. Cell Microbiol. 2007;9:1621–1629. doi: 10.1111/j.1462-5822.2007.00938.x. [DOI] [PubMed] [Google Scholar]
- 13.Schroeder JI, Allen GJ, Hugouvieux V, Kwak JM, Waner D. Guard cell signal transduction. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:627–658. doi: 10.1146/annurev.arplant.52.1.627. [DOI] [PubMed] [Google Scholar]
- 14.Fan LM, Zhao Z, Assmann SM. Guard cells: a dynamic signaling model. Curr Opin Plant Biol. 2004;7:537–546. doi: 10.1016/j.pbi.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 15.Gudesblat GE, Torres PS, Vojnov AA. Xanthomonas campestris overcomes Arabidopsis stomatal innate immunity through a DSF cell-to-cell signal-regulated virulence factor. Plant Physiol. 2009;149:1017–1027. doi: 10.1104/pp.108.126870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boller T, He SY. Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science. 2009;324:742–744. doi: 10.1126/science.1171647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang W, He SY, Assmann SM. The plant innate immunity response in stomatal guard cells invokes G-protein-dependent ion channel regulation. Plant J. 2008;56:984–996. doi: 10.1111/j.1365-313X.2008.03657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ali R, Ma W, Lemtiri-Chlieh F, Tsaltas D, Leng Q, von Bodman S, et al. Death don’t have no mercy and neither does calcium: Arabidopsis CYCLIC NUCLEOTIDE GATED CHANNEL 2 and innate immunity. Plant Cell. 2007;19:1081–1095. doi: 10.1105/tpc.106.045096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bray-Speth E, Melotto M, Zhang W, Assmann SM, He SY. Crosstalk in pathogen and 41 hormonal regulation of guard cell signaling. In: Yoshioka K, Shinozaki K, editors. Signal crosstalk in plant stress response. Ames: John Wiley and Sons, Ltd.; 2009. pp. 96–112. [Google Scholar]
- 20.Bender CL, Alarcon-Chaidez F, Gross DC. Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol Mol Biol Rev. 1999;63:266–292. doi: 10.1128/mmbr.63.2.266-292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uppalapati SR, Ishiga Y, Wangdi T, Kunkel BN, Anand A, Mysore KS, et al. The phytotoxin coronatine contributes to pathogen fitness and is required for suppression of salicylic acid accumulation in tomato inoculated with Pseudomonas syringae pv. tomato DC3000. Mol Plant Microbe Interact. 2007;20:955–965. doi: 10.1094/MPMI-20-8-0955. [DOI] [PubMed] [Google Scholar]
- 22.Brooks DM, Hernandez-Guzman G, Kloek AP, Alarcon-Chai-dez F, Sreedharan A, Rangaswamy V, et al. Identification and characterization of a well-defined series of coronatine biosynthetic mutants of Pseudomonas syringae pv. tomato DC3000. Mol Plant Microbe Interact. 2004;17:162–174. doi: 10.1094/MPMI.2004.17.2.162. [DOI] [PubMed] [Google Scholar]
- 23.Mittal S, Davis KR. Role of the phytotoxin coronatine in the infection of Arabidopsis thaliana by Pseudomonas syringae pv. tomato. Mol Plant Microbe Interact. 1995;8:165–171. doi: 10.1094/mpmi-8-0165. [DOI] [PubMed] [Google Scholar]
- 24.Budde IP, Ullrich MS. Interactions of Pseudomonas syringae pv. glycinea with host and nonhost plants in relation to temperature and phytotoxin synthesis. Mol Plant Microbe Interact. 2000;13:951–961. doi: 10.1094/MPMI.2000.13.9.951. [DOI] [PubMed] [Google Scholar]
- 25.Mino Y, Matsushita Y, Sakai R. The effect of coronatine on stomatal opening in leaves of broadbean and Italian ryegrass. Ann Phytopathol Soc Jp. 1987;53:53–55. [Google Scholar]
- 26.Liu J, Elmore JM, Fuglsang AT, Palmgren MG, Staskawicz BJ, Coaker G. RIN4 functions with plasma membrane H+-ATPases to regulate stomatal apertures during pathogen attack. PLoS Biol. 2009;7:e1000139. doi: 10.1371/journal.pbio.1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan J, Zhang C, Gu M, Bai Z, Zhang W, Qi T, et al. The Arabidopsis CORONATINE INSENSITIVE 1 protein is a jasmonate receptor. Plant Cell. 2009;21:2220–2236. doi: 10.1105/tpc.109.065730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Staswick PE, Tiryaki I. The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell. 2004;16:2117–2127. doi: 10.1105/tpc.104.023549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao Y, Thilmony R, Bender CL, Schaller A, He SY, Howe GA. Virulence systems of Pseudomonas syringae pv. tomato promote bacterial speck disease in tomato by targeting the jasmonate signaling pathway. Plant J. 2003;36:485–499. doi: 10.1046/j.1365-313x.2003.01895.x. [DOI] [PubMed] [Google Scholar]
- 30.Fonseca S, Chico JM, Solano R. The jasmonate pathway: the ligand, the receptor and the core signalling module. Curr Opin Plant Biol. 2009;12:539–547. doi: 10.1016/j.pbi.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 31.Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG. COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science. 1998;280:1091–1094. doi: 10.1126/science.280.5366.1091. [DOI] [PubMed] [Google Scholar]
- 32.Xu L, Liu F, Lechner E, Genschik P, Crosby WL, Ma H, et al. The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell. 2002;14:1919–1935. doi: 10.1105/tpc.003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, et al. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature. 2007;448:661–665. doi: 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- 34.Chini A, Fonseca S, Fernandez G, Adie B, Chico JM, Lorenzo O, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448:666–671. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- 35.Chung HS, Howe GA. A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the jasmonate ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell. 2009;21:131–145. doi: 10.1105/tpc.108.064097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chini A, Fonseca S, Chico JM, Fernandez-Calvo P, Solano R. The ZIM domain mediates homo- and heteromeric interactions between Arabidopsis JAZ proteins. Plant J. 2009;59:77–87. doi: 10.1111/j.1365-313X.2009.03852.x. [DOI] [PubMed] [Google Scholar]
- 37.Pauwels L, Barbero GF, Geerinck J, Tilleman S, Grunewald W, Perez AC, et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature. 2010;464:788–791. doi: 10.1038/nature08854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci U S A. 2008;105:7100–7105. doi: 10.1073/pnas.0802332105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Melotto M, Mecey C, Niu Y, Chung HS, Katsir L, Yao J, et al. A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine-and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 2008;55:979–988. doi: 10.1111/j.1365-313X.2008.03566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Freeman BC, Beattie GA. Bacterial growth restriction during host resistance to Pseudomonas syringae is associated with leaf water loss and localized cessation of vascular activity in Arabidopsis thaliana. Mol Plant Microbe Interact. 2009;22:857–867. doi: 10.1094/MPMI-22-7-0857. [DOI] [PubMed] [Google Scholar]
- 41.Guimaraes RL, Stotz HU. Oxalate production by Sclerotinia sclerotiorum deregulates guard cells during infection. Plant Physiol. 2004;136:3703–3711. doi: 10.1104/pp.104.049650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Allegre M, Daire X, Heloir MC, Trouvelot S, Mercier L, Adrian M, et al. Stomatal deregulation in Plasmopara viticola-infected grapevine leaves. New Phytol. 2007;173:832–840. doi: 10.1111/j.1469-8137.2006.01959.x. [DOI] [PubMed] [Google Scholar]
- 43.Blatt MR, Grabov A, Brearley J, Hammond-Kosack K, Jones JD. K+ channels of Cf-9 transgenic tobacco guard cells as targets for Cladosporium fulvum Avr9 elicitor-dependent signal transduction. Plant J. 1999;19:453–462. doi: 10.1046/j.1365-313x.1999.00534.x. [DOI] [PubMed] [Google Scholar]
- 44.McDonald KL, Cahill DM. Evidence for a transmissible factor that causes rapid stomatal closure in soybean at sites adjacent to and remote from hypersensitive cell death induced by Phytophthora sojae. Physiol Mol Plant Pathol. 1999;55:197–203. [Google Scholar]
- 45.Emi T, Kinoshita T, Shimazaki K. Specific binding of vf14-3-3a isoform to the plasma membrane H+-ATPase in response to blue light and fusicoccin in guard cells of broad bean. Plant Physiol. 2001;125:1115–1125. doi: 10.1104/pp.125.2.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee S, Choi H, Suh S, Doo IS, Oh KY, Choi EJ, et al. Oligogalacturonic acid and chitosan reduce stomatal aperture by inducing the evolution of reactive oxygen species from guard cells of tomato and Commelina communis. Plant Physiol. 1999;121:147–152. doi: 10.1104/pp.121.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Doyle MP, Erickson MC. Summer meeting 2007 - the problems with fresh produce: an overview. J Appl Microbiol. 2008;105:317–330. doi: 10.1111/j.1365-2672.2008.03746.x. [DOI] [PubMed] [Google Scholar]
- 48.Warriner K, Namvar A. The tricks learnt by human enteric pathogens from phytopathogens to persist within the plant environment. Curr Opin Biotechnol. 2010;21:131–136. doi: 10.1016/j.copbio.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 49.Kroupitski Y, Golberg D, Belausov E, Pinto R, Swartzberg D, Granot D, et al. Internalization of Salmonella enterica in leaves is induced by light and involves chemotaxis and penetration through open stomata. Appl Environ Microbiol. 2009;75:6076–6086. doi: 10.1128/AEM.01084-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thilmony R, Underwood W, He SY. Genome-wide transcriptional analysis of the Arabidopsis thaliana interaction with the plant pathogen Pseudomonas syringae pv. tomato DC3000 and the human pathogen Escherichia coli O157:H7. Plant J. 2006;46:34–53. doi: 10.1111/j.1365-313X.2006.02725.x. [DOI] [PubMed] [Google Scholar]