Abstract
Early detection of ovarian cancer is difficult owing to the lack of specific and sensitive tests available. Previously, we found expression of nectin 4 to be increased in ovarian cancer compared with normal ovaries. Reverse transcriptase–polymerase chain reaction (RT-PCR) and quantitative RT-PCR validated the overexpression of nectin 4 messenger RNA in ovarian cancer compared with normal ovarian cell lines and tissues. Protein levels of nectin 4 were elevated in ovarian cancer cell lines and tissue compared with normal ovarian cell lines as demonstrated by Western immunoblotting, flow cytometry, and immunohistochemical staining of tissue microarray slides. Cleaved nectin 4 was detectable in a number of patient serum samples by enzyme-linked immunosorbent assay. In patients with benign gynecologic diseases with high serum CA125 levels, nectin 4 was not detected in the majority of cases, suggesting that nectin 4 may serve as a potential biomarker that helps discriminate benign gynecologic diseases from ovarian cancer in a panel with CA125.
Keywords: Nectin 4, Ovarian cancer, Serum biomarker, Tissue microarray, Diagnosis, Detection
Ovarian cancer is the seventh leading cause of cancer-related death in women1; in the United States, more than 20,000 women were diagnosed with ovarian cancer, resulting in more than 14,000 deaths.2 Survival is greatly increased when ovarian cancer is diagnosed in early stages; however, approximately 70% of cases are diagnosed at stage III or IV, when the 5-year survival rate decreases dramatically.3 Subtle symptoms and the lack of a specific, sensitive diagnostic test contribute to the delay in diagnosis, highlighting the need for improved screening methods and novel targeting strategies for treatment.4–6 Currently, an elevated serum CA125 level is used to detect recurrent ovarian cancer; however, its use in screening the general population is precluded owing to a lack of specificity and modest sensitivity in the detection of early-stage disease.
Our group previously identified nectin 4 gene up-regulation in ovarian cancer tissues as part of an effort to identify novel ovarian cancer biomarkers.6 Nectins are a family of immunoglobulin-like cell adhesion molecules important in the formation and maintenance of adherens junctions and tight junctions. All nectins share a similar structure: 3 immunoglobulin-like extracellular loops, a single transmembrane region, and a short cytoplasmic domain that can bind to afadin.7–10 Nectins function by first forming homo cis-dimers on the cell surface and then trans-dimers on adjacent cells in a homophilic and a heterophilic manner. The specificity of binding is different for each nectin.9,11,12 For example, nectin 4 can form trans-homodimers and trans-heterodimers with nectin 1 but not with nectin 2 or 3.13
Four nectin proteins have been described; nectin 1 and 2 are broadly expressed in adult tissues, while nectin 3 is expressed mainly in the testes and placenta.11,13–15 Expression of nectin 4 is normally restricted to the placenta but has been reported in ductal breast carcinoma and lung adenocarcinomas. 13,16,17 Fabre-Lafay et al16,18 and Takano et al17 demonstrated that the ectodomain of nectin 4 can be cleaved from the cell surface by the metalloproteinase ADAM17 (A Disintegrin And Metalloproteinase 17) and detected in serum by enzyme-linked immunosorbent assay (ELISA) in cases of breast and lung cancer.
In this comprehensive study, we sought to validate the overexpression of nectin 4 in ovarian cancer tissues and cell lines compared with their normal ovarian counterparts using a variety of techniques, including reverse transcriptase–polymerase chain reaction (RT-PCR), quantitative RT-PCR, flow cytometry, Western immunoblotting, immunohistochemical analysis, and ELISA. We also sought to determine whether overexpression, at the tissue or serum level, correlated with relevant clinical outcomes. Tissue microarrays (TMAs) composed of 500 cases of clinically annotated ovarian cancer allowed us to determine the expression of nectin 4 in various subtypes, stages, and grades of ovarian cancer and to evaluate correlations with recurrence and survival. Finally, we used an ELISA to quantify nectin 4 levels in the serum samples of cases of ovarian cancer and normal and benign gynecologic cases.
Materials and Methods
Reagents
Cell culture media and supplements were purchased from Invitrogen (Carlsbad, CA) unless otherwise stated. Chemicals were purchased from Sigma Chemical (St Louis, MO) unless otherwise stated.
Monoclonal antibodies used were mouse antihuman nectin 4 (clone 337516; R&D Systems, Minneapolis, MN), mouse antihuman nectin 4 (clone N4.61; provided by M.L.), mouse antihuman nectin 1 (clone CK8; Invitrogen), mouse antihuman ADAM17 (clone 111623; R&D Systems), and normal mouse IgG (clone 3-5D1-C9, AbCam, Cambridge, MA).
Polyclonal antibodies used included biotin-conjugated goat antihuman nectin 4 (BAF2659, R&D Systems), horseradish peroxidase–conjugated rabbit antimouse IgG (ab5762, AbCam), and biotin-conjugated goat antimouse IgG F(ab')2 fragment (Jackson ImmunoResearch, West Grove, PA).
Cell Lines
Ovarian cancer cell lines SKOV3, ES2, OVCAR3, HEY, C13, OV2008, OVCA429, OVCA433, A2780s, and A2780cp (provided by Barbara Vanderhyden, PhD, University of Ottawa, Ottawa, Canada); NIH:OVCAR5 (provided by Judah Folkman, MD, Harvard Medical School, Boston, MA); CAOV3 (provided by Robert Bast Jr, MD, University of Texas, Houston); and MA148dsRed2 (provided by Sundaram Ramakrishnan, PhD, University of Minnesota, Minneapolis) were maintained as previously described.19–22 SKOV3, ES2, and OVCA429 cell lines were derived from clear cell carcinomas; OV2008 and C13 cells were derived from endometrioid tumors; OVCAR3, OVCAR5, OVCA433, CAOV3, HEY, MA148, and A2780s/cp cell lines were derived from serous adenocarcinomas.
Immortalized normal ovarian surface epithelial (NOSE) cell lines 1816-575, 1816-686, HIO117, HIO135, IMCC3, IMCC5, and 3173-11 (provided by Patricia Kruk, PhD, University of South Florida, Tampa) and IOSE-80 (provided by Nelly Auersperg, MD, PhD, University of British Columbia, Vancouver, Canada) were also maintained as described.23,24 Cells were maintained in a humidified chamber at 37°C with 5% carbon dioxide and were routinely subcultured with trypsin/EDTA.
Tissue Samples
Snap-frozen and formalin-fixed, paraffin-embedded (FFPE) tissue blocks were obtained from the University of Minnesota Tissue Procurement Facility after institutional review board approval. RNA was isolated from snap-frozen tissues, while FFPE tissues were used to optimize immunohistochemical staining. The five ovarian cancer tissues were of the serous subtype. The 7 normal tissues were benign leiomyoma (n = 3), endometriosis (n = 1), benign peritubal cyst (n = 1), and normal ovary (n = 2). All tissue samples underwent strict quality control measures before use in these studies. Namely, tumors were diagnosed by a pathologist at the time of surgery using Tissue Tek OCT (Sakura Finetek, Torrance, CA)–embedded tissue. The following day, the FFPE H&E-stained slides were reviewed by a pathologist (S.E.P.) to confirm the accuracy of the diagnosis. A third pathologist reviewed the quality control H&E-stained slides of all University of Minnesota Tissue Procurement Facility cases to confirm the diagnosis of the samples before distribution to researchers. In addition, a pathologist (S.E.P) reviewed the slides while scoring the immunohistochemical staining.
Blood and Ascites Samples
Staff at the University of Minnesota Tissue Procurement Facility ensured that all patients signed an institutional review board–approved consent form before surgery. Blood was collected immediately before surgery from women with abdominal masses suspected to be ovarian cancer. Blood samples were collected from 51 women with serous ovarian cancer (5 stage 1, 7 stage 2, 28 stage 3, 10 stage 4, and 1 unstaged; 4 Silverberg grade 1, 9 Silverberg grade 2, 37 Silverberg grade 3, and 1 ungraded), 14 women with clear cell ovarian cancer (8 stage 1, 1 stage 2, 4 stage 3, and 1 not staged; 14 Silverberg grade 3), 18 women with low malignant potential (LMP) ovarian tumors (10 LMP serous, 3 LMP mucinous, and 5 LMP with no subtype noted), and 51 women with benign gynecologic disease (12 benign endometriosis, 10 benign mucinous cystadenoma, 6 fibroma, 6 benign serous cystadenoma, 17 benign disease not specified). Ascites fluid was collected from 10 women with serous ovarian cancer.
Blood and ascites samples were processed by standard protocols, divided into aliquots, and stored at −80°C. Before use in this study, the pathology reports for the blood and ascites samples were reviewed by a gynecologist/oncologist (M.A.G or P.A.A.) to ensure that the samples were placed into the correct category.
Reverse Transcriptase–Polymerase Chain Reaction
Total RNA was extracted from cell lines and ovarian tissue samples using the RNeasy Mini kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. A 189-base-pair sequence corresponding to nectin 4 was amplified with the following primers: forward, 5'-CAAAATCTGTGGCACATTGG-3'; and reverse, 5'-GCTGACATGGCAGACGTAGA-3'. One-step RT-PCR was performed with the RT-PCR Access kit (Promega, Madison, WI), with conditions as follows: 45 minutes at 45°C; 1 cycle of 94°C, 2 minutes; 55°C, 1 minute; 68°C, 1 minute; 25 cycles of 94°C for 30 seconds, 55°C for 1 minute, and 68°C for 1 minute; and a final extension at 68°C for 7 minutes. Expression of nectin 4 in MCF7 breast cancer cell lines has been reported and was used as a positive control.18 Expression of β-actin in the samples confirmed that messenger RNA (mRNA) was not degraded and that similar amounts of mRNA were loaded.
Quantitative RT-PCR
Real-time quantification of nectin 4 was performed using the SYBRGreen assay (Bio-Rad Laboratories, Hercules, CA) and the iQ5 Real-Time PCR thermocycler (Bio-Rad). We amplified 2 µL of complementary DNA in a 25-µL reaction containing 13 µL of iQ SYBRGreen Supermix (Bio-Rad), 1 µL each of nectin 4 forward and reverse primers (forward, TGCTCAAGTGCCTGAGTGAA; reverse, AGACGTAGATGCCGCTGTG), and 8 µL of nuclease-free water. Following an initial denaturation step of 95°C for 3 minutes, 40 cycles of PCR were performed under the following conditions: 95°C, 10 seconds (denaturation), and 57°C, 30 seconds (annealing/extension). Product size was verified by agarose gel electrophoresis. Threshold cycle (Ct) values were calculated according to the iQ5 real-time detection software. Standard curves for nectin 4 and β-actin were generated by plotting Ct vs the log of the initial starting amount of RNA in nanograms.25 Each sample was normalized to the amount of β-actin mRNA present in the sample, and the relative amount of each sample was determined as a fold-change increase over the lowest expressing cell line (1816-575).
Flow Cytometry
Cells (1 × 106) were incubated with 1 to 2.5 µg of mouse anti–nectin 4 (clone 337516), mouse anti–nectin 1, mouse antihuman ADAM17, or a control mouse IgG for 30 minutes at 4°C. Cells were washed and then incubated with biotin-conjugated goat antimouse IgG F(ab')2 fragment for 30 minutes at 4°C. After washing, cells were incubated with allophycocyanin-conjugated streptavidin for 20 minutes at 4°C. After washing, cells were resuspended and run on a FACSCalibur (Becton Dickinson, Franklin Lakes, NJ) and analyzed using CellQuest Pro software (Becton Dickinson), gating on live cells by forward and side scatter.
Western Immunoblotting
Total protein extracts were derived from confluent monolayers of cells in 50 mmol/L tris(hydroxymethyl)aminomethane (Tris), 150 mmol/L sodium chloride, 1 mmol/L EDTA, 1% Triton X-100, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS), protease inhibitor cocktail (Roche Applied Science, Basel, Switzerland), and 1 mmol/L phenylmethylsulfonyl fluoride and then stored at –80°C. Fifty micrograms of total cell lysate were separated on a 10% SDS Tris-hydrochloride polyacrylamide gel and then blotted onto a polyvinylidene difluoride membrane (GE Healthcare Limited, Piscataway, NJ). Membranes were blocked with 5% powdered milk (Roundy’s, Milwaukee, WI) in PBS, as previously described,26,27 and then incubated in 1 µg/mL mouse antihuman nectin 4 (clone N4.61) overnight, followed by a 2-hour incubation in horseradish peroxidase–conjugated rabbit antimouse antibody diluted 1/5,000. Protein was visualized using the Super Signal West Femto kit (Thermo-Fisher Scientific, Rockford, IL) according to the manufacturer’s instructions. Membranes were exposed to autoradiography film (Midwest Scientific, Valley Park, MO) and developed.
Tissue Microarrays
TMA slides containing 0.6-mm duplicate core samples for 500 ovarian cancer cases were provided by the Cheryl Brown Ovarian Cancer Outcomes Unit (University of British Columbia, Vancouver, Canada). Cases included in the TMA were chosen based on having been optimally cytoreduced at initial surgery with no macroscopic residual disease remaining. Owing to these criteria, a significant proportion of early-stage cases were present on the TMA relative to the general population. None of the patients received neoadjuvant therapy, and all received platinum-based chemotherapy following surgery. The 500 cases included on the TMA were collected up to 18 years before this analysis. H&E-stained slides for all cases were reviewed by a gynecologic pathologist (C.B.G.) to confirm diagnosis, stage, tumor cell type, and grade before TMA inclusion to ensure that the current diagnostic criteria for subclassification of ovarian cancer based on cell type were uniformly applied.28,29 Samples displaying multiple cell types (mixed tumors) were excluded from the study. Details regarding the cohort used for these TMAs are provided in Table 1 and in Gilks et al.30 Patients were followed up for a median of 4.6 years (0.1–18 years) after the initial surgery.
Table 1.
Subtype, Stage, Silverberg Grade, and Nectin 4 Score of Tissue Microarrays*
| Subtype | Median Age, y (Range) |
Median Preoperative Serum CA125, U/mL (Range) |
Stage | N | Nectin 4 Positive |
Silverberg Grade |
N | Nectin 4 Positive |
Nectin 4 Positive Overall |
|---|---|---|---|---|---|---|---|---|---|
| Serous (n = 212) | 59.6 (33.5–86.0) | 108 (0–23,000) | I | 50 | 29 (58) | 1 | 12 | 4 (33) | 107 (50.5) |
| II | 93 | 47 (51) | 2 | 56 | 30 (54) | ||||
| III | 69 | 31 (45) | 3 | 144 | 73 (50.7) | ||||
| Endometrioid (n = 125) | 54.1 (29.4–88.1) | 130 (8–13,000) | I | 69 | 40 (58) | 1 | 82 | 49 (60) | 72 (57.6) |
| II | 50 | 29 (58) | 2 | 35 | 19 (54) | ||||
| III | 6 | 3 (50) | 3 | 8 | 4 (50) | ||||
| Clear cell (n = 132) | 55.0 (28.1–89.0) | 64 (4–7,750) | I | 68 | 34 (50) | 1 | 0† | 0 (0) | 55 (41.7) |
| II | 56 | 17 (30) | 2 | 0† | 0 (0) | ||||
| III | 8 | 4 (50) | 3 | 132 | 55 (41.7) | ||||
| Mucinous (n = 31) | 56.4 (25.4–76.7) | 45 (7–650) | I | 18 | 6 (33) | 1 | 11 | 2 (18) | 9 (29) |
| II | 12 | 2 (17) | 2 | 18 | 6 (33) | ||||
| III | 1 | 1 (100) | 3 | 2 | 1 (50) | ||||
| Total (n = 500) | 56.6 (25.4–89.0) | 98 (0–23,000) | I | 205 | 109 (53.2) | 1 | 105 | 55 (52.4) | 243 (48.6) |
| II | 211 | 95 (45.0) | 2 | 109 | 55 (50.5) | ||||
| III | 84 | 39 (46) | 3 | 286 | 133 (46.5) |
Data are given as number (percentage) unless otherwise indicated.
All clear cell carcinomas are considered high grade.
Immunohistochemical Staining of Tissues
Slides were dried, and tissue sections were deparaffinized and rehydrated as previously described.26 Staining was performed manually without antigen-retrieval procedures. Endogenous peroxidase activity was blocked by incubation in 0.3% hydrogen peroxide, and then slides were incubated with 10 µg/mL mouse antihuman nectin 4 (clone 337516) or normal mouse IgG diluted in 1:5 Sniper/PBS (Biocare, Concord, CA) overnight at 4°C. Slides were incubated with horseradish peroxidase–conjugated rabbit antimouse polyclonal secondary antibody for 20 minutes. Staining was visualized with Vulcan Fast Red (Biocare), and hematoxylin was used as a counterstain. Slides were dehydrated through a series of ethanol and xylene washes, and then coverslipped with VectaMount (Vector Laboratories, Burlingame, CA).
Tissues were graded on a 4-point scale by a pathologist (S.E.P.) in a blinded manner. Sections with no staining were scored as 0, those with less than 10% of cancer cells staining were scored as +1, tissues with 10% to 50% of the cancer cells staining were scored as +2, and tissues with more than 50% of the cancer cells staining were scored as +3. For most of the analysis, the data were binarized to create 2 groups, in which all positive scores (+1, +2, and +3) were grouped together and compared with the negatively scored tissues.
TMA Statistical Analysis
Differential expression for nectin 4 across the histopathologic subtypes was assessed with the Pearso n χ2 statistic. Univariable relapse-free survival for the entire cohort and each histopathologic subtype was examined with Kaplan-Meier survival curves. Results significant in univariable analysis were subjected to multivariable relapse-free survival using the Cox proportional hazards test. The level of significance for all comparisons was a P value less than .05. All statistical calculations were computed with JMP, version 6.0.3 (SAS Institute, Cary, NC).
Enzyme-Linked Immunosorbent Assay
A sandwich ELISA was used to detect soluble nectin 4 in patients’ serum and ascites samples. Ninety-six well tissue culture plates were coated with 10 µg/mL of mouse antihuman nectin 4 monoclonal antibody (clone N4.61). After blocking the wells with PBS containing 1% bovine serum albumin (R&D Systems), 100 µL of serum or ascites was incubated for 12 hours at 4°C and washed, and then 0.5 µg/mL biotinylated goat antihuman nectin 4 antibody was added. Streptavidin-peroxidase in PBS–bovine serum albumin was incubated for 1 hour at 37°C. Next, 100 µL of peroxidase substrate was added (One Step ABTS, Pierce, Rockford, IL), and optical density was read at 405 nm. Three to five wash steps were performed between incubations with PBS containing 0.5% polysorbate (Tween) 20. Analyses were done in duplicate. The nectin 4 concentration was calculated using serial dilution of recombinant human nectin 4-Fc protein as previously described.14
ELISA Statistical Analysis
Serum nectin 4 levels were compared among serous, clear cell, LMP, and benign gynecologic disease by using the Kruskal-Wallis test. Subsequent pairwise comparisons were conducted using the Wilcoxon rank sum test.
Receiver-operating characteristic (ROC) curves were constructed for nectin 4 alone, CA125 alone, and the combination of CA125 and nectin 4. The Youden index was used to place the optimal threshold point for nectin 4 to distinguish between a serous ovarian cancer mass and benign gynecologic disease on the curve; the corresponding sensitivity and specificity of the test were then determined. Exact P values for the classification table of the test results by the known status were then calculated.
Results
Nectin 4 mRNA Is Overexpressed in Ovarian Cancer
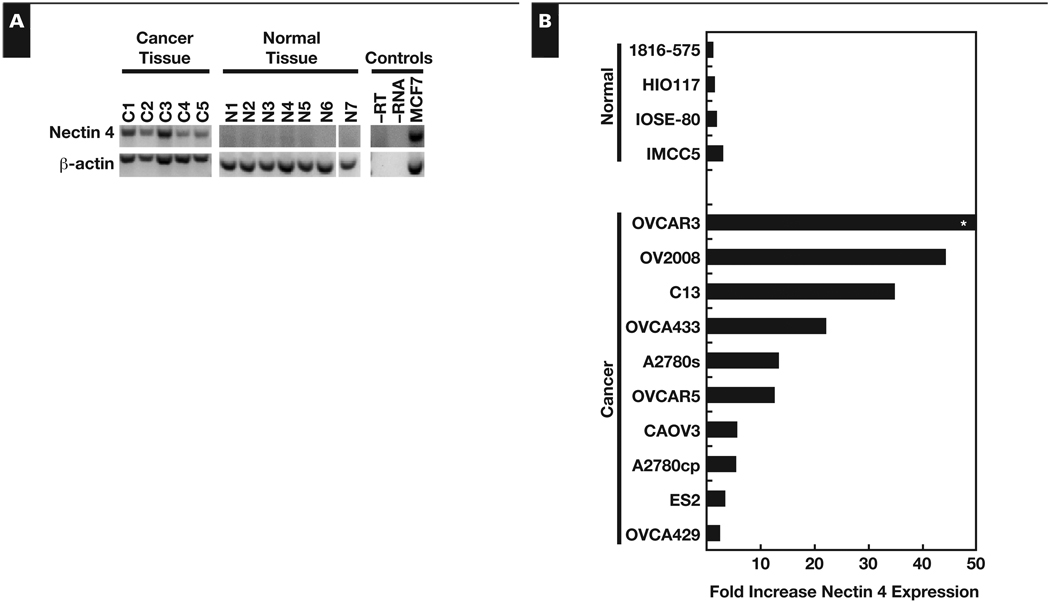
When analyzed by RT-PCR, nectin 4 mRNA was present in all 5 of the serous ovarian cancer tissue samples and absent from all 7 normal ovarian tissues Figure 1A, confirming our previous microarray data.6 Nectin 4 mRNA was detected in 5 of 13 ovarian cancer cell lines, while none of the 8 immortalized NOSE cell lines expressed substantial levels of nectin 4 (data not shown). Expression of nectin 4 in the MCF7 breast cancer cell line was used as a positive control (Figure 1A). Expression of β-actin in the samples confirmed that mRNA was not degraded and that equal amounts of mRNA were loaded (Figure 1A).
Figure 1.
Nectin 4 gene overexpression in ovarian cancer. A, Reverse transcriptase–polymerase chain reaction (RT-PCR) of nectin 4 in ovarian cancer (C1–C5) and normal ovarian tissue (N1–N7). MCF7 cell line, positive control. −RT, no reverse transcriptase; −RNA, no RNA. B, Quantitative RT-PCR analysis of nectin 4 expression in immortalized NOSE and ovarian cancer cell lines. Results expressed as fold-change compared with the lowest expressing cell line (1816-575). *OVCAR3 messenger RNA was increased 454-fold over the lowest expressing cell line.
Quantitative RT-PCR was performed on multiple ovarian cancer and immortalized NOSE cell lines Figure 1B. The mRNA levels were expressed as a fold-increase over the lowest expressing cell line (1816-575). Levels of nectin 4 mRNA averaged 60-fold higher (range, 2.3–453.6) in the ovarian cancer cell lines compared with the 1816-575 cell line. In immortalized NOSE cells, nectin 4 mRNA expression averaged only 1.8-fold higher (range, 1.5–2.8) than the lowest expressing cell line.
Nectin 4 Protein Is Overexpressed in Ovarian Cancer
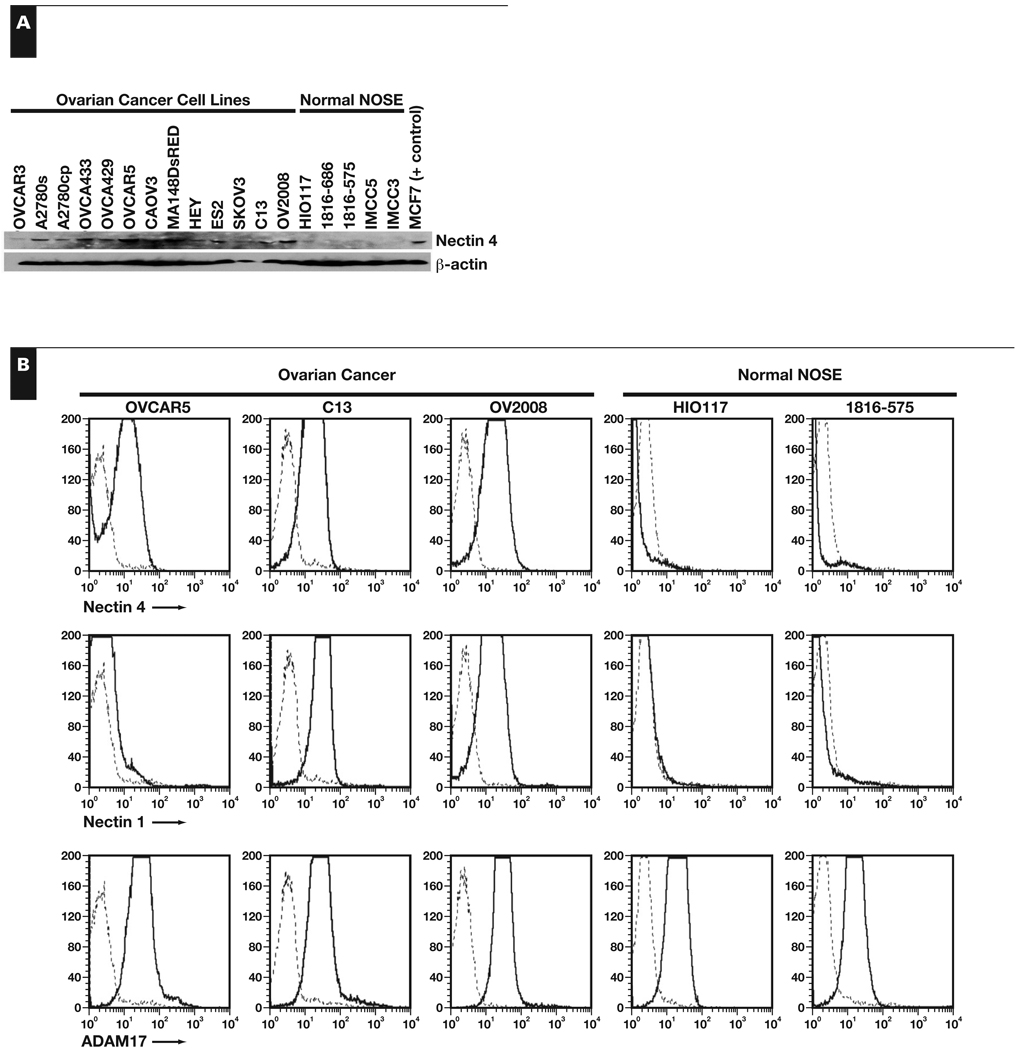
Nectin 4 protein levels in ovarian cancer cell lines were compared with immortalized NOSE cell lines by Western immunoblotting and flow cytometric analysis. A band of approximately 66 kDa corresponding to nectin 4 was present in the positive control MCF7 cell extracts Figure 2A. Nectin 4 protein was visibly expressed by all 13 of the ovarian cancer cell lines tested but absent or expressed at very low levels in the 5 NOSE cell lines tested (Figure 2A). By flow cytometric analysis, nectin 4 protein was detected on the surface of OVCAR5, C13, and OV2008 ovarian cancer cells, but not on the immortalized NOSE cell lines HIO117 or 1816-575 Figure 2B, confirming our RT-PCR and Western immunoblotting results.
Figure 2.
Nectin 4 protein expression in ovarian cancer. A, Western immunoblotting of ovarian cancer cell lines and immortalized normal ovarian surface epithelial (NOSE) cell lines. Nectin 4 appears as an approximately 66-kDa band. β-Actin was used as a loading control. B, Flow cytometric analysis of ovarian cancer and immortalized NOSE cell lines for nectin 4 (top row), nectin 1 (middle row), and ADAM17 (bottom row). Nectin 1 expression indicates that trans-heterodimers may form between nectin 4– and nectin 1–expressing cells. Expression of ADAM17 indicates that nectin 4 may be cleaved from the surface of the cells and may be detectable in patient serum samples. Dashed line, IgG control stain; solid line, nectin 4, nectin 1, or ADAM17.
We also investigated the expression of nectin 1 and ADAM17 on these cell lines by flow cytometry. Nectin 1, which can trans-dimerize with nectin 4 on adjacent cells, was present on 2 of the 3 ovarian cancer cell lines tested, but not on either of the immortalized NOSE cell lines (Figure 2B). ADAM17, a metalloproteinase that can cleave the ectodomain of nectin 4 from the surface of the cell, releasing a soluble fragment, was detected on all of the cell lines tested. These results suggest that nectin 4 may be released into the blood of patients with ovarian cancer and, thus, may serve as an accessible biomarker for the disease.
Nectin 4 Is Expressed in the Tissues of Patients With Ovarian Cancer
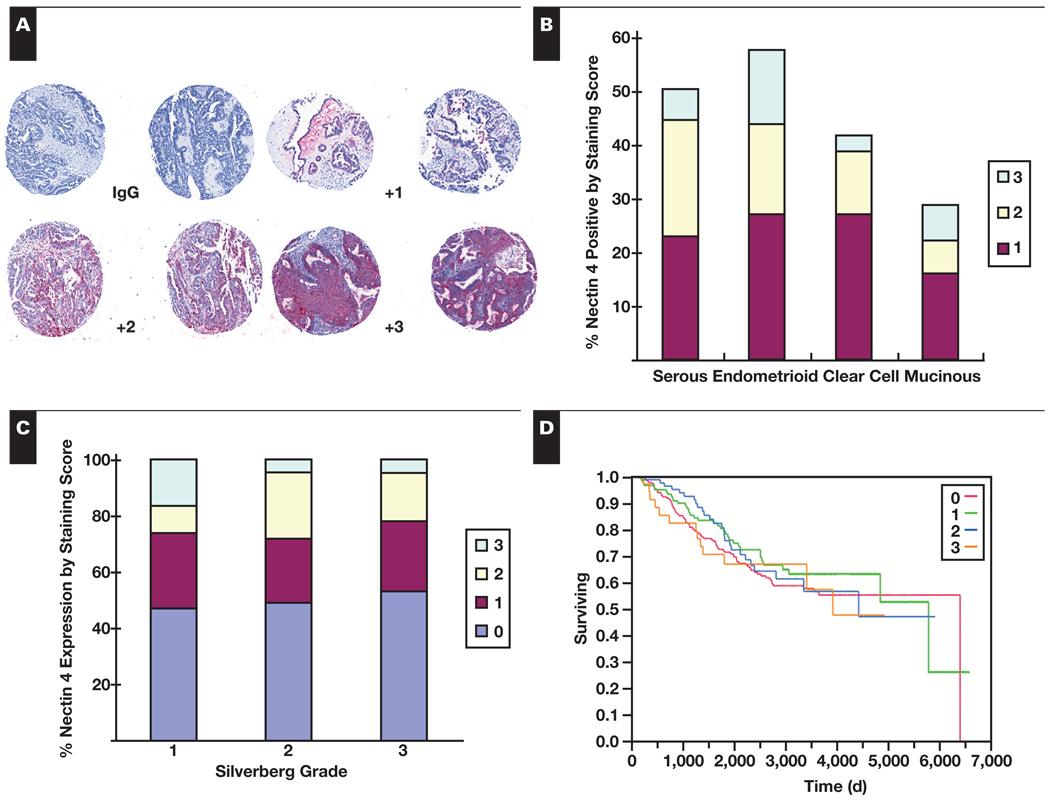
Immunohistochemical staining of TMAs allowed for simultaneous analysis of 500 patient specimens Figure 3A. Overall, 48.6% of the tissues in the TMA stained positively for nectin 4. Staining was uniformly cytoplasmic with no nuclear staining observed. Nectin 4 protein expression was differentially expressed (P = .008) across the histologic subgroups of ovarian cancer, with the highest expression in endometrioid carcinomas (57.6% of tumors positive), followed by serous (50.5%), clear cell (41.7%), and mucinous (29.0%) Figure 3B (Table 1).
Figure 3.
Immunohistochemical staining for nectin 4 of ovarian cancer tissue microarrays. A, Staining and grading examples of tissue microarray (TMA) samples. IgG, control primary stain. +1, <10% of cancer cells staining; +2, 10%–50% of cancer cells staining; and +3, >50% of cancer cells staining. B, Percentage of TMA cases positive for nectin 4 by ovarian cancer subtype. C, Percentage of TMA cases positive for nectin 4 by Silverberg grade. D, Kaplan-Meier survival curve of TMA patients measured in days. Red, staining score 0; green, staining score +1; blue, staining score +2; orange, staining score +3.
Silverberg grade 1 tumors showed a significantly higher percentage of intense nectin 4 staining (ie, 16% of the grade 1 tumors received scores of +3 compared with only 5% of the grade 2 or grade 3 tumors; P = .0026) when positive scores were not binarized Figure 3C. However, when binarized as “any staining” vs “no staining,” Silverberg grade 1, 2, and 3 tumors each were approximately 50% positive for nectin 4. Nectin 4 expression was not statistically different for disease stage for any histologic subgroups of ovarian cancer when positive scores were binarized (Table 1). Overall, expression of nectin 4 did not alter relapse-free survival (not shown) or overall survival Figure 3D, regardless of subtype, stage, or grade.
Nectin 4 Is Elevated in the Serum of Patients With Ovarian Cancer
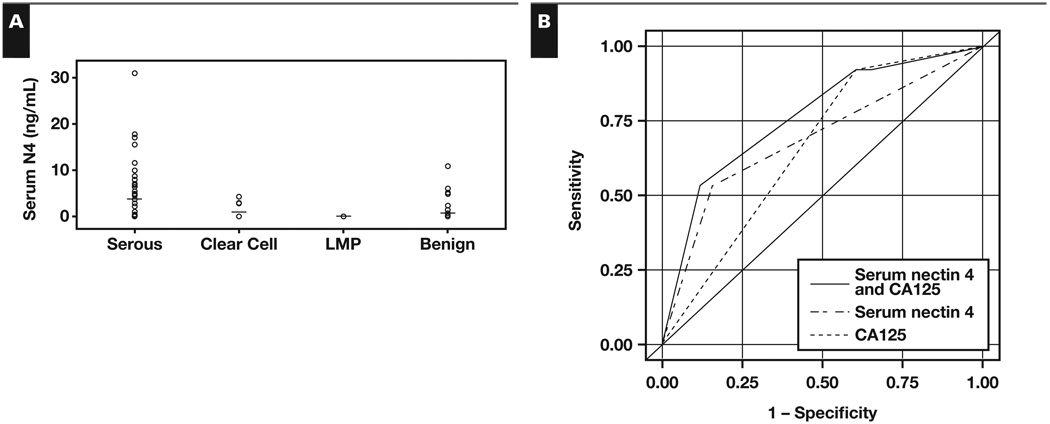
Serum samples from 134 women were analyzed by ELISA to quantify cleaved nectin 4. ROC curves were constructed, and the Youden index was used to place the optimal threshold value to distinguish between serous ovarian cancer and benign samples. The threshold value for serum nectin 4 levels was 0 ng/mL; any samples above 0 ng/mL were considered positive and indicative of serous ovarian cancer. Nectin 4 was detected in serum from 27 (53%) of 51 patients with serous ovarian cancer, 4 (29%) of 14 with clear cell cancer, and 8 (16%) of 51 with benign gynecologic diseases. It is interesting that none of the serum samples from patients with LMP tumors demonstrated elevated protein levels. The mean concentration of serum nectin 4 was significantly higher in patients with serous ovarian cancer (3.7 ng/mL) compared with patients with benign disease (0.6 ng/mL) and LMP tumors (0 ng/mL) Figure 4A and Table 2 (P = .001) but not in patients with clear cell cancer (0.9 ng/mL). Nectin 4 levels were significantly different among the groups (P < .001) using the Kruskal-Wallis test. Subsequent comparisons using the Wilcoxon rank sum test determined the difference detected was between the serous and both the benign groups and LMP groups.
Figure 4.
Nectin 4 enzyme-linked immunosorbent assay analysis. A, Serum nectin 4 levels (ng/mL) separated by diagnosis. B, Receiver operating characteristic (ROC) curves for nectin 4 alone (dash-dot line), CA125 alone (dashed line), or nectin 4 and CA125 combined (solid line). Area under the ROC curve for nectin 4 and CA125, 0.7620; for nectin 4, 0.0863; and for CA125, 0.6569.
Table 2.
Nectin 4 Enzyme-Linked Immunosorbent Assay Analysis of Patient Serum Samples
| Diagnosis | No. Positive |
Mean Nectin 4 (ng/mL) |
Mean CA125 (U/mL) |
Specificity Sensitivity (%) |
PPV (%) |
NPV (%) |
(%) |
|---|---|---|---|---|---|---|---|
| Serous (n = 51) | 3.699 (0–31.0) | 2,246.2 (6–22,780) | |||||
| Nectin 4 | 27 | 53 | — | 77 | — | ||
| CA125 | 47 | 92 | — | 60 | — | ||
| Nectin 4 and CA125 | 27 | 53 | — | 82 | — | ||
| Clear cell (n = 14) | 0.944 (0–4.23) | 222.6 (13-1,504) | |||||
| Nectin 4 | 4 | 29 | — | 33 | — | ||
| CA125 | 11 | 79 | — | 26 | — | ||
| Nectin 4 and CA125 | 3 | 21 | — | 33 | — | ||
| LMP (n = 18) | 0 (0-0) | 264.6 (6.6-1,539) | |||||
| Nectin 4 | 0 | — | 100 | — | 43 | ||
| CA125 | 14 | — | 22 | — | 50 | ||
| Nectin 4 and CA125 | 0 | — | 100 | — | 43 | ||
| Benign (n = 51) | 0.629 (0–10.83) | 151.4 (4-1,984) | |||||
| Nectin 4 | 8 | — | 84 | — | 64 | ||
| CA125 | 31 | — | 39 | — | 83 | ||
| Nectin 4 and CA125 | 6 | — | 88 | — | 65 |
LMP, low malignant potential; NPV, negative predictive value; PPV, positive predictive value.
When 0 ng/mL was used as a threshold value, the area under the curve (AUC) in ROC analysis for serous samples was 68.6% Figure 4B (P = .0001). Nectin 4 was 53% sensitive and had a positive predictive value (PPV) of 77% for serous ovarian cancer and was 84% specific and had a negative predictive value (NPV) of 64% for benign gynecologic disease (Table 2). No associations between stage (P = .44) or grade (P = .38) and nectin 4 concentration were detected.
In the same serous samples, the threshold value for CA125 was 30 U/mL per standard protocol, and the AUC of ROC analysis for CA125 was 66% (Figure 4B; P = .0003). While CA125 was more sensitive (92%) in detecting serous ovarian cancer, specificity was reduced greatly, at 39% (Table 2). The PPV and NPV for CA125 were 60% and 83%, respectively (Table 2). When nectin 4 and CA125 were combined, the AUC for ROC analysis increased to 76% (Figure 4B; P = .0013), with a 53% sensitivity and 88% specificity (Table 2). The PPV of the combined markers was 82% and the NPV was 65% (Table 2).
Nectin 4 was detected in all of the serous ovarian cancer ascites fluid samples tested (mean, 158 ng/mL; range, 13.7–270.2 ng/mL). Each of the 10 ascites fluid samples had been procured from patients whose corresponding serum samples were also tested by ELISA. It is interesting that only 6 of the 10 serum samples had detectable levels of nectin 4 (range, 0–31.0 ng/mL). The concentration of nectin 4 was significantly higher in each patient’s ascites fluid compared with the serum sample, suggesting that the soluble form of nectin 4 that is being detected by this ELISA may be degraded or cleared from the blood more readily than from the ascites fluid.
Discussion
In this study, we have demonstrated increased mRNA and protein expression of nectin 4 in ovarian cancer tissues and cell lines compared with their normal ovarian counterparts using several techniques. These results provide the first validation of our previously published gene microarray data.6 Immunohistochemical staining of TMAs allowed for the study of 500 patients’ tissues at one time; overall, 48.6% of the tissues on the TMA stained positively for nectin 4. Low-grade (Silverberg grade 1) tumors had a greater percentage of intensely positive samples (Figure 3C). This trend toward more intense staining and increased expression in low-grade cancers may indicate the feasibility of using nectin 4 to help predict the aggressiveness of ovarian cancer at early time points.
We have also demonstrated that the cleaved ectodomain of nectin 4 can be detected in the serum samples of patients with ovarian cancer. By ELISA, nectin 4 was detected in the serum of 53% of the serous ovarian cancer cases tested. It is interesting that this correlates closely with the TMA results for 50% of serous samples staining positively for nectin 4, indicating that the ELISA may be able to detect all or nearly all positive samples. Using nectin 4 in conjunction with CA125 for the diagnosis of ovarian cancer increases the PPV and specificity compared with CA125 alone, which may help in the triage of patients with a “suspicious” pelvic mass, as recently described with 4 other biomarkers.31 For example, women with elevated serum levels of CA125 could next be tested for elevated serum levels of nectin 4; this could help discriminate ovarian cancer from benign gynecologic disease. This would allow appropriate triage of women with pelvic masses; ie, women with ovarian cancer would benefit from a gynecologic-oncology surgeon performing the primary surgery, while patients with benign disease could be served by an obstetric-gynecologic surgeon with no impact on patient outcome. One caveat, however, is that ELISA detection of nectin 4 may not distinguish between primary ovarian cancers and other primary cancers (eg, breast or lung) that have metastasized to the pelvic region. Future studies are planned to determine the usefulness of nectin 4 as a biomarker for ovarian cancer by testing serum samples from a large cohort of the general population and from patients with a known abdominal mass.
The role of nectin 4 in the development and propagation of ovarian cancer is unknown. The recent discovery of nectin 4 in subsets of breast and lung cancer, along with demonstration that serum levels of nectin 4 correlated with the number of metastases and therapeutic efficacy in breast cancer, suggests that nectin 4 may have a conserved role in cancer biology.17,18
We identified significantly more patients with ovarian cancer with elevated serum levels of nectin 4 than had previously been reported (53% vs 4% by Fabre-Lafay et al18). The reason for the difference in detection rates is unclear, but case selection and detection bias based on the ELISA technique are likely impacting factors. Among the ovarian cancer serum samples tested by Fabre-Lafay et al,18 only 9 were of the serous subtype, and these included 3 low-grade tumors. In addition, we used a different detection antibody for our ELISA, which was more sensitive compared with the previously published ELISA method (data not shown).
Nectin 4 has also been detected in the serum of patients with non–small cell lung cancer (NSCLC).17 Takano et al17 developed an ELISA using different monoclonal antibodies against nectin 4 and established a threshold value of 1.0 ng/mL for serum nectin 4 in a panel of healthy people and patients with chronic obstructive pulmonary disease or NSCLC. They found that high serum nectin 4 levels correlated with poor survival in the patients with NSCLC. In addition, expression of nectin 4 increased lamellipodia formation, resulting in an increase in invasive potential in lung cancer cell lines, while short interfering RNAs directed against nectin 4 suppressed cell growth.
Besides modulating growth and invasive potential, expression of nectin 4 may increase the survival of ovarian cancer cells when treated with chemotherapeutic agents. Ovarian cancer cells often form spheroids that appear more resistant to chemotherapy; nectin 4 is important in cell-cell adhesion, and spheroids formed from nectin 4–expressing cells may be better able to exclude chemotherapy agents from the cells in the center of the spheroid, promoting resistance. Thus, in addition to determining expression of nectin 4, we also investigated expression of nectin 1 on ovarian cancer cells and immortalized NOSE cells. Nectin 1 was present on 2 of the 3 ovarian cancer cell lines tested. Nectin 4 interacts in trans with nectin 1 or nectin 4 on adjacent cells. Trans-heterophilic interactions between nectin 1 and nectin 4 are stronger than trans-homophilic interaction; ovarian cancer cells expressing both nectin 1 and nectin 4 may be able to form compact, tight spheroids that aid in survival due to decreased chemosensitivity.32,33
Why nectin 4 is shed from the surface of cells remains unknown, and functionality studies were beyond the scope of this study. However, given the apparent relationship between nectin 4 expression and metastases in breast and lung cancers, multiple hypotheses can be considered. It is possible that increased expression of nectin 4 contributes to the formation of ovarian cancer spheroids, which in turn have been demonstrated to be relatively resistant to chemotherapy. Reversal of this phenotype may occur once the cells come in contact with an appropriate metastatic site, whereby nectin 4 may be cleaved from the surface, allowing for the cells to better attach to and invade the site. In this study, ADAM17, a metalloproteinase that cleaves the ectodomain of nectin 4 from the cell surface, was detected on ovarian cancer cell lines. The presence of ADAM17 on the ovarian cancer cell surface increases the possibility that nectin 4 may be shed from the cell surface and then be detectable in the ascites fluid or blood of patients with ovarian cancer.
We have described the first comprehensive study of the expression of nectin 4 in ovarian cancer. Nectin 4 is overexpressed at the mRNA and protein levels in ovarian cancers and rarely overexpressed in healthy nonpregnant patients, suggesting potential use as a serum biomarker.13 As with most biomarkers, nectin 4 was not able to detect all ovarian cancers. However, several patients with benign gynecologic disease and high CA125 levels had no detectable levels of serum nectin 4, leading to an increase in specificity over CA125 alone. This finding indicates that nectin 4 may help discriminate ovarian cancer from benign disease when used in conjunction with CA125 in a panel for diagnosis. Identification of a complementary biomarker detecting nectin 4–negative tumors could greatly increase the sensitivity, resulting in improved diagnosis. Functional studies are needed to fully determine the impact of nectin 4 expression on ovarian cancer progression.
Acknowledgments
We thank Kristin Boylan, PhD, University of Minnesota, Minneapolis, for helpful suggestions and review of the manuscript; Colleen Forster, University of Minnesota, for help with immunohistochemical staining protocols; Kaylee Schwertfeger, PhD, and Haojie Huang, PhD, University of Minnesota, for use of laboratory equipment; the Cheryl Brown Ovarian Cancer Outcomes Unit for providing the TMAs; Sarah Bowell, Diane Rauch, Marissa Makey, and Carter Schmidt, University of Minnesota Tissue Procurement Facility, for procuring patient serum and ascites samples; and the University of Minnesota Flow Cytometry Core.
Supported by grant R01-CA106878 from the National Institutes of Health/National Cancer Institute, Bethesda, MD; and by the Minnesota Ovarian Cancer Alliance, Minneapolis, and CanCurables Foundation, Edina, MN.
References
- 1.Boyle P, Levin B. World Cancer Report 2008. Lyon, France: International Agency for Research on Cancer; 2008. [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 4.Verheijen RHM, von Mensdorff-Pouilly S, van Kamp GJ, et al. CA 125: fundamental and clinical aspects. Semin Cancer Biol. 1999;9:117–124. doi: 10.1006/scbi.1998.0114. [DOI] [PubMed] [Google Scholar]
- 5.Bast RC, Jr, Urban N, Shridhar V, et al. Early detection of ovarian cancer: promise and reality. Cancer Treat Res. 2002;107:61–97. doi: 10.1007/978-1-4757-3587-1_3. [DOI] [PubMed] [Google Scholar]
- 6.Hibbs K, Skubitz KM, Pambuccian SE, et al. Differential gene expression in ovarian carcinoma: identification of potential biomarkers. Am J Pathol. 2004;165:397–414. doi: 10.1016/S0002-9440(10)63306-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sakisaka T, Takai Y. Biology and pathology of nectins and nectin-like molecules. Curr Opin Cell Biol. 2004;16:513–521. doi: 10.1016/j.ceb.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 8.Sakisaka T, Ikeda W, Ogita H, et al. The roles of nectins in cell adhesions: cooperation with other cell adhesion molecules and growth factor receptors. Curr Opin Cell Biol. 2007;19:593–602. doi: 10.1016/j.ceb.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 9.Yasumi M, Shimizu K, Honda T, et al. Role of each immunoglobulin-like loop of nectin for its cell-cell adhesion activity. Biochem Biophys Res Commun. 2003;302:61–66. doi: 10.1016/s0006-291x(03)00106-2. [DOI] [PubMed] [Google Scholar]
- 10.Yamada A, Fujita N, Sato T, et al. Requirement of nectin, but not cadherin, for formation of claudin-based tight junctions in annexin II-knockdown MDCK cells. Oncogene. 2006;25:5085–5102. doi: 10.1038/sj.onc.1209525. [DOI] [PubMed] [Google Scholar]
- 11.Irie K, Shimizu K, Sakisaka T, et al. Roles and modes of action of nectins in cell-cell adhesion. Semin Cell Dev Biol. 2004;15:643–656. doi: 10.1016/j.semcdb.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 12.Fabre S, Reymond N, Cocchi F, et al. Prominent role of the Ig-like V domain in trans-interactions of nectins: nectin3 and nectin4 bind to the predicted C-C'-C''-D beta-strands of the nectin1 V domain. J Biol Chem. 2002;277:27006–27013. doi: 10.1074/jbc.M203228200. [DOI] [PubMed] [Google Scholar]
- 13.Reymond N, Fabre S, Lecocq E, et al. Nectin4/PRR4, a new afadin-associated member of the nectin family that transinteracts with nectin1/PRR1 through V domain interaction. J Biol Chem. 2001;276:43205–43215. doi: 10.1074/jbc.M103810200. [DOI] [PubMed] [Google Scholar]
- 14.Nakanishi H, Takai Y. Roles of nectins in cell adhesion, migration and polarization. Biol Chem. 2004;385:885–892. doi: 10.1515/BC.2004.116. [DOI] [PubMed] [Google Scholar]
- 15.Reymond N, Borg J-P, Lecocq E, et al. Human nectin3/PRR3: a novel member of the PVR/PRR/nectin family that interacts with afadin. Gene. 2000;255:347–355. doi: 10.1016/s0378-1119(00)00316-4. [DOI] [PubMed] [Google Scholar]
- 16.Fabre-Lafay S, Garrido-Urbani S, Reymond N, et al. Nectin-4, a new serological breast cancer marker, is a substrate for tumor necrosis factor-α-converting enzyme (TACE)/ADAM-17. J Biol Chem. 2005;280:19543–19550. doi: 10.1074/jbc.M410943200. [DOI] [PubMed] [Google Scholar]
- 17.Takano A, Ishikawa N, Nishino R, et al. Identification of nectin-4 oncoprotein as a diagnostic and therapeutic target for lung cancer. Cancer Res. 2009;69:6694–6703. doi: 10.1158/0008-5472.CAN-09-0016. [DOI] [PubMed] [Google Scholar]
- 18.Fabre-Lafay S, Monville F, Garrido-Urbani S, et al. Nectin-4 is a new histological and serological tumor associated marker for breast cancer. BMC Cancer. 2007;7:73–89. doi: 10.1186/1471-2407-7-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaw TJ, Senterman MK, Dawson K, et al. Characterization of intraperitoneal, orthotopic, and metastatic xenograft models of human ovarian cancer. Mol Ther. 2004;10:1032–1042. doi: 10.1016/j.ymthe.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 20.Burleson KM, Hansen LK, Skubitz APN. Ovarian carcinoma spheroids disaggregate on type I collagen and invade live human mesothelial cell monolayers. Clin Exp Metastasis. 2004;21:685–697. doi: 10.1007/s10585-004-5768-5. [DOI] [PubMed] [Google Scholar]
- 21.Skubitz APN, Campbell KD, Goueli S, et al. Association of β1 integrin with protein kinase activity in large detergent resistant complexes. FEBS Lett. 1998;426:386–391. doi: 10.1016/s0014-5793(98)00380-9. [DOI] [PubMed] [Google Scholar]
- 22.Subramanian IV, Bui Nguyen TM, Truskinovsky AM, et al. Adeno-associated virus-mediated delivery of a mutant endostatin in combination with carboplatin treatment inhibits orthotopic growth of ovarian cancer and improves long-term survival. Cancer Res. 2006;66:4319–4328. doi: 10.1158/0008-5472.CAN-05-3297. [DOI] [PubMed] [Google Scholar]
- 23.Nicosia SV, Wilbanks GD, Saunders B, et al. Cytology of human ovarian surface epithelial brushings. Cancer. 2004;102:1–10. doi: 10.1002/cncr.20001. [DOI] [PubMed] [Google Scholar]
- 24.Kruk PK, Maines-Bandiera S, Auersperg N. A simplified method to culture human ovarian surface epithelium. Lab Invest. 1990;63:132–136. [PubMed] [Google Scholar]
- 25.Fronhoffs S, Totzke G, Stier S, et al. A method for the rapid construction of cRNA standard curves in quantitative real-time reverse transcription polymerase chain reaction. Mol Cell Probes. 2002;16:99–110. doi: 10.1006/mcpr.2002.0405. [DOI] [PubMed] [Google Scholar]
- 26.DeRycke MS, Andersen JD, Harrington KM, et al. S100A1 expression in ovarian and endometrial endometrioid carcinomas is a prognostic indicator of relapse-free survival. Am J Clin Pathol. 2009;132:846–856. doi: 10.1309/AJCPTK87EMMIKPFS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andersen JD, Boylan KLM, Xue FS, et al. Identification of candidate biomarkers in ovarian cancer serum by depletion of highly abundant proteins and differential in-gel electrophoresis. Electrophoresis. 2010;31:599–610. doi: 10.1002/elps.200900441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soslow RA. Histologic subtypes of ovarian carcinoma: an overview. Int J Gynecol Pathol. 2008;27:161–174. doi: 10.1097/PGP.0b013e31815ea812. [DOI] [PubMed] [Google Scholar]
- 29.McCluggage WG. My approach to and thoughts on the typing of ovarian carcinomas. J Clin Pathol. 2008;61:152–163. doi: 10.1136/jcp.2007.049478. [DOI] [PubMed] [Google Scholar]
- 30.Gilks CB, Ionescu DN, Kalloger SE, et al. Tumor cell type can be reproducibly diagnosed and is of independent prognostic significance in patients with maximally debulked ovarian carcinoma. Hum Pathol. 2008;39:1239–1251. doi: 10.1016/j.humpath.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 31.Fung ET. A recipe for proteomics diagnostic test development: the OVA1 Test, from biomarker discovery to FDA clearance. Clin Chem. 2010;56:327–329. doi: 10.1373/clinchem.2009.140855. [DOI] [PubMed] [Google Scholar]
- 32.Herrmann R, Fayad W, Schwarz S, et al. Screening for compounds that induce apoptosis of cancer cells grown as multicellular spheroids. J Biomol Screen. 2008;13:1–8. doi: 10.1177/1087057107310442. [DOI] [PubMed] [Google Scholar]
- 33.Shield K, Ackland ML, Ahmed N, et al. Multicellular spheroids in ovarian cancer metastases: biology and pathology. Gynecol Oncol. 2009;113:143–148. doi: 10.1016/j.ygyno.2008.11.032. [DOI] [PubMed] [Google Scholar]