Abstract
Trypanosoma brucei brucei has two distinct developmental stages, the procyclic stage in the insect and the bloodstream stage in the mammalian host. The significance of each developmental stage is punctuated by specific changes in metabolism. In the insect, T. b. brucei is strictly dependent on mitochondrial function and thus respiration to generate the bulk of its ATP, whereas in the mammalian host it relies heavily on glycolysis. These observations have raised questions about the importance of mitochondrial function in the bloodstream. Peculiarly, akinetoplastic strains of Trypanosoma brucei evansi that lack mitochondrial DNA do exist in the wild and are developmentally locked in the glycolysis-dependent bloodstream stage. Using RNAi we show that two mitochondrion-imported proteins, mitochondrial RNA polymerase and guide RNA associated protein 1, are still imported into the nucleic acids-lacking organelle of T. b. evansi, making the need for these proteins futile. We also show that, like in the T. b. brucei procyclic stage, the mitochondria of both bloodstream stage of T. b. brucei and T. b. evansi import various tRNAs, including those that undergo thiolation. However, we were unable to detect mitochondrial thiolation in the akinetoplastic organelle. Taken together, these data suggest a lack of connection between nuclear and mitochondrial communication in strains of T. b. evansi that lost mitochondrial genome and that do not required an insect vector for survival.
Keywords: Trypanosoma, tRNA, protein import, mitochondrion, kinetoplast
INTRODUCTION
Trypanosoma brucei brucei is an evolutionary ancestral parasite, transmitted from one mammalian host to another by the blood-sucking tse-tse fly. While T. b. brucei is responsible for n’gana of livestock, African sleeping sickness of humans is caused by its subspecies T. b. rhodesiense and T. b. gambiense. Two other subspecies (T. b. evansi and T. b. equiperdum) are causative agents of surra and dourine, serious diseases afflicting mostly horses, water buffaloes and camels [1,2].
The single mitochondrion of T. b. brucei undergoes dramatic changes in the course of the life cycle. In the reticulated cristae-rich organelle of the procyclic stage parasitizing the tse-tse vector, functional cytochrome c-containing respiratory complexes capable of energy transduction are present [3]. The mitochondrion also contains alternative terminal oxidase (TAO), incomplete Krebs cycle and acetate:succinate CoA transferase cycle and other metabolic pathways. For energy production this active mitochondrion can utilize glycolytic substrates as well as various amino acids [4]. However, the bloodstream stage responsible for the disease in its vertebrate hosts harbors a mitochondrion, which is dramatically different from its procyclic counterpart by being repressed in numerous functions. The organelle is reduced in size, contains just a few cristae and lacks both Krebs cycle and respiratory complexes III and IV [5]. TAO is the only terminal oxidase [6] and membrane potential is upheld by the reverse action of the ATPase complex [7]. Yet even this mitochondrion is far from dormant, as replication and transcription of its kinetoplast (k) DNA, and extensive RNA editing and RNA processing remain fully active and essential [1,8–10]. This dual metabolism, in distinct life cycle stages, of a mitochondrion that occurs in a single copy per cell, makes it a very interesting and tractable model for exploring mechanisms that govern the switch from an active to an inactive mode.
Another approach that is particularly suitable for examining stage-specific differences of the mitochondrion involves comparisons of subspecies of T. b. brucei, which differ in terms of their kDNA content. One of these strains, T. b. equiperdum, has lost significant portions of its mitochondrial genome, while the organelle of some strains of T. b. evansi is totally devoid of kDNA [11–13]. These trypanosomes are therefore incapable of transmission by tsetse flies, and are locked into the bloodstream stage. Although their spreading among ungulate hosts relies solely on mechanical means of transmission, the lost dependence on the insect vector paradoxically allowed spreading of these trypanosomes outside of Africa [11,12,14]. Without the kDNA-encoded subunits a switch to the metabolically fully active procyclic organelle is impossible and T. b. evansi permanently depends on the glucose-rich environment of the vertebrate blood [11]. One can therefore postulate that any process found in its mitochondrion is genuinely associated with the bloodstream stage.
In a previous study, we have demonstrated that the akinetoplastic mitochondrion keeps importing proteins that are involved in kDNA replication, mitochondrial RNA editing and RNA processing [13]. While this occurrence is indeed not surprising for the organelle of the T. b. brucei bloodstream stage, it seems to be a counterintuitive phenomenon for T. b. evansi, as one would not predict efficient import of these proteins into a mitochondrion permanently devoid of DNA and RNA. As we show in this work, proteins involved in RNA synthesis and processing are still imported into the akinetoplastic mitochondrion of T. b. evansi, but in contrast to T. b. brucei, are non-essential. We also show for the first time that tRNAs are imported into and thiolated within the mitochondrion of T. b. brucei bloodstream stage, and imported into but not thiolated within the mitochondrion of T. b. evansi. This further supports the conclusion that subunits of respiratory complexes are required for neither tRNA [15], nor protein import.
RESULTS AND DISCUSSION
Proteins required for the bloodstream stage of T. b. brucei are non-essential for T. b. evansi
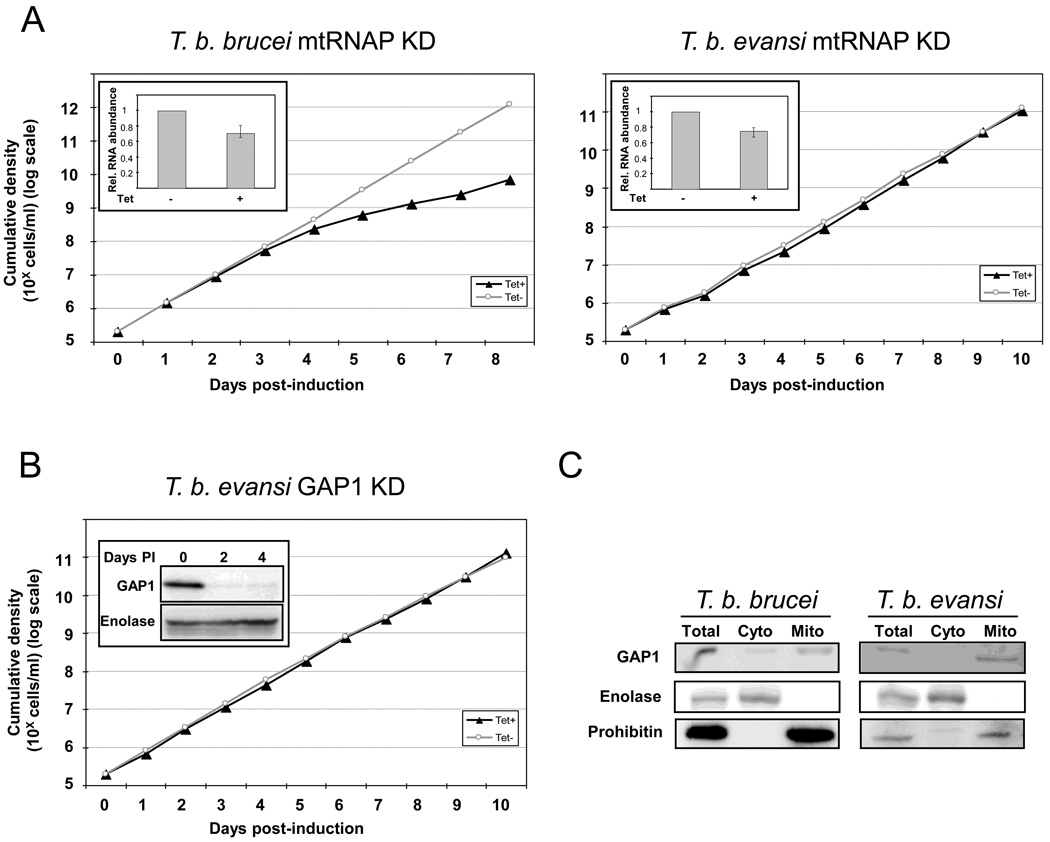
Despite its functional down-regulation and morphological reduction, numerous proteins are still imported into the mitochondrion of the bloodstream stage of T. b. brucei. Indeed, the RNA editing machinery, composed of proteins imported from the cytosol, is essential both at this and the procyclic stage [8]. However, at least one key protein component, RNA editing ligase 1, of the editing machinery is dispensable in the mitochondrion of T. b. evansi [7]. It was of interest to test whether a ubiquitous protein, such as the mitochondrial RNA polymerase (mtRNAP) is also non-essential in these cells. To test this, we used dyskinetoplastic strain of T. b. evansi genetically engineered for overexpression of double-stranded RNA to facilitate RNA interference (RNAi) [7]. A p2T7-177 construct containing the same mtRNAP gene fragment used previously for RNAi in the T. b. brucei procyclic stage [16] was electroporated into both the T. b. brucei bloodstream stage and T. b. evansi and transformants were selected using phleomycin. As no antibody is available against mtRNAP, the efficiency of RNAi was confirmed by quantitative real time PCR as described previously [10,13], with cDNA reverse-transcribed from RNA isolated from knockdown cells lines grown in medium that contained or lacked the RNAi-induction agent tetracycline (Fig. 1A, insets). Three days after RNAi induction similar downregulation of mtRNAP mRNA, namely 71 and 75% of the non-induced mRNA levels, was observed in T. b. brucei and T. b. evansi RNAi-silenced cells, respectively, This level of mtRNAP mRNA reduction was sufficient to inhibit growth of the bloodstream stage of T. b. brucei (Fig. 1A), which was expected as its expression was earlier shown to be essential in the procyclic stage [16]. However, ablation of the target mRNA in the dyskinetoplastic cells did not have any impact on their growth (Fig. 1A). Therefore, in contrast to T. b. brucei, mtRNAP is dispensable in T. b. evansi.
Figure 1. Mitochondrial RNA polymerase (mtRNAP) and guide RNA associated protein 1 (GAP1) are not essential for T. b. evansi.
(A) A comparison of the growth effect of RNAi-silencing of mitochondrial RNA polymerase in T. b. brucei bloodstream stage and T. b. evansi. The cummulative density of trypanosomes is depicted on the y-axis on a log scale and the days after tetracycline (Tet) induction are shown on the x-axis. Growth of RNAi-induced cells is plotted with the black line and full triangles, while the non-induced controls are shown as a grey line with open circles. Growth of T. b. brucei cells is in the left panel and T. b. evansi is in the right panel. Insets depict graphs showing the relative abundance of mtRNAP mRNA from cells grown three days in the presence of the RNAi-induction agent tetracycline (tet+) as compared to cells grown for the same time in medium lacking the drug (tet-). The abundance was determined by quantitative real-time (q) PCR using as a template cDNA generated by random hexamer primers from total RNA isolated from both samples using the following primer pair (forward 5'-CAGCATGAAGATCTCGGTGA-3' and reverse 5'-CGTACAATGGCTTCCCAGTT-3'). The relative mRNA levels were normalized using qPCR data obtained with the previously described primers amplifying β-tubulin [10, 13] from the same cDNA. All qPCR reactions were done in triplicate and in parallel with a negative control to ensure that the signal was not due to genomic DNA contamination. Standard deviations for the determined relative levels of mtRNAP mRNA in tet+ as compared to tet- samples are shown by error bars.
(B) RNAi-silencing of GAP1 does not influence cell division. Graph plotted and labeled as described for Fig 1A. Inset shows a Western blot of cells grown in the absence (0) and presence of Tet for two (2) and four (4) days. The membrane was immunodecorated with either anti-GAP1 (top) or anti-enolase antibodies (bottom), and visualized via peroxidase-conjugated secondary antibodies against rabbit IgGs.
(C) GAP1 is imported into the mitochondria of T. b. brucei (left) and T. b. evansi (right). Digitonin fractions were obtained as described in the text and 10 µg of protein from total, cytosolic (Cyto) and mitochondrial (Mito) fractions were separated by SDS-PAGE and transferred onto PVDF membrane, which was immunodecorated and visualized with antibodies against GAP1 (top), enolase (middle), and prohibitin (bottom) as described in Fig 1B.
This result led us to a similar question with the guide (g) RNA associated protein 1 (GAP1), which is essential for both the procyclic and bloodstream stages of T. b. brucei [10]. After electroporation with a linearized p2T7-177 vector containing a previously used fragment of the GAP1 gene [10], clonal cell lines were selected and RNAi induced as described above. At two and four days after RNAi induction, total cell lysates were collected and separated by SDS-PAGE and transferred to PVDF membranes, which were subsequently immunoprobed with anti-GAP1 antibodies, as well as with antibodies against cytosol-specific enolase. Under these conditions GAP1 expression is efficiently down-regulated when compared to the non-induced control (Fig. 1B; inset). Despite this down-regulation (Fig. 1B), no effect on cell growth was observed in the GAP1-silenced T. b. evansi cells, even after 10 days following RNAi induction. This result is in contrast to GAP1 down-regulation in T. b. brucei bloodstream cells, which exhibit growth inhibition after three days of RNAi induction [10]. We can therefore conclude that the elimination of mtRNAP and GAP1, two essential proteins in the T. b. brucei bloodstream stage, has no effect in the dyskinetoplastic T. b. evansi.
To confirm that GAP1 is imported into the mitochondrion of bloodstream T. b. brucei and T. b. evansi, as has been shown in the procyclic stage of the former sub-species, digitonin fractionation was performed [17,18] to separate the cell contents into their mitochondrial and cytosolic constituents using protocols slightly modified for a given stage. Proteins from total, cytosolic and mitochondrial fractions (10 µg/lane) were used for Western analysis as described above. The purity of fractions was assayed using antibodies against enolase and prohibitin [19], which are specific for cytosol and mitochondrion, respectively (Fig. 1C). An immunopositive signal was detected with the anti-GAP1 specific antibodies in mitochondrial and total cell fractions in both cell-types (Fig. 1C), indicating its import into the organelle in the dyskinetoplastic trypanosomes despite its apparent redundancy (Fig. 1B).
GAP1, like the RNA editing ligase, is a highly specialized and rather unique mitochondrial protein [8,10,20]. However, mtRNAP is a virtually ubiquitous component of the aerobic eukaryotic mitoproteomes, with undisputed essentiality [16]. Therefore, its apparent futile import into a mitochondrion devoid of a genome in T. b. evansi has important consequences. Firstly, it is obvious that these proteins do not have any moonlighting functions other than in RNA metabolism. Second, the protein import machinery is totally independent on the genomic status of the organelle, as it apparently imports dozens or even hundreds of different proteins with no function in this mitochondrion and thus produced and imported in vain. It is puzzling that in an organism with an eight hour generation time, which thrives in an environment where it is under constant selective pressure from the host immune response and from competition for nutrients with other trypanosomes, no mechanism exists that would prevent this obviously wasteful behavior. It is also plausible that stringent regulation of protein-specific import may be more energy demanding than futile import of the non-essential proteins.
tRNAs are imported into the T. b. brucei bloodstream mitochondrion
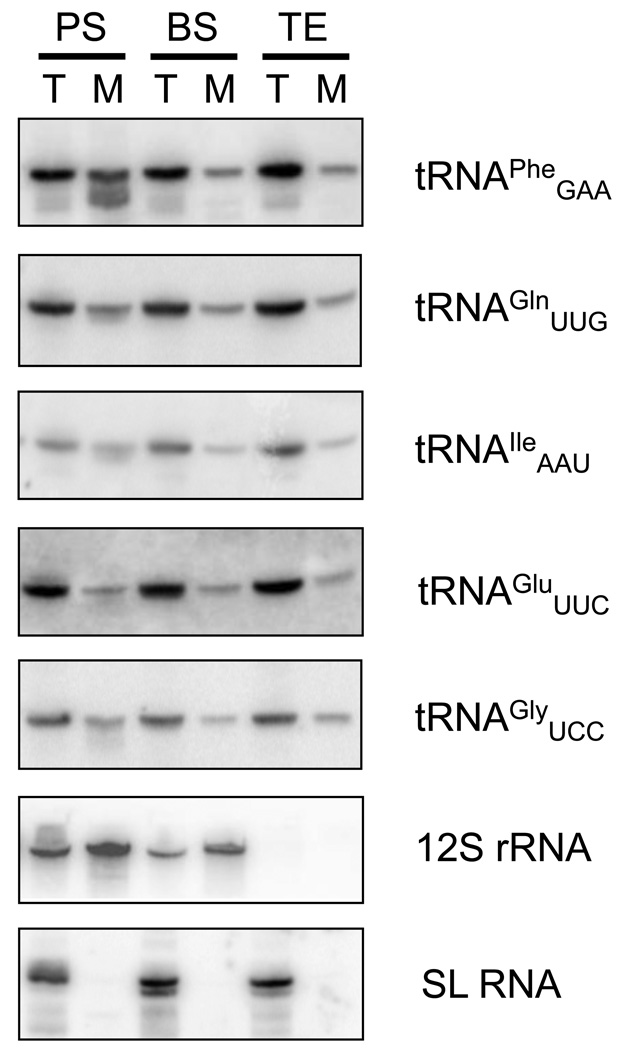
It is now widely accepted that in procyclic T. b. brucei, due to the absence of tRNA genes in the kDNA network, all tRNAs have to be imported from the cytosol into the mitochondrion for translation to occur [21]. However, nothing is known about this process in the bloodstream stage. It is expected that despite significant differences in mitochondrial metabolism between both stages, protein translation and therefore tRNA import are still required. To date, indirect evidence is available for mitochondrial translation of at least one protein in this life cycle stage [7,22]. Furthermore, nothing is known about the state of tRNA import into the akinetoplastic strains. Logically in these strains, the lack of mitochondrial genome obviates the need for organellar translation and conversely tRNA import. Before addressing tRNA import in T. b. evansi, we decided to first clarify the situation in the bloodstream stage of T. b. brucei. To this end, approximately 109 T. b. brucei cells were purified from the blood of an intraperitoneally-infected rat using a DEAE column. Similar amounts of procyclic T. b. brucei were also obtained by cultivation in SDM-79 medium. Mitochondria were then isolated from the procyclic and bloodstream cells by digitonin fractionation. The relative purity of the resulting mitochondrial and cytosolic fractions was confirmed by the aforementioned Western blot analysis using the compartment-specific antibodies against GAP1 and prohibitin (mitochondrial markers) and enolase (cytosolic markers) (Fig. 1C). Next, using guanidinium extraction, total and mitochondrial RNAs were purified from cell lysates and digitonin-extracted organelles, respectively. Northern blot analysis detected individual RNA species by probing with specific labeled oligonucleotides (Fig. 2). Mitochondrial-encoded 12S rRNA and the spliced leader RNA, used as mitochondrion and cytosol specific controls, respectively, verified the purity of the analyzed fractions. The set of five probes specific for individual tRNAs (tRNAPhe, tRNAGle, tRNAIle, tRNAGlu and tRNAGly) showed that all these species are present in the organelle of either stage, at comparable amounts (Fig. 2), demonstrating that tRNA import occurs in bloodstream trypanosomes.
Figure 2. tRNAs are imported into the T. b. evansi mitochondrion.
Total and/or mitochondrial RNA was purified using guanidinium extraction. Mitochondria were isolated from 4 × 108 cells using fractionation with 0.05 % final concentration of digitonin in the SoTE buffer. Obtained vesicles were subsequently treated with 2 µg/ml RNAse A in the SoTE buffer to remove contaminating cytosolic RNA. Samples of total (10 µg) and mitochondrial RNA (2.5 µg) were separated on denaturing 8 % polyacrylamide gel with 8 M urea and electrobloted to Zeta probe membranes, which were subsequently probed with [32 P]5´ end-labeled oligonucleotides specific for a given RNA. After the run, total (T) and mitochondrial (M) RNAs from procyclic (PS) and bloodstream stages of T. b. brucei (BS) and T. b. evansi (TE), blotted and visualized with radioactively labeled oligonucleotides specific for tRNAPhe, tRNAGle, tRNAIle, tRNAGlu and tRNAGly. 12S rRNA and spliced leader (SL) RNA were used as controls specific for mitochondrial and cytosolic fractions, respectively.
tRNAs are imported into the T. b. evansi mitochondrion
The only T. b. evansi strain engineered for inducible expression of transgenes is dyskinetoplastic, which means that it contains residual kDNA minicircles [7], whereas the strain for which tRNA import and modification has been herein assayed is the akinetoplastic 805 strain originally isolated from a water buffalo in China [13]. This strain completely lost its mitochondrial genome and consequently does not contain any RNA molecules derived from organellar transcription. Since neither translation nor tRNAs are required in this mitochondrion, it was reasonable to assume that their import does not occur. To corroborate this, a rat was inoculated with the 805 strain and parasites were purified at a terminal stage of infection. Total as well as mitochondrial RNAs obtained by digitonin fractionation were analyzed by Northern blot hybridization (Fig. 2). A spliced leader RNA-specific oligonucleotide probe showed that the mitochondrial fractions were devoid of cytosolic contamination, while the lack of any signal obtained with a 12S rRNA-specific probe confirmed that kDNA is absent from the 805 strain (Fig. 2). However, contrary to our expectations, signals were obtained by subsequent hybridization with five probes specific for individual aforementioned tRNAs, demonstrating that despite the lack of a mitochondrial genome, tRNA import is unabated in the akinetoplastic trypanosomes.
tRNAs are thiolated only in the T. b. brucei mitochondrion
It was shown recently that tRNAs are thiolated in the procyclic T. b. brucei but this modification plays no role in tRNA import [15,23]. These observations significantly differ from a previous report obtained with Leishmania tarentolae, a flagellate related to the genus Trypanosoma, where tRNA thiolation may serve as a negative determinant for mitochondrial tRNA import [24]. In light of the current results, we tested whether tRNA thiolation also occurs in the bloodstream stage of T. b. brucei and T. b. evansi.
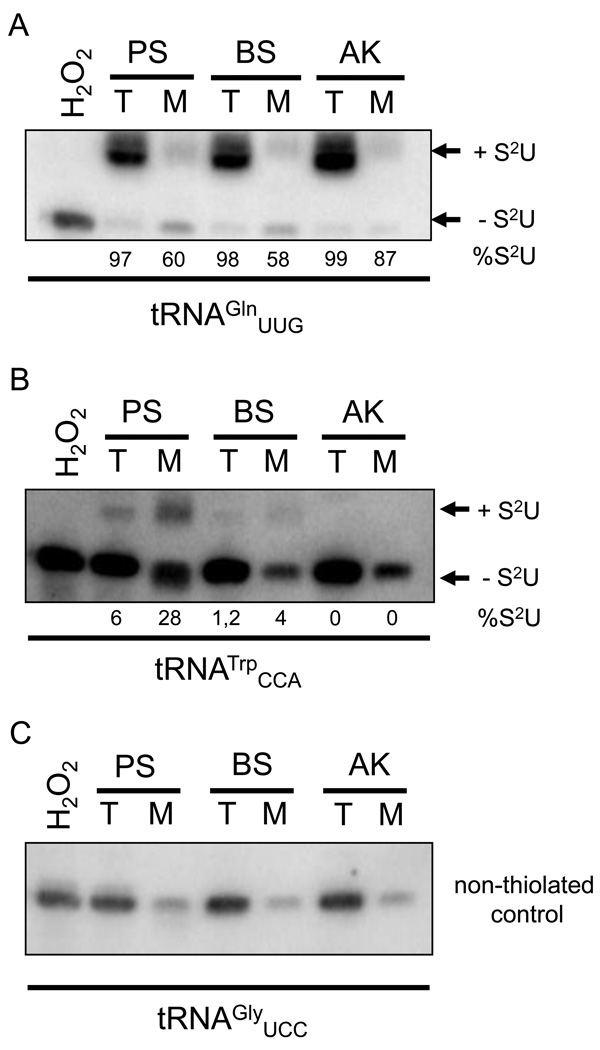
Thiolation of different tRNAs was analysed by APM gels as previously described [25]. These gels permit separation of thiolated and non-thiolated tRNAs on the basis of differential mobility during electrophoresis (Fig. 3). Northern blots of tRNAs obtained from total and mitochondrial cell fractions of T. b. brucei (both the procyclic and bloodstream stage) and T. b. evansi, resolved in the presence of APM, showed that cytosolic tRNAGln was thiolated to similar levels in all fractions tested (Fig. 3A). This tRNA species is known to be thiolated solely in the cytosol. We also analyzed the thiolation levels of tRNATrp, the only tRNA known to be thiolated exclusively in the mitochondrion following import [15,23,25]. Although this tRNA is highly thiolated in the mitochondrion of procyclic T. b. brucei, its thiolation is somewhat lower in the organelle of the bloodstream stage. With the method used, which is able to detect down to ~0.5% of thiolated species, and even with double amount of mitochondrial RNA loaded (5 µg; data not shown), we were unable to detect the band corresponding to the thiolated tRNATrp in T. b. evansi (Fig. 3B).
Figure 3.
tRNAs are thiolated in cytosol but not in the mitochondrion of T. b. evansi. Total (T) and mitochondrial (M) RNA from procyclic (PS) and bloodstream stages of T. b. brucei (BS), and T. b. evansi (TE) was separated in APM-containing polyacrylamide gels. Northern blots were probed for tRNAGln (A) and tRNATrp (B). Bands corresponding to thiolated (+S2U) and non-thiolated tRNAs (-S2U) are indicated. H2O2 was used as a control to show complete oxidation of thiolation, eliminating the mobility shift. The tRNAGly (C) served as a loading control. Percent thiolation (% S2U) was calculated by dividing the signal from the shifted band by the total signal of shifted (thiolated) and nonshifted and multiplied by 100.
The fact that thiolation occurs in the bloodstream stage testifies indirectly to the presence of cysteine desulfurase, an enzyme that is essential for Fe-S cluster assembly [18] and thiolation of both cytosolic and mitochondrial tRNAs in procyclic T. b. brucei [25]. Moreover, this cysteine desulfurase also has a limited selenocysteine lyase activity in the insect stage trypanosomes [26], yet its function(s) in the mammalian stage remain(s) to be established. The thiolation status of mitochondrial tRNA is revealing in several respects. As has been shown previously, the T. b. brucei bloodstream stage lacks any respiratory complexes in its mitochondrion [3–5], yet this has apparently no impact on tRNA import. Therefore, we posit that the Rieske Fe-S protein, as well as other subunits of respiratory complexes, do not play any role in the import of the tRNAs in T. b. brucei regardless of developmental stage, a conclusion contradicting results published for the Leishmania species [27]. In further support of this notion, the absence of the Rieske Fe-S protein and a subunit of respiratory complex IV has been confirmed in numerous T. b. evansi strains [13]. Furthermore, our data strongly suggest the absence of the interdependence between mitochondrial translation and tRNA import. The latter process apparently operates by a default mechanism, regardless of the situation “within” the organelle in terms of DNA content and translation requirements. It is also worth noting that thiolation is the only process, with the exception of the status of kDNA, which differs, at least to some extent, between the T. b. brucei bloodstream stage and T. b. evansi. As apparent from their comparison, thiolation of mitochondrial tRNAs, consistently weaker in the T. b. brucei bloodstream stage as compared to the insect stages, was undetectable in the akinetoplastic T. b. evansi (Fig. 3B). We propose that this is caused by a progressive degeneration of the apparently dysfunctional T. b. evansi mitochondrion, which imports tRNAs to no purpose. It has been suggested elsewhere that T. b. evansi may not be monophyletic, but rather an assembly of strains that for one reason or another started loosing their capacity to faithfully replicate, transcribe or edit their organellar nuclear acids [11–13], with mitochondrial tRNA thiolation now added to the list of possible triggers of dys/akinetoplastidy.
CONCLUSIONS
Our results further strengthen the argument that T. b. evansi represents the bloodstream stage of T. b. brucei that lost (part of) its kinetoplast DNA. We propose that organelles of both trypanosomes, likely having a relationship reminiscent of that between the yeast Saccharomyces cerevisiae and its petite mutants, are an ideal object for study of interactions between the mitochondrion and the nucleus. Our data further stress the impression that the organelle is much more independent of the nucleus than appreciated so far.
ACKNOWLEDGEMENTS
We thank Achim Schnaufer (University of Edinburgh) for sharing T. b. evansi RNAi cell line. This work was supported by the Grant Agency of the Czech Republic 204/09/1667 and 560/3092, the Ministry of Education of the Czech Republic (2B06129, LC07032 and 6007665801), and the Praemium Academiae award to J.L., and NIH grant GM084065 to J.D.A.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Schnaufer A, Domingo GJ, Stuart K. Natural and induced Dyskinetoplastic trypanosomatids: how to live without mitochondrial DNA. Int J Parasitol. 2002;32:1071–1084. doi: 10.1016/s0020-7519(02)00020-6. [DOI] [PubMed] [Google Scholar]
- 2.Claes F, Buscher P, Touratier L, Goddeeris BM. Trypanosoma equiperdum, master of disguise or historical mistake. Trends Parasitol. 2005;21:316–321. doi: 10.1016/j.pt.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 3.Besteiro S, Barrett MP, Rivière L, Bringaud F. Energy generation in insect stages of Trypanosoma brucei: Metabolism in flux. Trends Parasitol. 2005;21:185–191. doi: 10.1016/j.pt.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 4.Tielens AGM, van Hellemond JJ. Surprising variety in energy metabolism within Trypanosomatidae. Trends Parasitol. 2009;25:482–490. doi: 10.1016/j.pt.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 5.Hannaert V, Bringaud F, Opperdoes FR, Michels PAM. Evolution of energy metabolism and its compartmentation in Kinetoplastida. Kinetopl Biol Dis. 2003;2:11. doi: 10.1186/1475-9292-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaudhuri M, Ott RD, Hill GC. Trypanosome alternative oxidase: from molecule to function. Trends Parasitol. 2006;22:484–491. doi: 10.1016/j.pt.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 7.Schnaufer A, Clark-Walker JD, Steinberg AG, Stuart K. The F1-ATP synthase complex in bloodstream stage trypanosomes has an unusual and essential function. EMBO J. 2005;24:4029–4040. doi: 10.1038/sj.emboj.7600862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schnaufer A, Panigrahi AK, Panicucci B, Igo RP, Jr, Wirtz E, Salavati R, Stuart K. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science. 2001;291:2159–2162. doi: 10.1126/science.1058955. [DOI] [PubMed] [Google Scholar]
- 9.Fisk JC, Ammerman ML, Presnyak V, Read LK. TbRGG2, an essential RNA editing accessory factor in two Trypanosoma brucei life cycle stages. J Biol Chem. 2009;283:23016–23025. doi: 10.1074/jbc.M801021200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hashimi H, Čičová Z, Novotná L, Wen Y-Z, Lukeš J. Kinetoplastid guide RNA biogenesis is dependent on subunits of the mitochondrial RNA binding complex 1 and mitochondrial RNA polymerase. RNA. 2009;15:588–599. doi: 10.1261/rna.1411809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jensen RE, Simpson L, Englund PT. What happens when Trypanosoma brucei leaves Africa. Trends Parasitol. 2008;24:428–431. doi: 10.1016/j.pt.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lun ZR, Lai DH, Li FJ, Lukeš J, Ayala FJ. Trypanosoma brucei: two steps to spread out from Africa. Trends Parasitol. 2010;26:434–437. doi: 10.1016/j.pt.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 13.Lai DH, Hashimi H, Lun ZR, Ayala FJ, Lukeš J. Adaptations of Trypanosoma brucei to gradual loss of kinetoplast DNA: T. equiperdum and T.evansi are petite mutants of T. brucei. Proc Natl Acad Sci USA. 2008;105:1999–2004. doi: 10.1073/pnas.0711799105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schnaufer A. Evolution of dyskinetoplastic trypanosomes: How, and how often? Trends Parasitol. 2010;26:557–558. doi: 10.1016/j.pt.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paris Z, Rubio MAT, Lukeš J, Alfonzo JD. Mitochondrial tRNA import in Trypanosoma brucei is independent of thiolation and the Rieske protein. RNA. 2009;15:1398–1406. doi: 10.1261/rna.1589109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grams J, Morris JC, Drew ME, Wang Z, Englund PT, Hajduk SL. A trypanosome mitochondrial RNA polymerase is required for transcription and replication. J Biol Chem. 2002;277:16952–16959. doi: 10.1074/jbc.M200662200. [DOI] [PubMed] [Google Scholar]
- 17.Charrière F, Helgadóttir S, Horn EK, Söll D, Schneider A. Dual targeting of a single tRNATrp requires two different tryptophanyl-tRNA synthesis in Trypanosoma brucei. Proc Natl Acad Sci USA. 2006;103:6847–6852. doi: 10.1073/pnas.0602362103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smíd O, Horáková E, Vilímová V, Hrdý I, Cammack R, Horváth A, Lukeš J, Tachezy J. Knock-downs of iron-sulfur cluster assembly proteins Isis and IscU down-regulate the active mitochondrion of procyclic Trypanosoma brucei. J Biol Chem. 2006;281:28679–28686. doi: 10.1074/jbc.M513781200. [DOI] [PubMed] [Google Scholar]
- 19.Týč J, Faktorová D, Kriegová E, Jirků M, Vávrová Z, Maslov DA, Lukeš J. Probing for primary functions of prohibitin in Trypanosoma brucei. Int J Parasitol. 2010;40:73–83. doi: 10.1016/j.ijpara.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 20.Amaro RE, Schnaufer A, Interthal H, Hol W, Stuart KD, McCammon JA. Discovery of drug-like inhibitors of an essential RNA-editing ligase in Trypanosoma brucei. Proc Natl Acad Sci USA. 2008;105:17278–17283. doi: 10.1073/pnas.0805820105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schneider A, Martin JA, Agabian N. A nuclear encoded tRNA of Trypanosoma brucei is imported into mitochondria. Mol Cell Biol. 1994;4:2317–2322. doi: 10.1128/mcb.14.4.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hashimi H, Benkovičová V, Čermáková P, Lai DH, Horváth A, Lukeš J. The assembly of F1F0-ATP synthase is disrupted upon interference of RNA editing in Trypanosoma brucei. Int J Parasitol. 2010;40:45–54. doi: 10.1016/j.ijpara.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 23.Bruske EI, Sendfeld F, Schneider A. Thiolated tRNAs of Trypanosoma brucei are imported into mitochondria and dethiolated after import. J Biol Chem. 2009;6284:36491–36499. doi: 10.1074/jbc.M109.064527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaneko T, Suzuki T, Kapushoc ST, Rubio MA, Ghazvini J, Watanabe K. Wobble modification differences and subcellular localization of tRNAs in Leishmania tarentolae: implication for tRNA sorting mechanism. EMBO J. 2003;22:657–667. doi: 10.1093/emboj/cdg066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolgamuth-Benedum JM, Rubio MAT, Paris Z, Long S, Poliak P, Lukeš J, Alfonzo JD. Thiolation controls cytoplasmic tRNA stability and acts as a negative determinant for tRNA editing in mitochondria. J Biol Chem. 2009;284:23947–23953. doi: 10.1074/jbc.M109.029421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poliak P, Van Hoewy D, Oborník M, Zíková A, Stuart KD, Tachezy J, Pilon M, Lukeš J. Functions and cellular localization of cysteine desulfurase and selenocysteine lyase in Trypanosoma brucei. FEBS J. 2010;277:383–393. doi: 10.1111/j.1742-4658.2009.07489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mukherjee S, Basu S, Home P, Dhar G, Adhya S. Necessary and sufficient factors for the import of transfer RNA into the kinetoplast mitochondrion. EMBO Rep. 2007;8:589–595. doi: 10.1038/sj.embor.7400979. [DOI] [PMC free article] [PubMed] [Google Scholar]