Abstract
We report a female patient with delayed growth and development, skeletal and cardiac defects, and a male XY sex chromosome complement with early failure of gonad development. SRY sequencing was normal. Array comparative genome hybridization (CGH) analysis revealed a gain in copy number in the subtelomeric region of the short arm of chromosome 16, encompassing a region of approximately 560 kb in size including GNG13 which may be involved in ovarian development. The proximal breakpoint of the duplication maps about 18 kb upstream of SOX8 and involves evolutionary conserved regulatory elements. SOX8, like SOX9, is a transcription factor expressed in many tissues, including neural crest, nervous system, muscle, cartilage, adrenal gland, kidney, and testis. There was no increase in GNG13 or SOX8 expression in the patient's lymphoblastoid line. It is possible that an alteration of SOX8 or/and GNG13 expression is responsible for the multiple congenital anomalies and sex reversal in our patient.
Key Words: Agonadism, Bicuspid aortic valve, Delayed development, GNG13, Sex determination, Skeletal deformation, SOX8
Introduction
Male gonad development in mammals is normally initiated by the SRY gene, which either activates expression of SOX9 [Sekido and Lovell-Badge, 2008], or suppresses the activity of SOX9 negative regulators [reviewed in Erickson, 2008]. SOX 9 acts as a testis-determining regulator and also has important functions in the development of bones [Akiyama et al., 2002], the heart [Montero et al., 2002] and other organs [Koopman, 2005]. Haploinsufficiency of SOX9 causes camptomelic dysplasia, a severe abnormality of bone and brain development in which two-thirds of XY patients present as females [Wagner et al., 1994] while duplication of SOX9 can cause 46,XX sex reversal [Huang et al., 1999; Bernard et al., 2003]. The related gene, SOX8, is strongly expressed in many tissues, including neural crest, nervous system, muscle, cartilage, bone, adrenal gland, kidney, heart valves, and testis [Koopman, 2005]. However, its role in sex determination and congenital malformation disorders is not resolved.
Most of the genes involved in sex determination, for example DAX1 and SOX9, have complex regulatory regions extended over a very long distance that must remain intact for appropriate expression. Moreover, some patients with sex reversal have chromosome rearrangements that affect the expression of the genes located up to 1 Mb away from the breakpoint [Wunderle et al., 1998; Pfeifer et al., 1999; Velagaleti et al., 2005; Smyk et al., 2007]. Several highly conserved tissue-specific elements were identified in the vicinity of SOX8 and SOX9, and Sox8 enhancer activities were tested in transgenic mice [Guth et al., 2009]. Alterations in regulatory sequences have been shown to affect Sox8 expression in various tissues including facial mesenchyme, the first branchial arch, peripheral nervous system and other neural crest derivatives as well as central nervous system, eye and limb [Guth et al., 2010].
We report a patient with defects in the major organs/tissues affected by SOX8 expression and with a large duplication immediately upstream of SOX8. The activation of Sox9/SOX9 by distant control elements causing 46,XY sex reversal is well known [Wunderle et al., 1998; Pfeifer et al., 1999; Bishop et al., 2000] and demonstrates the essential role of SOX9 in sex determination. The role of SOX8 in human sexual development has yet to be determined. We believe that this patient's phenotype could result from the duplication involving putative regulatory elements of SOX8 causing abnormal expression of SOX8 during embryogenesis with a dominant negative effect. Alternatively, the duplication of GNG13, a gene encoding a guanine-nucleotide binding protein (a class of protein not previously implicated in sex determination) which is expressed in the very early development of the ovary [Fujino et al., 2007], might inhibit testicular development leading to agonadism, and the other findings could be co-incidental.
Case Report
Family History
The patient's family history was negative for related problems. The father is deceased.
Birth and Early History
Birth weight was 3.64 kg, but head circumference was greater than 95%. Because of a large head size at birth, a CAT scan was performed and no hydrocephalus was detected, but there was a question of cerebral atrophy at that time. A subsequent MRI showed large ventricles, cerebral cortex atrophy and a porencephalic cyst about 3 × 2.5 cm in diameter. A karyotype analysis was performed because of the birth defects and was found to be 46,XY despite her unambiguous female genitalia. Her feeding was poor, and she had a gastrostomy tube placed at 3 weeks of age (which remained until 10 years of age). At 8 months, a gonadectomy was attempted, but no real gonads or uterus were found. At 18 months, she had tear duct tubes placed for dacryostenosis. She had bilateral consecutive exotropia and had bilateral eye surgery at age 8 years.
Physical Exam
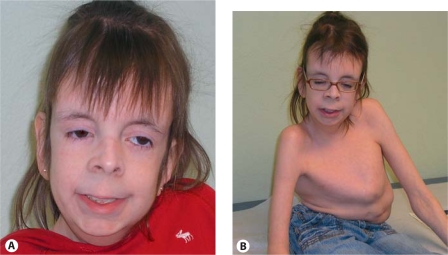
At 14 years of age, she is less than 5th percentile for height and weight while head circumference is now 75th percentile. She shows a scaphocephalic cranium with prominent sutural ridges, eyes that are deeply sunken, right eyelid ptosis, large posteriorly rotated ears with extra folds, a small upturned nose, a long philtrum that is quite smooth, a thin upper lip, and a high palate and small uvula (fig. 1A). In addition to the central skeletal findings visible in figure 1B, there are overlapping fingers and toes, and the hands show asymmetrical changes, with the right showing subluxation of the interphalangeal joints. Neurologically, tone is more decreased on the right side than the left and muscles are atrophied; the patient is wheelchair bound. Language is reasonably developed, but reading and arithmetical skills are rudimentary. The neurologists’ interpretation was that there was an in utero insult resulting in the porencephaly, large ventricles and cerebral atrophy with subsequent cerebral palsy (this seems insufficient to explain the degree of skeletal, the cardiac and the sex reversal problems).
Fig. 1.
Patient at 14 years of age. A Facial features with right ptosis, sunken eyes, small upturned nose, long philtrum, and thin upper lip visible. B Severe scoliosis remains despite multiple orthopedic procedures.
Cardiac
An echocardiogram demonstrated an abnormal aortic valve which appeared to have 3 leaflets, although it had a very prominent posterior sinus of Valsalva and a small posterior leaflet. The right coronary cusp was slightly thickened. There was dilatation at the level of the sinuses of Valsalva and ascending aorta with a very mild amount of aortic regurgitation. The majority of the multiple cardiology reports refer to the aortic valve as bicuspid with a very small third cusp.
Skeletal
At age 5½ years, she had right hip surgery for developmental dysplasia of the hip. At 6 years of age, she had a severe type of scoliosis with about a 50-degree curvature without apparent changes in bone density. A T6-L2 diskectomy with vertebral body fusion, using a morselized graft for posterior spinal fusion and instrumentation from T12–L3 was performed. Despite this, she continues to have a severely deformed chest with marked pectus carinatum and distortion (fig. 1B).
Laboratory
Multiple complete blood counts were normal. Bone age was within 2 standard deviations of chronological age. Thyroid testing was normal. IGFBP-3 was abnormal at one time, but normal at 8 years of age. IGFBP-1 was moderately elevated at 11 years of age. There was no evidence of masculinization and no sex steroid measurements have been performed.
Material and Methods
DNA samples from the proband and her mother were obtained after acquiring informed consents approved by the Institutional Review Board for Human Subject Research at Baylor College of Medicine.
Cytogenetic and FISH Analyses
Chromosome and FISH analyses were performed on peripheral blood lymphocytes using standard procedures. FISH analyses with the bacterial artificial chromosome (BAC) clones were performed using standard procedures. Briefly, the BAC clones of interest (RP11-26D18 and RP11-728H8) were grown in TB media with 20 μg/ml chloramphenicol. DNA was extracted from the BAC clones (Eppendorf Plasmid Mini Prep kit, Hamburg, Germany) and directly labeled with SpectrumOrange™ dUTP by nick translation (Abbot/Vysis, Downer Grove, Ill., USA) according to the manufacturers’ instructions.
BAC Array Analysis
DNA was extracted from whole blood using the Puregene DNA extraction kit (Gentra, Minneapolis, Minn., USA) according to the manufacturer's instructions. The comparative genome hybridization (CGH) analysis was conducted on a clinically available microarray (Version 6 BAC array) containing 1,472 BAC and PAC clones designed to cover genomic regions of 150 known genomic disorders, all 41 subtelomeric regions and 43 pericentromeric regions with minimum backbone coverage of every chromosome at the 650-band level of cytogenetic resolution (http://www.bcm.edu/geneticlabs/?pmid=16207). Procedures for DNA labeling and comparative genomic hybridization were described previously [Yu et al., 2003]. The fluorescent signals on the slides were scanned into image files using an Axon microarray scanner and ScanArray software (GenePix 4000B from Axon Instruments, Union City, Calif., USA). For each sample, 2 experiments were performed with reversal of the dye labels for the control and test samples, and the data from both dye-reversed hybridizations were integrated to determine inferences for each case. Microarray image files were quantified using GenePix Pro 4.0 software. The quantitation data were subjected to normalization and integration for all clones analyzed from a single patient sample as described [Shaw et al., 2004]. In addition, a single clone T-statistic and permutation-based p value were also computed, providing further criteria to determine whether a clone deviates significantly from the mean.
High-Resolution Oligonucleotide Microarray Analysis
Human whole genome oligonucleotide microarray kits 244K (Agilent Technologies, Inc., Calif., USA) were used to analyze DNA from the proband and her mother to further refine the identified genomic gain. The procedures for DNA digestion, labeling and hybridization were performed according to the manufacturer's instructions with some modifications [Probst et al., 2007].
RT-PCR
RNA was prepared from the patient's and 2 control lymphoblastoid lines using gDNA eliminator spin columns to eliminate possibly contaminating DNA. cDNA was prepared and PCR was performed at limiting conditions to amplify SOX8 and GNG13 mRNA. The primers for SOX8 were ones previously used by Schlierf et al. [2007]: forward 5′-TGTCTCCTTGCTGGCAGAGT-3′ and reverse 5′-GAGCAACGAGCAACGTGATG-3′ and have a 191-bp product. The primers for GNG13 were: forward 5′-TGCTGAAGTGGATCGAGGAC-3′ and reverse 5′-GTCTCACAGGATGGTGTGAG-3′ and have a 141-bp product.
Real-Time RT-PCR
These above primers were used to quantify the amount of SOX8 and GNG13 mRNA by real-time RT-PCR. The timed increase in PCR product was monitored with iTaq SYBR Green Supermix with ROX. GAPDH was used as a control with primers: forward 5′-TGTTGCCATCAATGACCCCTT-3′ and reverse 5′-CTCCACGACGTACTCAGCG-3′. Expression was analyzed using the 2s-ΔΔ CT method of comparing fold difference to the endogenous control [Livak and Schmittgen, 2001].
Results
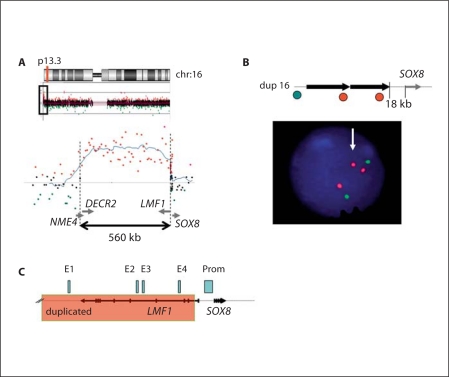
Karyotype analyses showed a normal male 46,XY complement despite external female genitalia. Additional FISH studies for all chromosome-specific subtelomeric regions, William's syndrome critical region and a methylation assay of the SNRPN gene were performed by previous attending physicians and were normal. No pathogenic mutations were found in the coding regions of the SRY gene. BAC array CGH analysis showed a gain in copy number of the 16p13.3 region encompassing an approximately 560-kb genomic interval [chr16:386,962–953,823; hg18] (fig. 2A). FISH analysis confirmed a duplication of the 16p13.3 region (fig. 2B). A review of the 34 genes in the duplicated region in GeneCard (which provides expression and some functional information) and GeneOntology (which provides richer functional information) found one other candidate gene for sex reversal: GNG13. Although found and first studied for its role in olfaction [Huang et al., 1999], Fujino et al. [2007] found that this guanine-nucleotide binding protein gene is expressed in the outer cortex of the developing ovary and implicated it in ovarian development. Interestingly, the duplication proximal breakpoint was located in intron 1 of the LMF1 gene, just 18 kb upstream of SOX8 (fig. 2C). The duplicated segment involves evolutionary conserved regulatory sequences E1, E3 and E4 of the SOX8 gene. The patient's mother was studied by whole genome array CGH analysis and showed negative results for the 16p13.3 gain. The patient's father is deceased. Duplications of 16p encompassing GNG13 and SOX8 have been reported as copy number variants (CNVs) (Database of Genomic Variants, GRCh 37, 2009), but the breakpoints location are different from those presented here.
Fig. 2.
A High-resolution array CGH showing the duplication within the 16p13.3 region, encompassing an ∼560-kb segment. Below, an enlargement of the breakpoint boundary detected by array CGH. The duplicated segment contains at least 29 known genes and putative gene products. The 5′ region of the SOX8 gene is located at the proximal breakpoint of the duplication. B A schematic view of the rearrangement. Black arrow represents the duplicated 16p13.3 segment. Green and red circles correspond to the FISH clones RP11-26D18 (green) and RP11-728H8 (red). Below, FISH analysis showing a duplication detected by clone RP11-728H8.C A magnified view of a duplication boundary upstream of the SOX8 gene. Black arrows show the location and direction of transcription of the LMF1 and SOX8 genes. Localization of enhancers E1–E4 (light blue rectangles) and the SOX8 promoter (Prom) is shown as proposed [Guth et al., 2010]. Pink shaded area indicates a duplicated segment with a proximal breakpoint within intron 1 of the LMF1 gene, 18 kb upstream of SOX8.
In addition, whole genome BAC array CGH using the patient's DNA revealed a few other regions with copy number gains or losses. A gain in copy number was observed for chromosome 1 (1q44-q44) and a copy number loss was observed for chromosome 8 (8q11-q11). The same changes in copy number for the above regions are also present in the mother. Therefore, these findings represent a familial copy number variant. A gain in copy number was observed for chromosome 9 (9p24.2-p24.2) and a copy number loss was observed for chromosome 20 (20q13.12-q13.12). Maternal microarray analyses revealed normal copy number for these regions; however, changes in these regions of the genome were found among healthy individuals and reported previously as benign CNV.
SOX8 and GNG13 Quantification in Lymphoblast Cell Lines
RT-PCR showed that the patient's level of SOX8 and GNG13 mRNAs were lower than or intermediate between that of 2 controls as determined by this semi-quantitative method (not shown). Real-time RT-PCR confirmed the semi-quantitative results with only very low levels of mRNAs detectable (less than 1% of GAPDH) (table 1). The precise parallel for the 2 genes, both by the semi-quantitative method and by real-time PCR based on a ratio to GAPDH mRNA levels, may suggest that the differences between the measured mRNA levels may have more to do with the cell lines than the particular levels of gene expression for these 2 genes.
Table 1.
Quantitative real-time, reverse transcription PCR results for SOX8 and GNG13 mRNA
Assigned value for comparison.
Discussion
While trisomy 16 causes early lethality, trisomy 16p is sometime viable and occasionally associated with sexual ambiguity [Stern and Mureh, 1975; Leschot et al., 1979; Leonard et al., 1992]. As array CGH has become more available, it has become clear that the short arm of chromosome 16, because of abundant low copy repeats (LCR) allowing nonhomologous recombination, is a ‘hot spot’ for CNV. A 1.5 Mb-fragment flanked by LCRs has shown reciprocal duplications and deletions in probands identified for mental retardation and/or autism [Ullmann et al., 2007]. Further study confirms a phenotype of microcephaly and mental retardation with variable epilepsy, cleft lip and midline facial defects with the deletion of this segment [Ballif et al., 2007] while the duplication is a normal variant [Hannes et al., 2008].
The duplication we report is much more distal on 16p and is immediately adjacent to SOX8. A review of genes in the duplication revealed GNG13 as a candidate for ovarian development. LMF1 (lipase maturation factor 1), the gene which is nearly completely duplicated, is primarily involved in triglyceride metabolism but contains SOX8 enhancers E1, E2, E3, and E4 [Guth et al., 2010].
The role of the SOX9 in sex differentiation is major [Qin and Bishop, 2005]. SOX8 is often characterized as redundant with SOX9. There is a very large overlap in the expression pattern of SOX8 with SOX9. In humans, the co-deletions of SOX8 with alpha globin loci may contribute to the mental retardation of the ATR-16 syndrome [Pfeifer et al., 2000] which does not include bone or sexual ambiguity problems. The knockout (KO) is viable but with failure-to-thrive [Sock et al., 2001]. Indeed, while initially fertile, there is progressive male infertility [O'Bryan et al., 2008]. There has been much interest in SOX8's potential role in sexual differentiation. It has been shown to be capable of in vitro of interaction with Sf1 to activate the Amh promoter [Schepers et al., 2003] and the double KO of Sox8 and Sox9 shows the synergistic action of the 2 [Chaboissier et al., 2004]. However, Sox8 cannot substitute for Sox9 when the latter is knocked out only in the gonadal primordium [Chaboissier et al., 2004] while studies of another conditional KO of Sox9 in the absence of Sox8 show that concerted Sox8 and Sox9 function in Sertoli cells is essential for the maintenance of testicular function [Barrionuevo et al., 2009].
There has been enormous recent progress in the study of distant-acting enhancers [reviewed in Visel et al., 2009], particularly relevant to studies of SOX9. The recent identification of embryonic enhancers for Sox8 by sequence conservation between man, mouse and chicken, confirmed by transgenic expression, found 4 important enhancers for non-gonadal expression in the 5′ 100-kb region of the gene [Guth et al., 2010]. Thus, we hypothesized that the duplication just upstream of SOX8 altered transcription of the gene. We tested this hypothesis in a lymphoblastoid line with negative results. However, a lymphoblastoid line is unlikely to reflect developmental expression in gonadal ridges, cartilage, brain, and heart.
We believe that it would be more likely that if the duplication is affecting SOX8 expression, it would be increasing, not decreasing expression. As mentioned above, the KO for Sox8 only shows failure-to-thrive (which our patient shows) and progressive male infertility. There was no problem with skeleton or heart in the mice. On the other hand, over-expression of SOX8 could function as a dominant negative with SOX9. The SOX9 has a dimerization domain which is required for chondrogenesis but not for sex determination [Bernard et al., 2003]. Lack of dimerization has been shown to cause camptomelic dysplasia and 2 46,XY cases had a male phenotype [Bernard et al., 2003; Sock et al., 2003]. However, if a significant amount of SOX8 dimerized with SOX9 and caused inefficient DNA binding, the patient's skeletal problems (not typical of camptomelic dysplasia, however) and bicuspid aortic valve might be explained while competition with SOX9 at the monomer site upstream of SF1, or elsewhere, might cause sex reversal with an early failure of gonad development resulting in both müllerian and wolffian derivatives. It is also possible that a more subtle relationship of this duplication to the phenotype is causal. Alterations near SOX9 have included quite different phenotypes than that of camptomelic dysplasia: Pierre-Robin sequence [Benko et al., 2009] and brachydactyly-anonychia [Kurth et al., 2009].
Although we focused on the highly studied SOX8 (with it potential roles in sex differentiation and bone, heart and brain development) as a target for a unitary pathological process, an alternative 2-hit explanation for out patient's phenotype can be provided. Duplication and increased expression of GNG13 might stimulate an early stage of ovarian development with subsequent failure of both testicular and ovarian formation and persistent müllerian and wolffian ducts. A separate, in utero insult resulting in a porencephalic cyst with resultant defects in neurological development and a deformed skeleton secondary to unbalanced muscle tension (as the neurologists opined) might be explanatory. The bicuspid aortic valve, present in about 2% of the population [Blase and Carabello, 2002], would be a chance co-occurrence under this hypothesis.
Acknowledgements
We thank Ivan Borbon for administrative assistance and the patient and her family for their co-operation. Dr. Erickson is supported by the Holsclaw Family Professorship in Human Genetics and Inherited Diseases.
References
- Akiyama H, Chaboissie MC, Martin JF, Schedl A, de Crombrugghe B. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 2002;16:2813–2828. doi: 10.1101/gad.1017802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballif BC, Hornor SA, Jenkins E, Madan-Khetarpal S, Surti U, et al. Discovery of a previously unrecognized microdeletion syndrome of 16p11.2–p12.2. Nat Genet. 2007;39:1071–1073. doi: 10.1038/ng2107. [DOI] [PubMed] [Google Scholar]
- Barrionuevo F, Georg I, Scherthan H, Lecureuil C, Guillou F, et al. Testis cord differentiation after the sex determination stage is independent of Sox9 but fails in the combined absence of Sox9 and Sox8. Dev Biol. 2009;327:301–312. doi: 10.1016/j.ydbio.2008.12.011. [DOI] [PubMed] [Google Scholar]
- Benko S, Fantes JA, Amiel J, Kleinjan DJ, Thomas S, et al. Highly conserved non-coding elements on either side of SOX9 associated with Pierre Robin sequence. Nat Genet. 2009;42:359–364. doi: 10.1038/ng.329. [DOI] [PubMed] [Google Scholar]
- Bernard P, Tang P, Liu S, Dewing P, Harley VR, et al. Dimerization of SOX9 is required for chondrogenesis, but not for sex determination. Hum Mol Genet. 2003;12:1755–1765. doi: 10.1093/hmg/ddg182. [DOI] [PubMed] [Google Scholar]
- Bishop CE, Whitworth DJ, Qin Y, Agoulnik AI, Agoulnik IU, et al. A transgenic insertion upstream of Sox9 is associated with dominant XX sex reversal in the mouse. Nat Genet. 2000;26:490–494. doi: 10.1038/82652. [DOI] [PubMed] [Google Scholar]
- Blase A, Carabello MD. Aortic stenosis. N Engl J Med. 2002;346:677–682. doi: 10.1056/NEJMcp010846. [DOI] [PubMed] [Google Scholar]
- Chaboissier MC, Kobayashi A, Vidal VI, Lützkendorf S, van de Kant HJ, et al. Functional analysis of Sox8 and Sox9 during sex determination in the mouse. Development. 2004;131:1891–1901. doi: 10.1242/dev.01087. [DOI] [PubMed] [Google Scholar]
- Erickson RP. The sex determination pathway. In: Epstein CJ, Erickson RP, Wynshaw-Boris A, editors. Inborn Errors of Development: The Molecular Basis of Inborn Errors of Development. New York: Oxford University Press; 2008. pp. 203–211. [Google Scholar]
- Fujino A, Pieretti-Vanmarcke R, Wong A, Donahoe PK, Arango NA. Sexual dimorphism of G-protein subunit Gng13 expression in the cortical region of the developing mouse ovary. Dev Dyn. 2007;236:1991–1996. doi: 10.1002/dvdy.21183. [DOI] [PubMed] [Google Scholar]
- Guth SI, Bösl MR, Sock E, Wegner M. Evolutionary conserved sequence elements with embryonic enhancer activity in the vicinity of the mammalian Sox8 gene. Int J Biochem and Cell Biol. 2010;42:465–471. doi: 10.1016/j.biocel.2009.07.008. [DOI] [PubMed] [Google Scholar]
- Hannes FD, Sharp AJ, Mefford HC, de Ravel T, Ruivenkamp CA, et al. Recurrent reciprocal deletions and duplications of 16p13.11: the deletion is a risk factor for MR/MCA while the duplication may be a rare variant. J Med Genet. 2008;46:223–232. doi: 10.1136/jmg.2007.055202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Wang S, Ning Y, Lamb AN, Bartley J. Autosomal XX sex reversal caused by duplication of SOX9. Am J Med Genet. 1999;87:349–353. doi: 10.1002/(sici)1096-8628(19991203)87:4<349::aid-ajmg13>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Huang L, Shanker YG, Dubauskaite J, Zheng JZ, Yan W, et al. Ggamma 13 colocalizes with gustducin in taste receptor cells and mediates IP3 responses to bitter denatorium. Nat Neurosci. 1999;2:1055–1062. doi: 10.1038/15981. [DOI] [PubMed] [Google Scholar]
- Koopman P. Sex determination: a tale of two Sox genes. Trends Genet. 2005;21:367–370. doi: 10.1016/j.tig.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Kurth I, Kloppocki E, Stricke S, van Oosterwijk J, Vanek S, et al. Duplications of noncoding elements 5′ of SOX9 are associated with brachydactyly-anonychia. Nat Genet. 2009;41:862–863. doi: 10.1038/ng0809-862. [DOI] [PubMed] [Google Scholar]
- Leonard C, Huret JL, Imbert MC, Lebouc Y, Selva J, et al. Trisomy 16p in a liveborn offspring due to maternal translocation t(16;21)(q11;p11) and review of the literature. Am J Med Genet. 1992;43:621–625. doi: 10.1002/ajmg.1320430324. [DOI] [PubMed] [Google Scholar]
- Leschot NJ, De Nef JJ, Geraedts JP, Becker-Bloemkolk MJ, Talma A, et al. Five familial cases with a trisomy 16p syndrome due to translocation. Clin Genet. 1979;16:205–214. doi: 10.1111/j.1399-0004.1979.tb00991.x. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time Quantitative PCR and the 2(-DeltaDelta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Montero JA, Giron B, Arrechedera H, Cheng YC, Scotting P, et al. Expression of Sox8, Sox9 and Sox10 in the developing valves and autonomic nerves of the embryonic heart. Mech Dev. 2002;118:199–202. doi: 10.1016/s0925-4773(02)00249-6. [DOI] [PubMed] [Google Scholar]
- O'Bryan MK, Takada S, Kennedy CL, Scott G, Harada S, et al. Sox8 is a critical regulator of adult Sertoli cell function and male fertility. Dev Biol. 2008;316:359–370. doi: 10.1016/j.ydbio.2008.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer D, Kist R, Dewar K, Devon K, Lander ES, et al. Campomelic dysplasia translocation breakpoints are scattered over 1 Mb proximal to SOX9: evidence for an extended control region. Am J Hum Genet. 1999;65:111–124. doi: 10.1086/302455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer D, Poulat F, Holinski-Feder E, Kooy F, Scherer G. The SOX8 gene is located within 700 kb of the tip of chromosome 16p and is deleted in a patient with ATR-16 syndrome. Genomics. 2000;63:108–116. doi: 10.1006/geno.1999.6060. [DOI] [PubMed] [Google Scholar]
- Probst FJ, Roeder ER, Enciso VB, Ou Z, Cooper ML, et al. Chromosomal microarray analysis (CMA) detects a large X chromosome deletion including FMR1, FMR2, and IDS in a female patient with mental retardation. Am J Med Genet A. 2007;143A:1358–1365. doi: 10.1002/ajmg.a.31781. [DOI] [PubMed] [Google Scholar]
- Qin Y, Bishop CE. Sox9 is sufficient for functional testis development producing fertile male mice in the absence of Sry. Hum Mol Genet. 2005;14:1221–1229. doi: 10.1093/hmg/ddi133. [DOI] [PubMed] [Google Scholar]
- Schepers G, Wilson M, Wilhelm D, Koopman P. SOX8 is expressed during testis differentiation in mice and synergizes with SF1 to activate the Amh promoter in vitro. J Biol Chem. 2003;278:28101–28108. doi: 10.1074/jbc.M304067200. [DOI] [PubMed] [Google Scholar]
- Schlierf B, Friedrich RP, Roerig P, Felsberg J, Reifenberger G, et al. Expression of SoxE and SoxD genes in human gliomas. Neuropathol Appl Neurobiol. 2007;33:621–630. doi: 10.1111/j.1365-2990.2007.00881.x. [DOI] [PubMed] [Google Scholar]
- Sekido R, Lovell-Badge R. Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature. 2008;453:930–934. doi: 10.1038/nature06944. [DOI] [PubMed] [Google Scholar]
- Shaw CJ, Shaw CA, Yu W, Stankiewicz P, White LD, et al. Comparative genomic hybridisation using a proximal 17p BAC/PAC array detects rearrangements responsible for four genomic disorders. J Med Genet. 2004;41:113–119. doi: 10.1136/jmg.2003.012831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyk M, Berg JS, Pursley A, Curtis FK, Fernandez BA, et al. Male-to-female sex reversal associated with an approximately 250 kb deletion upstream of NR0B1 (DAX1) Hum Genet. 2007;122:63–70. doi: 10.1007/s00439-007-0373-8. [DOI] [PubMed] [Google Scholar]
- Sock E, Schmidt K, Hermanns-Borgmeyer I, Bösl MR, Wegner M. Idiopathic weight reduction in mice deficient in the high-mobility-group transcription factor Sox8. Mol Cell Biol. 2001;21:6951–6959. doi: 10.1128/MCB.21.20.6951-6959.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sock E, Pagon RA, Keymolen K, Lissens W, Wegner M, et al. Loss of DNA-dependent dimerization of the transcription factor SOX9 as a cause for campomelic dysplasia. Hum Mol Genet. 2003;12:1439–1447. doi: 10.1093/hmg/ddg158. [DOI] [PubMed] [Google Scholar]
- Stern LM, Mureh AR. Pseudohermaphroditism with clinical features of trisomy 19 in an infant trisomic for parts of chromosomes 16 and 18: 47,XY, der(18),t(16,18)(p12;q11)mat. J Med Genet. 1975;12:305–307. doi: 10.1136/jmg.12.3.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullmann R, Turner G, Kirchhoff M, Chen W, Tonge B, et al. Array CGH identifies reciprocal 16p13.1 duplications and deletions that predispose to autism and/or mental retardation. Hum Mutat. 2007;28:674–682. doi: 10.1002/humu.20546. [DOI] [PubMed] [Google Scholar]
- Velagaleti GV, Bien-Willner GA, Northup JK, Lockhart LH, Hawkins JC, et al. Position effects due to chromosome breakpoints that map approximately 900 kb upstream and approximately 1.3 Mb downstream of SOX9 in two patients with campomelic dysplasia. Am J Hum Genet. 2005;76:652–662. doi: 10.1086/429252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Rubin EM, Pennacchio LA. Genomic views of distant-acting enhancers. Nature. 2009;461:199–205. doi: 10.1038/nature08451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner T, Wirth J, Meyer J, Zabel B, Held M, et al. Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell. 1994;79:1111–1120. doi: 10.1016/0092-8674(94)90041-8. [DOI] [PubMed] [Google Scholar]
- Wunderle VM, Critcher R, Hastie N, Goodfellow PN, Schedl A. Deletion of long-range regulatory elements upstream of SOX9 causes campomelic dysplasia. Proc Natl Acad Sci USA. 1998;95:10649–10654. doi: 10.1073/pnas.95.18.10649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Ballif BC, Kashork CD, Heilstedt HA, Howard LA, et al. Development of a comparative genomic hybridization microarray and demonstration of its utility with 25 well-characterized 1p36 deletions. Hum Mol Genet. 2003;12:2145–2152. doi: 10.1093/hmg/ddg230. [DOI] [PubMed] [Google Scholar]