Abstract
Translational medicine applications would like to leverage the biological and biomedical ontologies, vocabularies, and data sets available on the semantic web. We present a general solution for RDF information set reuse inspired by database views. Our view definition language, vSPARQL, allows applications to specify the exact content that they are interested in and how that content should be restructured or modified. Applications can access relevant content by querying against these view definitions. We evaluate the expressivity of our approach by defining views for practical use cases and comparing our view definition language to existing query languages.
Keywords: translational medicine applications, RDF views, ontologies, vocabularies
1. Introduction
The semantic web seeks to enable computers to automatically associate and process well-defined information on the web. To that end, a number of biological and biomedical information sets have been developed for or converted to semantic web formats. These information sets include vocabularies, ontologies, and data sets; they may be available in basic RDF [1] or languages with higher-level semantics, such as OWL [2].
Translational medicine applications want to leverage the biomedical information sets available on the semantic web. For example, radiologists may want an application for viewing and annotating medical images. The application can aid annotation by offering suggestions for visible anatomical parts from the Foundational Model of Anatomy (FMA) [10]. Additionally, if suspicious nodes are identified on an image, the application can offer suggestions from NCI Thesaurus [21] for annotating the nodes.
This paper asks the question: how can we make it easier for applications to leverage the information sets available on the semantic web? In leveraging these information sets, applications must contend with their size, content depth and breadth, and rate of change.
Many biomedical information sets are of significant size. For example, Reactome [22] as distributed through NeuroCommons contains 3.6M triples, the FMA 3.0 in OWL contains 1.7M triples, and UniProt [5] v13.4 contains 1.7B triples. Additionally, medical applications may require information from multiple information sets, such as the FMA/NCI example above. The combined information sets may be too large for applications to easily manipulate.
Applications may only need a portion of the content contained in an information set, which are likely to have a wider scope and greater depth than that needed by any particular application. Applications determine the set of relevant content needed from an information set; the set could be a list of concepts, a subgraph of related terms, a sub-ontology, or any other subset of information. Techniques ( [9], [41], [39], [43]) have been developed for some of these scenarios and are discussed in related work.
While leveraging biomedical information sets, applications may wish to augment, modify, or restructure selected content. For example, an application may seek to define a new relationship between selected concepts. Alternatively, an application may disagree with a portion of the information set (e.g. the content is too coarsely specified) and need to “fix” it.
In addition to specifying relevant content, applications must accommodate updates to information sets. If an application includes a materialized subset, new materialized subsets may need to be derived and distributed every time an update occurs. An application that queries the information set accesses the most up-to-date content; however, the queries must be written to restrict responses to the application-specified content.
We propose a general solution for information set reuse. We validate our approach with eight different use cases over one or more of four biomedical datasets. Our approach is inspired by database views. A database view identifies a subset of a dataset that can be accessed through the view; queries on the view can only access the specified subset. However, a database view may export a restructured or modified version of the data; the exported view is not limited to faithfully replicating a subset of the underlying information.
We present a view definition language vSPARQL (briefly described in [8]) that allows the specification of views for semantic web content. vSPARQL is used to specify both the selection of the information that can be accessed through the view and how the selected information is reorganized and/or modified through the view. Applications can submit queries against these views to access the most up-to-date versions of the information sets upon which they depend.
Our view definition language allows for the reuse of RDF graphs. It is able to define views over information sets in RDF, OWL (all flavors), and potentially other language formats layered on top of RDF. The decision to manipulate RDF graphs restricts our view definitions to facts that are explicitly stated in the information set; if the information set uses a higher-level language with formally defined logical semantics, we do not use logical inference to infer new facts. For example, our language cannot perform all of the reasoning tasks (e.g. classification) available to tools that leverage the higher-level semantics of languages like OWL. The view definition is not limited by the information set’s underlying semantics; for example, a view can define new associations between concepts that are not supported by the original information set.
Research has been conducted using formal logics to infer new facts from formally defined information sets. We consider this research to be orthogonal to the general problem of enabling a specified subset of content to be reused by an application. For example, reasoners can infer new facts from an available information set; these new facts can be materialized and combined with the original information set through a view.
Our view definition language, vSPARQL, is an extension of SPARQL [11]. vSPARQL was developed to address the needs of practical use cases that could not be met by SPARQL or other semantic web query languages. The language extensions include virtual graphs, recursion, and the ability to construct new resource identifiers, for both querying and node creation, from those existing in input data sources. We demonstrate the expressiveness of our language by using it to derive a set of views for our eight motivating use cases. A GUI application http://ontviews.biostr.washington.edu:8080/VSparQL_Service/GUI/vsparql_oh_degrafa.html has been developed to simplify the creation of view definitions.
This work makes several contributions. We have identified the functionality required for a general solution to enable reuse of information sets on the semantic web. The required functionality has been used to define and implement a view definition language as a set of extensions to SPARQL. We have evaluated the expressivity of our solution on eight examples of information set reuse, and we have compared the functionality of our solution to that of existing semantic web query languages.
vSPARQL has been developed to be used in a clearinghouse similar to Bio-Portal [4], which contains a collection of ontologies and views defined over them. For our prototype, all of the information sets are either local, downloadable as files from the web, or accessible in an SDB [19] datastore accessible via JDBC. Although important, we do not address the difficult problem (exacerbated by the presence of blank nodes) of accessing data through remote SPARQL servers.
The rest of the paper is organized as follows. Section 2 describes our eight motivating examples of application reuse of semantic web information sets. Section 3 identifies the requirements of our view definition language, which is presented in Section 4. Section 5 discusses our implementation and a GUI application for defining views. We evaluate our solution by using it to derive views, described in Section 6, for our eight use cases. We compare our approach to related work and evaluate our language against our use cases and several semantic web query languages in Section 7. We end with a discussion of future work before concluding in Section 9.
2. Motivation: Use Cases
Can we make it easier for many different applications to leverage biomedical information sets on the semantic web? To address this question, we consider eight application specific use cases for leveraging large semantic web information sets. These use cases reuse the information sets in various ways, including list, subgraph, and sub-ontology extractions; modifications of extractions; and generation of new facts, including combining content across multiple information sets. In the past, several of these use cases were addressed through custom programs written against the Protege [12] API; unfortunately, these programs are not generalizable and they needed to be rerun when the information set was updated.
2.1. Use Cases
The use cases we consider in this work are over four information sets: NCI Thesaurus [21], Reactome [22], Ontology of Physics for Biology [13], and the Foundational Model of Anatomy [10]. We briefly discuss each information set before describing the use cases.1
The NCI Thesaurus (NCIt) is an open-source vocabulary containing information about cancer, including information for clinical care and basic and translational research. It contains over 34,000 concepts and is available in OWL format.
Reactome is an open-source curated database of biological pathways. It contains information on humans and 22 other non-human species. Reactome is available at NeuroCommons [23]; it consists of an OWL schema and associated data and contains more than 3.6 million RDF triples.
The Ontology of Physics for Biology (OPB) is a ontology containing concepts from classical physics necessary for representing, annotating, and encoding quantitative models of biological processes. It is developed in OWL and contains approximately 2,000 RDF triples.
The Foundational Model of Anatomy (FMA) is a reference ontology representing the structure of the human body. The FMA contains approximately 75,000 classes, over 120,000 terms and 168 relationship types to represent a model of anatomy. The FMA is developed in Protege [12] frames. The FMA utilizes meta-modeling extensively; classes are directly related, via properties, to other classes. Thus, its direct export to OWL, which preserves these relationships, puts the OWL version of FMA into OWL Full [37]. This version contains 1.7M triples.
Eight different use cases were defined over these four information sets. The majority of our use cases (6 of the 8) were motivated by actual requests from collaborating researchers in biomedical informatics, biosimulation, and knowledge-based image segmentation ( [45], [46], [47], [48], [49]). While there were no specific requests for the remaining use cases (2 of 8), they illustrate requested functionality in a view generation mechanism.
For ease of understanding, below we group the use cases into three categories: extraction, extraction and modification, and multiple information sets as input.
Extraction
Three use cases seek to extract one or more subgraphs from an information set without introducing new or modifying existing triples.
Mitotic cell cycle: From Reactome, extract the mitotic cell cycle, all of its component processes, and their accompanying labels. This extraction will be used by an application for exploring processes and their subprocesses. Although the data leverages an OWL specification, only process data should be included in the view.
Organ spatial location: From the FMA, extract the spatial information that can be used by image recognition software to automatically identify objects in medical images of the gastrointestinal tract. The result should only include the organs found in the gastrointestinal tract and their associated orientation and location properties.
NeuroFMA ontology: From the FMA, generate a subset ontology that contains all of the information regarding neuro-anatomy. It should contain all the neural structures from the FMA, their attributes, and the properties connecting them.
Extraction and Modification
Four use cases require the introduction of new facts or the modification of facts extracted from an input information set.
NCI Thesaurus simplification: Create a modified version of the NCI Thesaurus (NCIt) that approximates the information displayed to users by the web-based NCI browser. In addition to the original content, this modified NCIt contains new high-level relationships that are abstracted away from their low-level OWL representation. While the constructs of the OWL language are designed for maximizing expressiveness while ensuring completeness and decidability of logical inference, it can be difficult for users to understand the relationships that they are describing. The new relationships more directly map to users’ understanding of the content.
Biosimulation model editor: From the FMA, identify the relationship graph between a set of concepts; the graph is displayed by a biosimulation model editing tool. The application displays the relationships between the concepts as a restructured, pared down hierarchy; the only nodes that should remain in the hierarchy are those representing leaf concepts and intermediate nodes with multiple children. The result changes as new concepts are added and removed from the set of interest. As the user selects concepts within the hierarchy, the application queries additional properties from the FMA and displays it.
Blood contained in the heart: From the FMA, generate a graph, to be used by physiology modelers, representing portions of blood found in the heart. The results include heart parts and the blood portions they contain. In the FMA, only spaces are allowed in the domain of the fma:contains property; in this use case, if a space contains blood, the structures that it is a part of should also be said to contain blood.
Radiologist liver ontology: From the FMA, generate a sub-ontology to be used by an application for annotating medical images of the liver. The sub-ontology contains all of the visible parts of the liver, their corresponding part subgraph, and their associated superclass hierarchy; no properties other than the part and superclass relationships should be included. Modify the structure of the subclass hierarchy to remove the concepts “Cavitated organ” and “Solid organ.”
Use of Multiple Input Information Sets
Our final use case leverages two different input information sets to create new facts.
Blood fluid properties: Combine information from two independent ontologies (FMA and OPB) to create new information for a biosimulation model editing application. Properties of fluids defined in the FMA should be combined with the kinetic properties of fluids defined in the OPB. For example, concepts like blood in aorta and blood in coronary artery in the FMA can be combined with flow, pressure,and viscosity in the OPB [13], resulting in new concepts like blood in aorta flow, blood in coronary artery viscosity, etc. The result contains the newly created resources and their properties which can be used to annotate computational models of physiology.
3. High-level requirements for reuse through views
The goal of this work is to enable different kinds of information set reuse, as demonstrated by our use cases. To that end, we have adapted the notion of database views to allow applications to replace inclusion of large semantic web datasets with remote querying through a view. A view definition language makes it possible to select (and possibly modify) the specific content needed by an application without materializing the information; queries are written against the view definition and evaluated against the original information set.
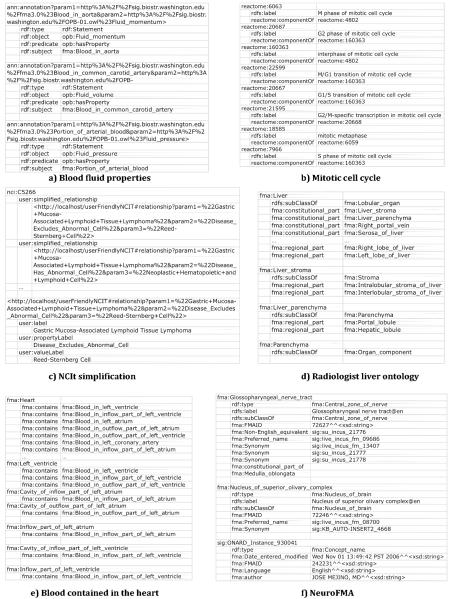
We have identified a set of criteria for a view definition language that permits applications to select and reorganize information to meet their particular needs. We identify the criteria below and indicate (with italics) specific functionality they encompass; the functionality requirements of our use cases are characterized in Figure 1.
Figure 1.
Functionality requirements of each of our motivating use cases.
Input should be RDF graphs
Our view definition language should be applicable to as many information sets on the semantic web as possible. We want it to apply to all languages that can be serialized as RDF, including all flavors of OWL and possibly future semantics web languages.
Because our view definition language is applied to RDF graphs, it may be harder to create views over information sets that use language formats with higher-level semantics, such as OWL. Our view definitions cannot leverage logically inferred facts that might be available in query languages specific to the high-level language format. Furthermore, the view definitions themselves become more complex and multi-layered as we use RDF-level language to traverse OWL restrictions, for example. However, not all information sets will use the same language format and we want to enable views over all RDF-based formats. Additionally, as large RDF stores of data (not ontologies or vocabularies) become more readily available, applications will need to carve out relevant and manageable subsets for reuse.
Output should be an RDF graph
This ensures that tools developed for manipulating our input graphs can be used on the output of a view definition (i.e. closure). Although the result of a view definition is RDF, the output can still contain constructs of higher-level languages built on top of RDF. For example, a view definition may produce an OWL ontology. Additionally, this approach does not require that the result of a view use the same language format as the input. For example, a view over an OWL information set may result in an RDF graph without any OWL constructs.
Because the output of a view definition is an RDF graph, our language can allow view definitions to be wrapped as intermediate results. Intermediate results may simplify a view definition by making it more modular, just as subroutines make it easier to write and maintain an application. If multiple graphs are being queried, allowing the user to query each source separately can make the query simpler to write.
View definitions should be able to include arbitrary facts from the input
Applications are responsible for identifying exactly which triples to include and exclude in a view definition. To that end, the view definition must explicitly indicate the triples to include and exclude; this is in contrast to techniques that use formal logics to determine which triples must be included in a view.
Our view definition language must support basic edge selection, which allows a view to indicate inclusion or exclusion of triples based upon individual triple values. However, because of their complexity, large information sets may have very complicated property paths between two arbitrary resources. For example, a user may want to find recursively all of the constitutional and systemic parts of the liver. Our view language should support the specification of paths of arbitrary length (e.g. [constitutional_part | systemic_part]+ ) for identifying triples to include in a view. Additionally, because an application may wish to extract large swaths of an input based upon the relationship between two resources, our language should support subgraph extraction over paths of arbitrary length. This would, for example, allow an application to identify and include the entire subgraph starting at organ and containing (recursively) all of its parts.
Views should be able to restructure, modify or augment selected facts
Views should not be limited to exposing just a subset of the original information set. Applications should be able to define new properties between resources in the input source, or introduce entirely new resources. New resources and properties can be introduced using static edge creation, through the introduction of hard-coded triples, or dynamic edge creation using the results of querying over the input source. Some applications may need to introduce anonymous nodes for inclusion in a view, while other applications may need to dynamically create nameable nodes or edges using the results of querying over the input source.
For some applications, the introduction of new triples may not be sufficient. These applications may wish to restructure the subgraphs that they extract from an information set. If these extractions are the result of recursively following a path, restructuring may require iterating over paths of arbitrary length to generate new relationships while eliminating others. For example, a view may wish to eliminate non-leaf nodes that have single children from the subgraph. Alternatively, a user may wish to combine two subgraphs by simultaneously iterating over the subgraphs, one edge at a time, and create an entirely new subgraph with elements of each.
Views should be able to leverage multiple input graphs
Our view definition language should allow for a single view that combines content from different graphs.
Views should be distinct from the information set
Our view definition language is intended to be used with large RDF-based information sets. By requiring our views to be separable from the data they reference, we ensure that each view definition does not require modification of a copy of the data.
4. vSPARQL: A view definition language
In this section we describe a view definition language for RDF, called vSPARQL, that supports all of the functionality needed for creating the example views described in Section 2. The language is a series of extensions to SPARQL, the semantic web query language. After describing the relevant aspects of the SPARQL query language, we describe our subquery, recursive subquery, and Skolem function extensions. These extensions provide the ability to: 1) dynamically generate new resource identifiers that can be used for both querying over graphs and generating new resources, 2) define virtual graphs, thus allowing users to explicitly indicate if blank nodes in intermediate result sets can be directly compared to blank nodes in their source graph, and 3) use recursive subqueries to iterate over paths of arbitrary length, including paths containing blank nodes.
4.1. Background: SPARQL
SPARQL is the W3C’s recommendation for querying RDF models. SPARQL queries consist of triple patterns that are evaluated against an underlying RDF graph to find matches. The following SPARQL query demonstrates several language features that we build upon for our ontology view definition language. From the FMA, the query creates a graph containing all of the regional parts of the liver.
CONSTRUCT { fma:Liver fma:regional_part ?c }
FROM NAMED <http://…/fma>
WHERE { GRAPH <http://…/fma> { fma:Liver fma:regional_part ?c } }
The CONSTRUCT keyword indicates that the output of this query is an RDF graph; within the brackets, a list of triple patterns dictate the set of triples that are to be contained in the output RDF. The WHERE clause consists of a set of triple patterns that are evaluated on the input graph to find matches. In this example, ?c is a variable that is bound when a match is found.
The FROM NAMED (or FROM) statement indicates the RDF graph that should be used as input. A query has a set of named graphs specified using the FROM NAMED directive; within a WHERE clause triple patterns can be evaluated against specific named graphs using the GRAPH keyword. A SPARQL query has a default graph which is defined by FROM statements; if the GRAPH keyword is not used, triple patterns are evaluated against this default graph.
In its unmodified form, SPARQL provides several pieces of functionality needed for our view definition language. We automatically get support for multiple sources via FROM and FROM NAMED, and closure, edge selection, static edge creation, dynamic edge creation, and blank node creation via CONSTRUCT.
4.2. vSPARQL
vSPARQL consists of several extensions to SPARQL. Figure 2 details the modifications to the SPARQL grammar which are discussed below.
Figure 2.
vSPARQL modifications to the SPARQL grammar. The bracketed numbers correspond to rules in the SPARQL grammar; modifications to the rules are italicized.
4.2.1. Skolem functions
vSPARQL uses Skolem functions and string manipulation to allow users to dynamically generate nameable (not anonymous) identifiers of both nodes and properties in views. These generated names can be used in both query patterns and CONSTRUCT templates to create new nodes.
In developing a view, users may want to create new entities based upon the information encoded in one or more existing information sets. Creating a new entity amounts to the creation of either 1) a blank node or 2) a new URI that uniquely identifies the new entity. SPARQL does not provide functionality for dynamically creating new URIs based upon query results; nodes’ attributes can be queried and/or modified, but new named nodes (URIs) cannot be created.
vSPARQL incorporates Skolem functions to enable the creation of new nameable identifiers. In general, Skolem functions have the properties such that, for any unique combination of Skolem function and parameters, the same result is always returned; additionally, no two distinct combinations of Skolem function and parameters will map to the same result. In vSPARQL, Skolem functions can occur anyplace in a query that expects a resource node – WHERE clauses, FILTER constraints, and CONSTRUCT templates.
Our language extension enables users to specify the URI of a Skolem function and the arguments that should be used to construct a new node. Skolem functions are specified by a URL with a list of zero or more arguments: [[<skolem_function_url>(arg1, …)]]
The nodes and properties created by Skolem functions are represented in RDF graphs by URLs; the arguments to a Skolem function are web encoded and appended to the URL similar to variables in a web form.
<skolem_function_url>?param1=arg1¶m2=arg2…
Constructing the URL of the new nodes in this manner has the advantage that the new node contains some basic provenance information. By web encoding the URIs of all parameters to the Skolem function, we maintain information about the derivation of the new node and its origins.
Other SPARQL extensions, such as SPARQL++ [25] have proposed the ability to create new nameable resources in CONSTRUCT templates through a combination of string manipulation functions. vSPARQL shares this capability but also adds the ability to use these generated nameable resources in query patterns. For example, the following query finds all of the matching rdf:Statements that have an rdf:object opb:Fluid_momentum; the query returns the first parameter used to generate the rdf:Statement’s annot_view identifier.
SELECT ?a
FROM <http://…/fma_opb>
{
[[annot_view:annotate(?a,?b)]] rdf:type rdf:Statement .
[[annot_view:annotate(?a,?b)]] rdf:object opb:Fluid_momentum .
}
Users may wish to have more control over the URLs that are created by Skolem functions. They may not wish to simply combine entities found in the input graph; they may wish to manipulate resource names queried from an input graph. For example, instead of using http://…/left_lower_leg as a parameter to a Skolem function, a user may simply wish to use “lower leg” as a parameter.
To accommodate user demands for creating new URLs, vSPARQL includes support for a string substitution function strSubst() that can be used within the specification of a Skolem function. strSubst() can be used to generate either the Skolem function URL or the individual function arguments. strSubst(str,regexp,repl) performs string substitution using regular expressions. The function has three inputs: str is the input string that is going to be manipulated; regexp is a regular expression that is evaluated against str; repl is the specification of the string that should be the output of the function. When regexp is evaluated against str, local variable bindings of $1,$2,$3, … are generated as specified by the regexp; these bindings are used to define repl, which is a concatenation of variable bindings and strings. The variable bindings from the regular expression are not visible outside of the repl specification. The output of strSubst() is the string corresponding to repl.
For example, strSubst(“http://…/left_lower_leg”, “(.*)left_(.*)”, “$1$2”) produces the string http://…/lower_leg. String concatenation of two variables $x and $y can be achieved by specifying strSubst(“”,””,”$x$y”).
4.2.2. Subqueries
Our view definition language uses subqueries to provide support for intermediate result sets. Named subqueries were previously proposed in [33], and non-named nested subqueries are supported in [25]. We describe them here to build upon them in defining subqueries that produce virtual graphs, defined in Section 4.2.3.
SPARQL queries specify their data source through the use of the FROM (and FROM NAMED) directive; a URL is required to indicate the RDF graph that is to be used as an input data source. We have extended FROM (and FROM NAMED) to allow temporary data sources to be created on-the-fly through the use of the CONSTRUCT query. The sketch of the syntax for subqueries is:
FROM <http://…/subquery> [ FROM NAMED <http://…/subquery> [
CONSTRUCT { … } CONSTRUCT { … }
FROM … FROM …
WHERE { … } WHERE { … }
] ]
The following example uses a subquery to create a temporary RDF graph which contains all of the FMA’s constitutional parts of the liver. As with any named graph, the GRAPH keyword can be used to apply triple patterns to the subquery-generated RDF graph.
SELECT *
FROM NAMED <http://…/liver_parts> [
CONSTRUCT { fma:Liver fma:constitutional_part ?part }
FROM <http://…/fma>
WHERE { fma:Liver fma:constitutional_part ?part }
]
WHERE { GRAPH <http://…/liver_parts> { ?a ?b ?c } }
In vSPARQL, a subquery’s default graph is empty unless the subquery itself contains a FROM statement. The subquery’s set of named graphs is empty; FROM NAMED statements can be used to add any of the subquery-generated graphs preceding the subquery’s definition. The variables used within a subquery are restricted in scope to the subquery. There are no restrictions on the number of levels of subqueries that can be nested.
The intermediate results generated by subqueries can be referenced in subsequent query clauses. These subqueries provide modularity for constructing large queries from smaller, simpler views. They enable separate input graphs to be queried individually and the results of those queries to be combined later. They also enable subqueries to be cascaded, making it possible to specify queries over existing views by simply wrapping the view as a subquery.
A more complicated example indicates how intermediate results can be used to exclude items from a graph. In this example, the subquery creates a temporary RDF graph called http://…/exclude_list that contains all of the FMA’s constitutional parts of the liver. (Note that there is no straightforward way to build a RDF collection within a query; therefore, in this example, we build an object list of the items to be excluded.) The intermediate results can be used with SPARQL’s OPTIONAL and bound() features to exclude all edges that have an object that is a constitutional part of the liver. Here ?s ?p ?c is bound optionally: if a binding is found, then the ?a ?b ?c tuple is not included (the ’!’ before ’bound’) in our result. (This is an especially useful pattern when a recursive query is used to define the http://…/exclude_list graph.)
CONSTRUCT { ?a ?b ?c }
FROM NAMED <http://…/exclude_list> [
CONSTRUCT { tmp:set tmp:member ?part }
FROM <http://…/fma>
WHERE { fma:Liver fma:constitutional_part ?part }
]
FROM <http://…/fma>
WHERE { ?a ?b ?c .
OPTIONAL { GRAPH <http://…/exclude_list> { ?s ?p ?c } }
FILTER (!bound(?s)) }
4.2.3. VirtualGraphs: Subqueries and blank nodes
A problem with subqueries is that the usefulness of the intermediate result sets generated by subqueries is diminished in the case of RDF blank nodes. Blank nodes in different RDF graphs are not equal. Thus, when a subquery is evaluated and a temporary RDF graph is created, the blank nodes in the output graph are not equal to the blank nodes queried from the subquery’s input graph. As a result, intermediate results that contain blank nodes cannot be directly compared with blank nodes in an input graph. This limitation is significant since we want to manipulate OWL ontologies, which often use blank nodes in defining constructs such as OWL restrictions and anonymous classes, and RDF graphs that may contain lists.
We have extended SPARQL with virtual graphs to allow blank nodes contained in the intermediate results generated by a subquery to be directly compared to the blank nodes in the input graph to the subquery. Users can define the results of a subquery to be a virtual graph using FROM NAMEDV. A FROM NAMEDV statement causes a subquery-generated virtual RDF graph to be added to a query’s set of named graphs. The virtual graph is identical to an RDF graph if it contains no blank nodes. However, if the virtual graph contains blank nodes that were queried from an input graph, pointers are kept to the blank nodes in the input graph instead of instantiating new graph-specific blank nodes. By keeping pointers to the input graph’s blank nodes, intermediate results containing blank nodes can be directly compared to a subquery’s input graph.
vSPARQL’s virtual graphs allow users to choose between two different types of blank node semantics when defining views. RDF blank nodes are unique within a specific graph; subqueries that wish to maintain RDF semantics can do so through the use of the FROM or FROM NAMED constructs. In contrast, vSPARQL’s FROM NAMEDV construct defines blank nodes that are unique to the original graph and virtual graphs derived from it. We have found this subtle distinction allows views to be defined that more closely match non-technical users’ expectations.
4.2.4. Recursive subqueries
vSPARQL allows recursive queries which can manipulate complex graphs. Instead of requiring the query writer to know the exact structure and depth of an input graph, our extension allows queries to be written that can follow paths based upon the graph’s structure and content. The results produced by recursive subqueries are virtual graphs.
Our recursive subquery extension builds upon our subquery extension; a recursive subquery generates a temporary graph that can be used in subsequent queries. A sketch of the syntax for a recursive query is
SELECT *
FROM NAMED <http://…/recursive_query> [
CONSTRUCT { … } # seed query
FROM <http://…/fma>
WHERE { … }
UNION
CONSTRUCT { … } # recursive query
FROM <http://…/fma>
FROM NAMED <http://…/recursive_query>
WHERE { GRAPH <http://…/recursive_query> { … }
… }
]
WHERE { … }
We have modeled our recursive extension on recursion in SQL.2 The first query clause is not recursive and provides the initial seeds for the new data source http://…/recursive_query. The second clause refers to this developing data source; on the first iteration the recursive data source is empty, but on subsequent iterations the data source contains triples that were added during the previous iteration. The second clause continues to be evaluated until the recursive data source reaches a steady state. Any number of seed and recursive subqueries are permitted within a recursive query.
The results of the subqueries within a recursive query are combined via a set union. This ensures that a triple will be added to the result set only once and that recursion will stop once a steady state has been reached.
Each of the subqueries in a recursive subquery have the same default properties as simple subqueries; subqueries can use a FROM NAMED statement to add the graph being generated by the recursive subquery. There is no limit on the number of levels of recursive subqueries that can be nested.
Recursive subqueries make it possible to generate queries over paths of arbitrary length. For example, the following query finds the subclasses of Organ that have constitutional parts and whose superclasses have constitutional parts.
SELECT ?sub
FROM NAMED <http://…/organ_with_parts> [
# build list of direct subclasses of Organ with constitutional parts
CONSTRUCT { tmp:set tmp:member ?sub }
FROM <http://…/FMA>
WHERE { ?sub rdfs:subClassOf fma:Organ .
?sub fma:constitutional_part ?part }
UNION
# add indirect subclasses of Organ that have constitutional parts
CONSTRUCT { tmp:set tmp:member ?next }
FROM <http://…/FMA>
FROM NAMED <http://…/organ_with_parts>
WHERE { GRAPH <http://…/organ_with_parts> { ?s ?p ?sub } .
?next rdfs:subClassOf ?sub .
?next fma:constitutional_part?part . }
]
WHERE { GRAPH <http://…/organ_with_parts> { ?x ?y ?sub } }
The seed query identifies the direct parts of Organ that have constitutional parts and adds them to the new data source http://…/organ_with_parts; on subsequent iterations, the items contained in the http://…/organ_with_parts data source are used to traverse one level further in the input graph.
In addition to being able to traverse an arbitrary graph, vSPARQL can extract subgraphs by following paths of arbitrary length. The following query is a modification of the previous example. This recursive subquery produces the entire subgraph of the subclasses of Organ that have constitutional parts and whose superclasses have constitutional parts; each subclass’ constitutional parts are also included in the output RDF graph.
CONSTRUCT { ?x ?y ?z }
FROM NAMED <http://…/organ_with_parts> [
# build graph of direct subclasses of Organ with constitutional parts
CONSTRUCT { ?sub rdfs:subClassOf fma:Organ .
?sub fma:constitutional_part ?part . }
FROM <http://…/FMA>
WHERE { ?sub rdfs:subClassOf fma:Organ .
?sub fma:constitutional_part ?part }
UNION
# extend with indirect subclasses of Organ with constitutional parts
CONSTRUCT { ?next rdfs:subClassOf ?sub .
?next fma:constitutional_part ?part . }
FROM <http://…/FMA>
FROM NAMED <http://…/organ_with_parts>
WHERE {
GRAPH <http://…/organ_with_parts>
{ ?sub rdfs:subClassOf ?super } .
?next rdfs:subClassOf ?sub . ?
next fma:constitutional_part ?part . }
]
WHERE { GRAPH <http://…/organ_with_parts> { ?x ?y ?z } }
vSPARQL’s recursive subqueries are not limited to traversing or extracting regular-expression-like paths. Recursive subqueries dictate the output they generate on each recursive iteration, and they can be used to iteratively manipulate paths of arbitrary length in a graph. Thus recursive subqueries can be used to produce output graphs that involve restructuring an input graph or combining, node-by-node, properties from two distinct graphs. As an example of vSPARQL’s iterative capabilities, the following recursive subquery restructures an extracted subgraph’s hierarchy by eliminating non-leaf nodes that do not have more than one child.
CONSTRUCT { ?a view:edge ?b }
FROM NAMED <extracted_hierarchy> [ … ]
FROM NAMED <restructure> [
# Identify all possible nodes for inclusion
CONSTRUCT { grph:a1 tmp:poss ?b1 . }
FROM <extracted_hierarchy>
WHERE { grph:Root grph:edge ?b1 }
UNION
CONSTRUCT { ?a1 view:edge ?b1 .
?b1 tmp:poss ?c1 . ?b1 tmp:poss ?c2 .
?a1 tmp:poss ?d1 . ?a1 view:edge ?b3 . }
FROM <extracted_hierarchy>
FROM NAMED <restructure>
WHERE {
{ # If a node has two children, include it in view
# Identify children as possible nodes for inclusion
GRAPH <restructure> { ?a1 tmp:poss ?b1 } .
?b1 grph:edge ?c1 . ?b1 grph:edge ?c2 . FILTER(?c1 != ?c2)
} UNION {
# If only one child, identify child as possible node
# for inclusion
GRAPH <restructure> { ?a1 tmp:poss ?b2 } .
?b2 grph:edge ?d1 .
OPTIONAL { ?b2 grph:edge ?d2 . FILTER(?d1 != ?d2) }
FILTER( !bound(?d2) ) } UNION {
# If a leaf node, include it in view
GRAPH <restructure> { ?a1 tmp:poss ?b3 } .
OPTIONAL { ?b3 grph:edge ?e1 }
FILTER( !bound(?e1) ) .
}
}
]
WHERE { GRAPH <restructure> { ?a view:edge ?b } }
When using recursive subqueries, users must take care to prevent infinite recursion. vSPARQL treats the results of a recursive query as a virtual graph, thus any blank nodes in our result set that are from our input graph are directly comparable with blank nodes in the input graph. A recursive query that extracts a subgraph containing blank nodes by using variable bindings in the CONSTRUCT template will terminate. However, because we allow value creation in CONSTRUCT templates, users that specify the creation of a new resource using a blank node or Skolem function can create recursive queries that do not terminate. vSPARQL provides warnings when blank nodes or Skolem functions are explicitly created in recursive CONSTRUCT templates.
vSPARQL allows value creation in recursive queries because users have requested the ability to “stitch” together two graphs by iterating over them. As a basic example, the following query combines two graphs by generating the Cartesian product at each level in the hierarchy. (Please note that for simplicity, this query only handles the case where all paths have the same depth; two more CONSTRUCT clauses would be needed to handle varying depth.)
FROM NAMED <stitch> [
CONSTRUCT {
[[combine:merge(ref:Liver,local:Liver)]] ref:part [[combine:merge(?fo,?lo)]] .
}
FROM NAMED <http://…/reference>
FROM NAMED <http://..local>
WHERE {
GRAPH <http://…/reference> { ref:Liver ref:part ?fo }
GRAPH <http://…/local> { local:Liver ref:part ?lo }
}
UNION
CONSTRUCT {
[[combine:merge(?fo,?lo)]] ref:part [[combine:merge(?fsub,?lsub)]] .}
FROM NAMED <stitch>
FROM NAMED <http://…/reference>
FROM NAMED <http://..local>
WHERE {
GRAPH <stitch> { [[combine:merge(?f,?l)]] ?c [[combine:merge(?fo,?lo)]] }
GRAPH <http://…/reference> { ?fo ref:part ?fsub }
GRAPH <http://…/local> { ?lo ref:part ?lsub }
}
]
vSPARQL limits the use of negation in recursive subqueries to stratified negation, which ensures that recursive subqueries that use negation cannot depend, directly or indirectly, on themselves. (Without this limitation, vSPARQL recursive queries with negation would have inflationary semantics.)
The use of subqueries to provide recursion has been proposed by SPARQL++ [25], NetworkedGraphs [34] and vSPARQL [8], as described above. To recap, vSPARQL allows recursion with stratified negation and allows value creation via blank nodes and Skolem functions.
SPARQL++ allows extended datasets to be defined that combine RDF data with, potentially recursive, CONSTRUCT queries. Within these CONSTRUCT queries, value creation can be achieved via blank nodes and built-in string manipulation functions provided strong safety requirements are met. The scope of blank nodes is defined by the graph it appears in. As a result, within an extended graph containing blank nodes, recursion will terminate. Because of the different scope, if an extended graph is recursively derived from another graph containing blank nodes the query may produce unexpected results as blank nodes in the derived graph will not be equal to those in the input graph.
NetworkedGraphs also allow graphs to be defined that combine RDF data and views over other graphs via, potentially recursive, CONSTRUCT queries; the result is a distributed “network” of graphs linked together via views. The language allows recursion with negation, leveraging well-founded semantics to ensure that a fixpoint is reached during evaluation. Value creation, via blank nodes, is not permitted in CONSTRUCT statements to ensure recursively defined views terminate. Because a CONSTRUCT query over a graph containing blank nodes will generate unique blank nodes – different from those in the input graph – for any blank nodes in its result set, a recursive query over a graph containing blank nodes may not produce the expected results. Similarly, joining results of two views derived from the same input graph may produce unexpected results if blank nodes are involved. 3
GLEEN:vSPARQL can leverage GLEEN’s [7] path expressions as syntactic sugar for some recursive subqueries. Instead of specifying full recursive subqueries, users can query using regular-expression-like paths between nodes in an input graph. GLEEN path expressions support the following operators: ’?’ (zero or one), ’*’ (zero or more), ’+’ (one or more), ’|’ (alternation), and ’/’ (concatenation). GLEEN can be used to determine the existence of a path between two nodes or to perform subgraph extraction as specified by a path expression.
GLEEN provides only a subset of the functionality available through recursive subqueries. For example, while GLEEN can use a path expression equivalent to “?sub rdfs:subClassOf+ fma:Organ” (find all of the subclasses of organ), it cannot be used to specify our second example in this section – there is no way in GLEEN to specify that each subclass of Organ also has constitutional parts. Using recursive subqueries, a user can specify additional constraints on nodes in the path.
5. Implementation
We have developed the vSPARQL language extensions on top of Jena’s ARQ [17](v2.3) query processor. View definitions over very large information sets can be evaluated through the use of our modified ARQ with SDB [19], a relational database engine for storing and querying (via SPARQL) RDF. The prototype uses a configuration file to provide a mapping between graph URIs and files or SDB repositories; remote files are downloaded from the web, and all SDBs, local and remote, are accessed using JDBC.
Recursive subqueries are rewritten for evaluation using the semi-naive evaluation algorithm [20], which repeatedly evaluates a query on incremental results until a fixpoint is reached.
6. Evaluation of Expressiveness
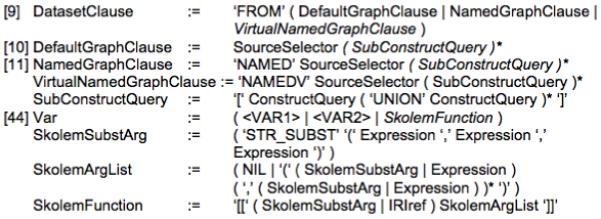
vSPARQL was designed to support a wide range of applications’ reuse of information sets on the semantic web. In this section, we evaluate our language’s expressiveness by creating view definitions for each of the use cases in Section 2. We briefly describe each of the view definitions; the view definitions’ statistics are presented in Figure 3. (Several of the views use GLEEN [7], thus reducing length of the view definition.)
Figure 3.
Statistics for use case view definitions written in vSPARQL. (*) Indicates number of edges added to input source in its entirety. Views were checked for correctness by: comparison to a gold standard, inspection by the requester/expert, or iterative development with a requester/expert.
http://sig.biostr.washington.edu/projects/ontviews/vsparql/jbi_use_cases.html has the complete text of all these queries, as well as a link to the query engine.
Mitotic cell cycle: From Reactome, this view uses a recursive subquery to extract theo ’componentOf’ hierarchy (and accompanying labels) for the mitotic cell cycle and its subprocesses.
Organ spatial location view: The view uses a recursive subquery to identify all of the parts of the Gastrointestinal tract and intersects it with all of the subclasses of Organ. For each identified organ, the view produces a subgraph specifying the organ’s orientation, containment, and several continuity properties. Figure 4 presents a portion of the materialized RDF graph generated by this view.
NeuroFMA ontology: Several recursive subqueries are used to extract all of the neural structures from the FMA, their attributes, and the properties connecting them. To ensure a proper ontology, all of the types, superclasses, and superproperties of the elements of the NeuroFMA ontology are recursively identified and included in the derived ontology. The NeuroFMA ontology contains approximately 2,300 classes and 40 properties used to represent more than 70,000 relationships.
This ontology was defined by an ontologist over the FMA. The ontology was originally derived from the frames representation via custom programming against the Protege API; the output was converted to OWL Full using Noy’s [37] conversion software. A corresponding view definition for the ontology was developed in vSPARQL. The output of the two derivations was compared using a modified version of RDFSync [38]; the two were found to be semantically equivalent. The materialized view can be loaded and explored in Protege; it has been uploaded as a view in BioPortal [4] as the NeuroFMA view at http://bioportal.bioontology.org/ontologies/39966#views
NCI Thesaurus simplification view: The view [7] creates direct properties that simplify the relationships found in the underlying OWL representation for Gastric Mucosa-Associated Lymphoid Tissue Lymphoma. The new properties are generated by recursively following paths of arbitrary length containing owl:intersectionOf, owl:equivalentClass, and RDF lists. For example, the restrictions on a node may be found using a path expression like ([rdfs:subClassOf]|[owl:equivalentClass])/([owl:intersectionOf]/[rdf:rest]*/[rdf:first]?)+ The hierarchy can be “flattened” by creating direct properties on the node. Skolem functions are used to combine the labels of identified properties to define the simplified relationship; Skolem functions were used instead of blank nodes to eliminate duplicates. The resulting view consists of the original ontology plus the newly generated direct properties.
Biosimulation annotation editor view: The view extracts the hierarchical tree connecting a list of specified concepts. A recursive subquery is used to restructure the hierarchy by eliminating non-leaf nodes that have only a single child. Figure 5 displays an example of the hierarchical tree before and after restructuring by the view. Restructuring the hierarchy eliminates 62 non-leaf nodes with only a single child.
Blood contained in the heart view: The view uses a recursive subquery to identify all of the parts of the heart; if a structure S has a part that is a space that contains a portion of blood pob, a triple is added to the view indicating that the structure S contains the portion of blood pob.
Radiologist liver ontology: The view [8] recursively follows several different paths to generate this view. Starting at the liver, the ’part’ property is recursively followed to determine the set of liver parts. This set is combined with the results of recursively following the rdfs:subclassOf hierarchy to determine all of the subclasses of “Cell” and “Cardinal cell part” – only liver parts that are not subclasses of Cell and Cardinal cell part are visible and included in the view. The output ontology is generated by extracting the rdfs:subclassOf hierarchy for each of the visible liver parts and combining it with the visible liver part hierarchy. Because the distinction is not wanted in the result, “Cavitated organ” and “Solid organ” are eliminated from the subclasses hierarchy and their subclasses are reassigned to “Organ.”
Blood fluid properties view: Recursion is used to identify portions of blood from the FMA and kinetic fluid properties from the OPB; the identified concepts are combined to generate new concepts using Skolem functions. For example, after querying to associate ?pob with a subclass of fma:Portion_of_blood and ?fluid_prop with a kinetic property of fluids, a new concept can be created that combines the two using Skolem functions: [[annot_view:annotation(?pob, ?fluid_prop)]]; this new node can be assigned properties from both the FMA and the OPB.
Figure 4.
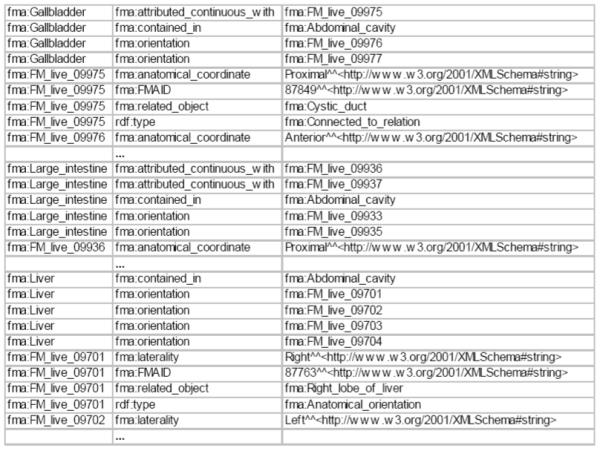
Excerpt of materialized organ spatial location view.
Figure 5.
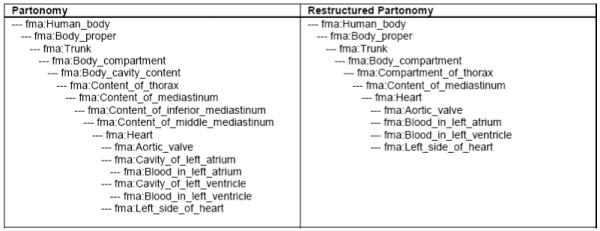
Comparison of snippet of materialized partonomy for biosimulation annotation editor. The left panel shows the unmodified hierarchy; the right panel shows the hierarchy where non-leaf nodes with a single child are removed.
6.1. Evaluating generated views
We were able to use vSPARQL to create view definitions for all of our motivating use cases. As shown in Figure 3, our view definitions ranged in length from 13 to 405 query lines. Performance ranged from a couple of seconds to a few minutes to materialize each of our views.
In vSPARQL, a view definition is deemed correct if the output contains the exact set of RDF triples desired by the user. Views that include extra facts are incorrect. Our generated views were checked for correctness in one of three ways: 1) we compared the results to a gold standard (e.g. existing artifact), 2) we had the results of the view inspected by the requester (or an expert if the view was not specifically requested), or 3) the view was iteratively created by a technical expert and the requesting expert.
Comparison of Query Languages over Use Cases
In this section, we evaluate several semantic web languages to determine if they support all of the functionality needed for creating views. We evaluate each language to determine if the language supports the functionality listed in Section 3.
Query languages
We compare the following RDF query languages: RQL+RVL [15], SPARQL [11], ARQ [17], SPARQLer [24], CPSPARQL [28], nSPARQL [29], SPARQL++ [25], NetworkedGraphs [34] and our view definition language vSPARQL. Excluding RQL+RVL, we have chosen to compare query languages based upon SPARQL because of the language’s W3C recommendation, wide adoption and active development. The languages cover the set of functionality proposed in SPARQL extensions. We have included RQL+RVL as it is an existing view language developed for RDF data. We briefly describe the high-level features of the languages we are considering before evaluating their functionality.
RQL is a declarative query language for RDF that allows querying over both resource descriptions and schemas. RVL [15] is a view definition language for creating virtual resource descriptions and schemas; RVL uses RQL as its query language.
SPARQL is the W3C’s recommended query language for RDF; it supports querying RDF graphs via graph patterns and using filters for checking values.
ARQ (v2.8.2) is a SPARQL query processor for Jena; it includes extensions for aggregation, property paths, sub-SELECT queries and explicit variable assignment.
SPARQLer is an extended version of SPARQL that supports path queries; path queries can be used to gather subgraphs out of an input graph.
PSPARQL [27] is an extension to SPARQL that supports querying with regular expression patterns for path expressions. CPSPARQL builds upon PSPARQL to permit constraints on path steps.
nSPARQL is an extension to SPARQL designed to allow sophisticated querying of RDF graphs using nested path expressions.
SPARQL++ is an extended version of SPARQL designed to allow mapping between RDF vocabularies; the extensions include nested queries, external functions, and aggregates. Extended graphs allow data to be combined with CONSTRUCT statements to add new facts to the graph.
NetworkedGraphs allows users to define RDF graphs by combining explicitly listed data and views over other RDF graphs, specified via SPARQL CONSTRUCT statements. Views are dynamically evaluated by querying remote SPARQL servers holding the necessary graphs. The remote graphs may themselves be defined by views over data at other machines, thus creating a network of graphs. To ensure decidability, NetworkedGraphs prohibits value creation via blank nodes in CONSTRUCT templates.
Language functionality
Figure 6 charts our required functionality for a view definition language for each of the semantic web languages under consideration. An asterisk is indicated in the figure to indicate instances where a language provides partial support of a piece of functionality.
Figure 6.
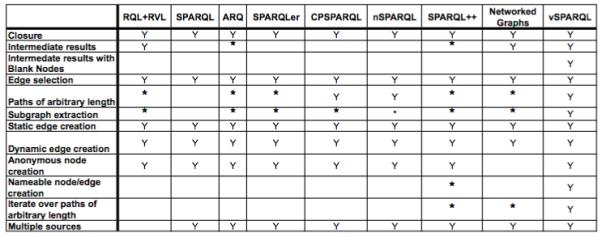
For each semantic language, the chart indicates which view definition functionality is and is not supported. Instances of “*” indicate that there is partial support, as explained in the text.
RQL expands an RDF graph’s subsumption hierarchy. This allows queries to be applied to paths of arbitrary length over RDFS’ subClass and subProperty hierarchies; users can also perform a subgraph extraction over these hierarchies. The language does not support querying for paths of arbitrary length for any other properties. RVL permits the definition of views over different namespaces, making possible multiple intermediate sets within a single view definition file.
Most of the languages we are comparing are extensions of SPARQL, and therefore they all inherit its basic functionality. SPARQL provides support for querying over multiple sources and simple paths that require knowledge of the underlying graph.
Several of the languages being compared support paths of arbitrary length without providing support for subgraph extraction. For simple paths (e.g. alternation), subgraph extraction can be performed using the following pattern. This query returns the Liver and the subgraph containing all of its regional and constitutional parts.
CONSTRUCT { ?a ?b ?c }
WHERE { fma:Liver (fma:regional_part|fma:constitutional_part)* ?a .
?a ?b ?c .
FILTER((?b=fma:regional_part)||(?b=fma:constitutional_part)) .
}
While ARQ supports paths of arbitrary length with its “property paths,” the types of paths that can be specified are limited by the language. The language supports typical regular expression behavior, such as concatenation, alternation, various path length specifications and reverse paths. Subgraph extraction is available for simple paths as described above. However the language does not support, for example, placing multiple constraints on the nodes along a path; you cannot require nodes in the path to have two different properties.
ARQ contains an extension for SELECT subqueries, thus providing a mechanism for intermediate results. However, the semantics of SELECT and CONSTRUCT queries are different; in a SELECT query that uses UNION or OPTIONAL, variables projected from the sub-SELECT may be unbound.
SPARQLer’s paths can be specified using regular expresssions; paths can be combined with CONSTRUCT to generate subgraphs. Although SPARQLer’s regular expression-like paths do not allow specification of additional constraints on the nodes of a path, the elements along a path are returned as a sequence whose elements can be queried. It is possible to use SPARQLer to test for the existence of a specific property or node along a path or to check the value of specific elements in the path sequence. However, without first using CONSTRUCT to generate a subgraph, for a path of arbitrary length it is not clear how to transform or modify elements of the path; SPARQLer does not support nested CONSTRUCT queries.
CPSPARQL’s constrained path expressions support a subset of the functionality provided by vSPARQL’s recursive subqueries. CPSPARQL allows constraints to be checked on individual elements of a path of arbitrary length. However, CPSPARQL does not provide the ability to iteratively restructure a graph. For simple path expressions (described above), subgraph extractions can be performed.
nSPARQL uses nested regular expressions for querying over an RDF graph. Nested subqueries and recursive subqueries in vSPARQL can be used to express the queries supported by nSPARQL’s nested regular expressions. The language does not support subgraph extraction, nor does it provide vSPARQL’s functionality for restructuring a graph by iterating over a path. Subgraph extractions for simple path expressions of arbitrary length can be performed as described above.
SPARQL++ supports a large portion of the functionality needed for a view definition language. Currently SPARQL++ supports string concatenation for the creation of nameable resources; this is not sufficient for users that wish to extract portions of or augment the URIs to create nameable nodes. However, SPARQL++ advocates for addition of external functions and could easily be extended to permit this behavior. While SPARQL++ allows dynamic creation of URIs in CONSTRUCT templates, it does not allow this functionality to be used within query WHERE clauses as allowed by vSPARQL’s Skolem functions; queries over a dynamically generated node cannot easily determine the parameters used to generate the node’s identifier.
SPARQL++’s nested CONSTRUCT queries are supported in FROM clauses, not FROM NAMED clauses; as a result, the intermediate result sets are immediately combined in to the outer query’s default graph. This small limitation prevents the language from using the intermediate results in operations such as conditional joins between intermediate result sets or excluding results.
However, SPARQL++ does allow CONSTRUCT queries to be combined with RDF triples to define extended graphs; the CONSTRUCT queries add new facts to the graph. (Note that SPARQL++’s extended graphs violate our requirement that queries be separate from data.) Thus, a new extended graph can be used to define intermediate results over a base graph. Unfortunately, the blank nodes contained in the derived intermediate graph will not be equal to the corresponding blank nodes in the original graph; this limits the ability to use intermediate results for operations like conditional joins with the original graph or other derived graphs.
CONSTRUCT statements can also be used to recursively add new facts to SPARQL++’s extended graphs. When a recursive CONSTRUCT is defined within a graph, SPARQL++ is able to maintain its finite semantics while recursing over a graph containing blank nodes. Alternatively, an extended graph can use CONSTRUCT statements over a base graph to recursively extract facts from the base graph. In this way, SPARQL++ can support subgraph extraction and iterating over a graph. Unfortunately if the derived graph recursively queries a base graph containing blank nodes, the blank nodes in the derived graph will not be equal to the corresponding blank nodes in the base graph and a recursive query may produce unexpected results.
NetworkedGraphs’s recursive capabilities provide support for paths of arbitrary length, subgraph extraction, and the ability to restructure by iterating over paths. However, these capabilities will only produce correct results when iterating over paths that do not contain blank nodes. Additionally, the language prohibits value creation via blank nodes (and presumably other forms of dynamically generating new nodes) in CONSTRUCT statements to ensure termination of recursive queries.
Although there is overlap in the functionality provided by the query languages, only vSPARQL provides all of the functionality required for defining views for our eight motivating use cases.
Query language vs. use case evaluation
Figure 7 indicates, based upon their description in the literature, which views can be expressed by our specified set of query languages. All of our view queries require either paths of arbitrary length or subgraph extraction over properies other than rdfs:subClassOf and rdfs:subPropertyOf. Neither RQL+RVL nor SPARQL support this functionality and therefore cannot express any of our views; we exclude these languages from the discussion below.
Figure 7.
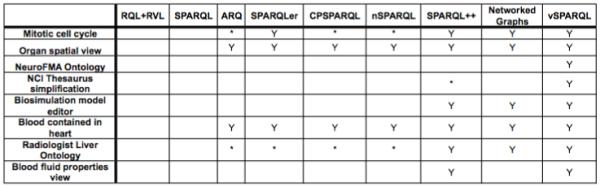
Use case views and the languages that can express them. “*” indicates that the language was able to support the view but that they do so under special circumstances described in the text.
All of the remaining languages were able to support paths of arbitrary length; as a result, they could all be used to define the Organ Spatial Location and the Blood Contained in the Heart views. Additionally, because the Mitotic Cell Cycle and Radiologist Liver Ontology views perform subgraph extraction over simple alternating paths, these views could be supported by ARQ, SPARQLer, CPSPARQL, and nSPARQL using the technique described in Section 7:Language functionality.
SPARQLer’s ability to support the Mitotic Cell Cycle and the Radiologist Liver Ontology highlights an important aspect of languages with support for querying and extracting paths from a graph. The Mitotic Cell Cycle simply extracts subgraph corresponding to the “componentOf” hierarchy for the mitotic cell cycle; the radiologist liver ontology example, however, requires a transformation on an extracted subgraph. Because SPARQLer does not support nested queries, we were not able to directly use the language’s support for extracting subgraphs; instead, we had to leverage the technique described in Section 7:Language functionality.
SPARQL++ is able to support all but one of our use case queries, with a few caveats related to blank nodes. In SPARQL++, to recurse over a set of data, a user may either embed CONSTRUCT statements in the same extended graph as the data or the user may create a new extended graph that contains CONSTRUCT queries on the original graph. If a query needs to recurse over blank nodes in the original graph, the recursive CONSTRUCT queries must be embedded with the data in the original graph; because of the scope of blank nodes, a derived extended graph cannot recurse over blank nodes in the original graph.
Intermediate result sets, subgraph extraction and iterating over paths of arbitrary length require a new extended graph to be derived from the original graph using CONSTRUCT statements. Thus, if these actions must be performed over data paths containing blank nodes, SPARQL++ cannot support this functionality. In general, CONSTRUCT queries to calculate paths of arbitrary length can either be combined with the original graph or defined in a derived extended graph; however, if the path must traverse blank nodes, it must be defined in the original graph. This restriction violates our requirement that view definitions be kept separate from data.
SPARQL++ is the only language that can support the NCI Thesaurus Simplification and the Blood Fluid Properties views, as it is the only language that supports dynamic nameable node creation.
At a quick glance, Figure 7 would seem to indicate that SPARQL++ is almost exactly what is needed for our view language. However, the results are a bit misleading. Only two of our use case views require traversal over or extraction of blank nodes: the NeuroFMA Ontology and the NCI Thesaurus simplification views. Of these two, the NCI Thesaurus simplification view can be achieved by embedding recursive CONSTRUCT statements with the graph; the NeuroFMA Ontology requires intermediate sets, subgraph extraction, and iteration over data containing blank nodes and therefore cannot be supported by SPARQL++. More generally, this indicates that SPARQL++ is a good view language for RDF data without blank nodes; unfortunately, OWL ontologies often use blank nodes to represent elements such as requirements or anonymous classes, and blank nodes are used to represent RDF collections.
NetworkedGraphs was able to support all use cases except those requiring dynamic node creation (NCI Thesaurus simplication and Blood Fluid Properties views), and those requiring recursion over paths containing blank nodes (NeuroFMA and NCI Thesaurus simplification views.)
Size of queries
When possible, our view definitions leveraged GLEEN’s path expressions instead of vSPARQL’s recursive subqueries. This decision makes the size of our view definitions comparable to the size of queries generated by ARQ, SPARQLer, CPSPARQL, and nSPARQL. The one exception to this is the Blood Contained in the Heart view. Due to an implementation limitation in GLEEN that prevents the combination of path expressions with both a variable subject and object, this view definition uses recursive subqueries. If path expressions were used instead, the view would be 12 lines.
We were able to write the Radiologist Liver Ontology, which requires a transformation on a subgraph extraction, without nested queries. However, because of the special cases that we needed to handle, the view definition without subqueries was 52 lines versus the 40 lines needed when subqueries were available; using subqueries, the view definition was much easier to write and understand.
Because they do not have a built-in path expression capability, SPARQL++ and NetworkedGraphs will produce queries/views that are longer than the other query languages.
In evaluating these queries, we found several takeaway points. Many views requiring subgraph extraction can be expressed using languages that support path expressions. A general recursive strategy ensures that we can perform both subgraph extraction and restructuring of graphs. Transforming the results of a subgraph extraction is much simpler when nested CONSTRUCTs can be used. Finally, an inability to handle blank nodes in intermediate graphs and recursive queries is a significant limitation.
Snippets of the materialized views for our queries can be seen in Figure 8.
Figure 8.
Snippets of six of the materialized views generated by our view definitions.
7. Comparisons to other work
Our work for defining views for the semantic web can be compared to work grouped into three categories: subset selection techniques, rule languages, other view approaches and query languages.
Subset selection / extraction
Three approaches have been developed for materialization of a subset of an information set for reuse: manual, structural, and logical.
Traditionally, manual techniques have been used to derive a relevant subset of an information set for an application’s use. A developer acquires a copy of the information set and then manually deletes, modifies, and adds facts until the application’s requirements are met. This approach requires significant user effort and must be repeated whenever the information set is updated.
Structural techniques such as PROMPT’s Traversal Views [9] and Web ontology segmentation [43] require a set of concepts and properties be specified for inclusion in the result. Starting from these key concepts, the output set is grown by recursively adding the specified properties and concepts until a fixed point is reached. [9] allows multiple distinct derivations to be unioned to produce a materialized result. Both of these approaches can be accommodated by our view definition language. However, our view definition language allows intermediate results such as [9]’s derivations to be conditionally combined or intersected. Unlike vSPARQL, these approaches are purely extraction techniques; any modification or restructuring of the information must be performed externally on a materialized copy of the output.
In contrast, logical techniques ( [39], [40], [41]) have been developed for deriving modules from OWL-DL ontologies. A signature is specified containing all of the key concepts that should be contained within a selected subset of the ontology; leveraging the high-level semantics of OWL-DL, the extractor grows the set of concepts and axioms needed in the output module. For concepts in the signature, the ontology module is guaranteed to capture the meaning of the concepts, such that an application that performs reasoning after importing the module would give the exact same results as an application that performs reasoning after importing the entire ontology.
Ontology module extractors are a powerful tool for extracting logically equivalent modules from an OWL-DL ontology. However, they are not suitable for all applications’ needs. The current approaches are limited to DL languages such as OWL-DL; some ontologies, such as the FMA, are expressed in OWLFull. Additionally, these techniques are specifically for deriving subsets of the original ontology that have the same meaning as the original for a specified signature. However, not all users want to extract ontologies; some users simply want to extract lists of terms or small connected subgraphs. Alternatively, some users want to modify or transform the data that they extract from an ontology. Module extraction does not allow you to specify changes to be made to the ontology before or during extraction; you need to modify the original data and then derive a module from the materialized modified ontology. Finally, module extraction techniques may produce modules that contain data that the user is not interested in. This could be because a non-minimal module was produced, or because the user would like to eliminate terms. For example, our liver radiologist application excludes all cellular and subcellular concepts. Forgetting ( [50], [51], [52]) can be can be used to eliminate concepts from modules if the set of terms to eliminate is known, as in the radiologist example. However, if a non-minimal module is produced, the user must inspect the extracted subset to determine the concepts to forget.
Our view definition language does not use formal logics to guarantee that a derived subset has the same properties as the original ontology. Instead, it allows the application developer to specify exactly which facts are relevant and how those facts should be arranged or augmented. Even when those facts are modified in the view, subsequent queries can still be answered against the original ontology, ensuring that information set updates are reflected in the query results.
Rule languages
Rule languages allow new facts to be inferred from existing facts in an information set. Several different rule languages like TRIPLE [30] and SWRL [32] have been developed for the semantic web. General rule languages, such as Jena’s [18] general rule language, can be used to derive new facts without SWRL’s semantic restrictions. vSPARQL’s subquery and recursive subquery features provide the functionality of a general rule language for RDF.
Other view approaches
We have already described several view languages, particularly SPARQL++ and NetworkedGraphs, in detail. In this section, we describe several other approaches for manipulating or transforming RDF data.
RVL and Lightweight Ontologies [16] both leverage RQL to provide declarative mechanisms for defining views over ontologies. The RDFS semantics of the underlying data are used to ensure that the defined views’s semantics match the data; new classes and properties can be defined in the view. RVL allows the restructuring of subsumption hierarchies.
Several projects are building upon the notion of pipes to provide easy-to-use mechanisms for transforming and aggregating RDF data. Semantic Web Pipes [53] provides specialized operators that can be graphically arranged for acyclic processing of data. Operators are provided for extracting data from Web content, performing SPARQL queries over data, and inference. Similarly, SPARQLMotion [54] provides a GUI editor for pipelining RDF data sources and transformation operations together. Banach [55] provides a set of operators that can be pipelined inside Sesame to transform RDF data. [56] proposes splitting mash-up development into two separate pieces – data-level and service-level – to simplify mash-up creation for users.
NRL [57] uses named graphs and views in the context of the Social Semantic Desktop. Graph views are used to define specialized semantics and assumptions over RDF named graphs. Graph roles declaratively assign meaning (e.g. ontology, knowledge base, data) to named graphs. However graph views are procedural, specifying the application or query/rules that need to be used to generate an output graph from an input graph.
Query languages
vSPARQL is designed based upon decades of query language research and development. SQL is the most well-known query language for relational databases. The language supports subqueries and recursive subqueries for gathering data from a database; additionally, SQL supports database views to restrict the set of data accessible to queries. Techniques have been developed for query rewriting and optimization so that very large data sets can be efficiently queried. Skolem functions have been incorporated into several different query languages such as Struql [44]. vSPARQL leverages the experience of these query languages to develop a view definition language specifically for the RDF data model.
Several different query languages have been proposed for querying within the semantic web, including RQL [14], RDQL [31], and SPARQL [11]. A number of proposals have been made for extending SPARQL’s functionality, including SPARQLer [24], ARQ [17], SPARQL++ [25], CPSPARQL [28], nSPARQL [29], and NetworkedGraphs [34]. We compared many of these query languages to vSPARQL in Section 6.1.
8. Discussion
We have proposed a general solution for enabling medical applications to leverage ontologies, vocabularies, and data sets available in RDF format. After detailing the reuse (or view) requirements, we introduced vSPARQL, a view definition language and implementation, and demonstrated through eight use cases that it meets our requirements. View mechanisms allow applications to choose, and potentially transform, the specific information that they desire from large knowledge and data sets.
Our evaluation of vSPARQL focused on its expressivity. Although we found that it was sufficiently expressive to generate views for all of our use cases, vSPARQL views can be difficult to write for users who are not computer scientists. Several users have likened the experience to trying to write SQL. In the future, we would like to develop user-friendly tools and a high-level language to generate view definitions. Additionally, we would like to improve vSPARQL performance and extend it to allow inclusion of other technologies.
8.1. Mapping to a user’s intuition
vSPARQL’s support for subqueries allows queries to be nested arbitrarily deep. In practice, however, we found that queries were cognitively simpler when they were flattened to form something analogous to a workflow. Workflow style queries are those where the results of all earlier subqueries are available as input to (in scope of) subsequent subqueries.
Until recently, ontology view extraction was largely performed adhoc. One-off programs were written to produce each new use case view or informaticists produced views by hand, pruning and augmenting knowledge sources as needed. When asked, view creators defined their process as a workflow. There remains a disconnect between the workflow operations as defined by the informaticist and the workflow operations in vSPARQL. For example, a single conceptual operation may map down to a sequence of query operations. We have begun the work of formally defining this mapping in order to support view definitions described at a more intuitive (and more terse) level.
At the same time, to ease the creation of vSPARQL queries we have developed a GUI application in Flex for creating workflow-style view definitions; it can be accessed at http://ontviews.biostr.washington.edu:8080/VSparQL_Service/GUI/vsparql_oh_degrafa.html The application allows the user to specify data sources, subqueries, recursive queries, and queries (e.g. select, ask); these individual components are connected graphically to control the data flow through the query. In addition to simplifying the user’s specification of a vSPARQL query syntactically, the GUI application allows a user to inspect the vSPARQL query and output results of individual subquery and recursive subquery blocks.
8.2. Optimization
Currently our vSPARQL query plans are chosen primarily by an optimizer built to handle regular SPARQL queries. While we have added some processing optimizations specific to vSPARQL, such as the semi-naive evaluation of recursive queries, further optimization is possible. We anticipate leveraging work in the database community (RDF-3X [35],vertical partitioning [36]) for efficient querying of RDF. Additionally, we are exploring the feasibility of optimizing at the level of the “more intuitive” workflow, as discussed in the previous section.
We are investigating the applicability of database query rewriting techniques for evaluating queries against vSPARQL views. While answering a query in the existing engine, vSPARQL first evaluates each subquery and materializes its results; however, the query may not require all of the subqueries to be evaluated before answering a query. Efficiency improvements could be realized simply by avoiding the materialization of statements not needed to answer a query.
8.3. Leveraging related work
The use cases in this paper illustrate a variety of application specific information extraction scenarios (i.e. input is an RDF graph, input is an OWL-DL ontology, output is an OWL Full ontology, output is a list of terms, output is not a proper subset of the input, etc.). We compared vSPARQL with several other view generation approaches and demonstrated that vSPARQL is a good general solution covering all of our use cases. However, in certain situations, leveraging other extraction mechanisms may simplify view definition. For example, if a view needs to preserve entailed facts from a source ontology, and if the source is amenable to logical reasoning (i.e. is in OWL Lite or OWL-DL) then a DL reasoner based extraction mechanism may be a better fit. We would like to expand on our workflow notion of view definitions to allow substitution of alternate methods at stages where they are deemed more appropriate by the view creator.
9. Conclusions
In this paper we have presented a general solution for reusing biomedical information sets available in the semantic web’s RDF format. We have described the requirements for a view definition language that permits applications to specify the (possibly modified) content that they wish to leverage from an information set; applications can query these view definitions to access the relevant content. We have described vSPARQL, a series of extensions to SPARQL that realize our view definition language. We have developed a prototype implementation of vSPARQL and a GUI editor for defining views. We have evaluated the expressivity of our view definition language by using it to create view definitions for eight use case examples. We have compared our language’s functionality to that of existing RDF query languages and found that none of these languages meet all of our requirements needed for a view definition language. These results suggest that vSPARQL has the potential to allow widespread reuse of semantic web resources.
10. Acknowledgements
This work was funded by NIH grant HL087706. We thank Nikki Dell for feedback on the manuscript, and Onard Mejino, Dan Cook, and Max Neal for help in defining the views.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
These information sets were dictated by our requested use cases. Our prototype builds on top of SDB which uses relational databases to store data and can therefore handle large information sets. Within a single SDB repository, joins between large information sets leverage the capabilities of relational databases; however, joins of large information sets across SDB stores can be timeconsuming
Recent versions of SQL allow recursive queries; these extensions are modeled after Datalog.
NetworkedGraphs is built on Sesame. Earlier versions of Sesame (pre-v2.2) did not handle blank node scope correctly; as a result, a recursion over graphs in a single repository would be correctly evaluated over graphs containing blank nodes. This is not true in later versions of Sesame. Recursion over graphs containing blank nodes in remote repositories may not produce expected results.
References
- [1].Resource Description Framework. http://www.w3.org/RDF/
- [2].OWL Web Ontology Language Guide. 2004 February 10; http://www.w3.org/TR/owl-guide/
- [3].The Open Biomedical Ontologies. http://www.obofoundry.org.
- [4].NCBO BioPortal. http://bioportal.bioontology.org/
- [5].UniProt http://www.uniprot.org/
- [6].Brinkley James F, Suciu Dan, Detwiler Landon T, Gennari John H, Rosse Cornelius. A framework for using reference ontologies as a foundation for the semantic web; Proceedings, American Medical Informatics Association Fall Symposium; 2006; pp. 96–100. [PMC free article] [PubMed] [Google Scholar]
- [7].Detwiler Landon T, Suciu Dan, Brinkley James F. Regular Paths in SparQL: Querying the NCI Thesaurus; Proceedings, American Medical Informatics Association Fall Symposium; 2008; pp. 161–165. [PMC free article] [PubMed] [Google Scholar]
- [8].Shaw Marianne, Detwiler Landon T, Suciu Dan, Brinkley James F. Generating Application Ontologies from Reference Ontologies; Proceedings, American Medical Informatics Association Fall Symposium; 2008; [PMC free article] [PubMed] [Google Scholar]
- [9].Noy Natalya F., Musen Mark A. Specifying Ontology Views by Traversal; Proceedings, International Semantic Web Conference; 2004.pp. 713–725. [Google Scholar]
- [10].Rosse C, Mejino JVL. A reference ontology for biomedical informatics: the Foundational Model of Anatomy. Journal of Biomedical Informatics. 2003;36:478–500. doi: 10.1016/j.jbi.2003.11.007. [DOI] [PubMed] [Google Scholar]
- [11].SPARQL Query Language for RDF. 2008 January 15; http://www.w3.org/TR/rdf-sparql-query/
- [12].The Protege Ontology Editor and Knowledge Acquisition System. http://protege.stanford.edu.
- [13].Cook Daniel L., Mejino Jose LV, Neal Maxwell L, Gennari John H. Bridging Biological Ontologies and Biosimulation: The Ontology of Physics for Biology; Proceedings, American Medical Informatics Association Fall Symposium; 2008; pp. 136–140. [PMC free article] [PubMed] [Google Scholar]
- [14].Karvounarakis Gregory, Alexaki Sofia, Christophides Vassilis, Plexousakis Dimitris, Scholl Michel. RQL: a declarative query language for RDF; Proceedings, 11th International Conference on World Wide Web; 2002.pp. 592–603. [Google Scholar]
- [15].Magkanaraki Aimilia, Tannen Val, Christophides Vassilis, Plexousakis Dimitris. Viewing the semantic web through RVL lenses; Proceedings, International Semantic Web Conference; 2003.pp. 96–112. [Google Scholar]
- [16].Volz R, Oberle D, Studer R. Implementing Views for Light-Weight Web Ontologies; 7th International Database Engineering and Applications Symposium; 2003. [Google Scholar]
- [17].ARQ - A SPARQL Processor for Jena. http://jena.sourceforge.net/ARQ.
- [18].Jena - A Semantic Web Framework for Java. http://jena.sourceforge.net.
- [19].SDB - A SPARQL Database for Jena. http://jena.sourceforge.net/SDB.
- [20].Ullman Jeffrey D. Principles of Database and Knowledge-Base Systems. Volume I: Classical Database Systems. Vol. 1988. Computer Science Press; Rockville, MD: pp. 126–128. [Google Scholar]
- [21].NCIthesaurus. http://nciterms.nci.nih.gov.
- [22].Reactome - a curated knowledgebase of biological pathways. doi: 10.1111/j.1538-7836.2012.04930.x. http://www.reactome.org. [DOI] [PMC free article] [PubMed]
- [23].The NeuroCommons Project http://neurocommons.org.
- [24].Kochut Krys J., Janik Maciej. SPARQLeR: Extended Sparql for Semantic Association Discovery; Proceedings, 4th European conference on The Semantic Web; 2007.pp. 145–159. [Google Scholar]
- [25].Polleres Axel, Scharffe Francois, Schindlauer Roman. SPARQL++ for Mapping between RDF Vocabularies; Proceedings, 6th International Conference on Ontologies, Databases, and Applications of Semantics; 2007. [Google Scholar]
- [26].Anyanwu Kemafor, Maduko Angela, Sheth Amit. SPARQ2L: Towards Support for Subgraph Extraction Queries in RDF Databases; Proceedings, 16th International World Wide Web Conference; 2007.pp. 797–806. [Google Scholar]
- [27].Alkhateeb Faisal, Baget Jean-Francois, Euzenat Jerome. RDF with regular expressions. INRIA; 2007. Research Report 6191. [Google Scholar]
- [28].Alkhateeb Faisal, Baget Jean-Francois, Euzenat Jerome. Constrained Regular Expressions in SPARQL. INRIA; 2007. Research Report 6360. [Google Scholar]
- [29].Perez Jorge, Arenas Marcelo, Gutierrez Claudio. nSPARQL: A Navigational Language for RDF; Proceedings, International Semantic Web Conference; 2008.pp. 66–81. [Google Scholar]
- [30].Miklos Zoltan, Neumann Gustaf, Zdun Uwe, Sintek Michael. Querying Semantic Web Resources Using TRIPLE Views; Proceedings, International Semantic Web Conference; 2003.pp. 517–532. [Google Scholar]
- [31].RDQL - A Query Language for RDF. 2004 January 9; http://www.w3.org/Submission/2004/SUBM-RDQL-20040109/
- [32].SWRL: A Semantic Web Rule Language Combining OWL and RuleML. 2004 May 21; http://www.w3.org/Submission/SWRL/
- [33].Schenk Simon. A SPARQL Semantics Based on Datalog; Proceedings, 30th German Conference on Advances in Artificial Intelligence; 2007.pp. 160–174. [Google Scholar]
- [34].Schenk Simon, Staab Steffen. Networked graphs: A declarative mechanism for SPARQL rules, SPARQL views and RDF data integration on the web; Proceedings, 17th International Conference on the World Wide Web; 2008. [Google Scholar]
- [35].Neumann Thomas, Weikum Gerhard. RDF-3X: A RISC-style engine for RDF; Proceedings, International Conference on Very Large Data Bases; 2008.pp. 647–659. [Google Scholar]
- [36].Abadi Daniel J., Marcus Adam, Madden Samuel R., Hollenbach Kate. Scalable semantic web data management using vertical partitioning; Proceedings, 33rd International Conference on Very Large Data Bases; 2007.pp. 411–422. [Google Scholar]
- [37].Noy Natalya F., Rubin Daniel L. Translating the Foundational Model of Anatomy into OWL; Web Semantics: Science, Services and Agents on the World Wide Web; 2008; pp. 133–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Tummarello Giovanni, Morbidoni Christian, Bachmann-Gmr Reto, Erling Orri. RDFSync: Efficient Remote Synchronization of RDF Models; Proceedings, International Semantic Web Conference; 2007.pp. 537–551. [Google Scholar]
- [39].Kontchakov R, Pulina L, Sattler U, Schneider T, Selmer P, Wolter F, Zakharyaschev M. Minimal Module Extraction from DL-Lite Ontologies using QBF Solvers; Proceedings 21st Int. Joint Conf. on Artificial Intelligence (IJCAI); 2009.pp. 836–841. [Google Scholar]
- [40].Grau Bernardo Cuenca, Horrocks Ian, Kazakov Yevgeny, Sattler Ulrike. Just the right amount: extracting modules from ontologies; Proceedings of the 16th international conference on World Wide Web; 2007.pp. 717–726. [Google Scholar]
- [41].Grau Bernardo Cuenca, Horrocks Ian, Kazakov Yevgeny, Sattler Ulrike. Modular Reuse of Ontologies: Theory and Practice. Journal of Artificial Intelligence Research (JAIR) 2008;31:273–318. [Google Scholar]
- [42].Konev Boris, Lutz Carsten, Walther Dirk, Wolter Frank. Semantic Modularity and Module Extraction in Description Logics; Proceedings of the 18th European Conference on Artificial Intelligence.2008. pp. 55–59. [Google Scholar]
- [43].Seidenberg Julian, Rector Alan. Web ontology segmentation: analysis, classification and use; Proceedings of the 15th international conference on World Wide Web.2006. pp. 13–22. [Google Scholar]
- [44].Fernandez Mary, Florescu Daniela, Kang Jaewoo, Levy Alon, Suciu Dan. Overview of Strudel - A Web-Site Management System. Networking and Information Systems Journal. 1998;1:115–140. [Google Scholar]
- [45].Chen JH, Shaprio LG. Medical Image Segmentation via min s-t Cuts with Side Constraints; International Conference on Pattern Recognition; 2008. [Google Scholar]
- [46].Teng C, Shaprio LG, Kalet I. Head and Neck Cancer Patient Similarity Based on Anatomical Structural Geometry; IEEE International Symposium on Biomedical Imaging.2007. [Google Scholar]
- [47].Gennari JH, Neal ML, Mejino JLV, Cook DL. Using Multiple Reference Ontologies: Managing composite annotations; Proceedings of the International Conference on Biomedical Ontology; 2009; pp. 83–86. in press. [Google Scholar]
- [48].Cook DL, Mejino JLV, Neal ML, Gennari JH. Composite Annotations: Requirements for Mapping Multiscale Data and Models to Biomedical Ontologies; Proceedings of the Engineering in Medicine and Biology Conference; 2009; in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Virtual Soldier Project http://www.virtualsolder.us/index.htm.
- [50].Either T, Ianni G, Schindlauer R, Tompits H, Wang Kewen. Forgetting in Managing Rules and Ontologies; Proceedings of the International Conference on Web Intelligence; 2006. [Google Scholar]
- [51].Qi G, Wang Y, Haase P, Hitzler P. A Forgetting-based Approach for Reasoning with Inconsistent Distributed Ontologies International Workshop on Ontologies: Reasoning and Modularity (WORM’08). [Google Scholar]
- [52].Lin Fangzhen, Reiter Ray. Forget It!; Proceedings of the AAAI Fall Symposium on Relevance; 1994. [Google Scholar]
- [53].Le-Phuoc D, Polleres A, Hauswirth M, Tummarello G, Morbidoni C. Rapid Prototyping of Semantic Mash-Ups through Semantic Web Pipes; 2009. Proceedings, International Conference on the World Wide Web. [Google Scholar]
- [54].SPARQLMotion. http://www.topquadrant.com/products/SPARQLMotion.html.
- [55].Banach http://simile.mit.edu/wiki/Banach.
- [56].Westerski A. Integrated environment for visual data-level mashup development; Proceedings, Web Information Systems Engineering; 2009. [Google Scholar]
- [57].Sintek M, van Elst L, Scerri S, Handschuh S. Distributed Knowledge Representation on the Social Semantic Desktop: Named Graphs, Views and Roles in NRL; Proceedings of the 4th European Semantic Web Conference; 2007. [Google Scholar]