Summary
We applied whole cell electron cryotomography to the archaeon Sulfolobus infected by Sulfolobus turreted icosahedral virus (STIV), which belongs to the PRD1-Adeno lineage of dsDNA viruses. STIV infection induced the formation of pyramid-like protrusions with sharply defined facets on the cell surface. They had a thicker cross-section than the cytoplasmic membrane and did not contain an exterior surface protein layer (S-layer). Intra-pyramidal bodies often occupied the volume of the pyramids. Mature virions, procapsids without genome cores, and partially assembled particles were identified, suggesting that the capsid and inner membrane co-assemble in the cytoplasm to form a procapsid. A two-class reconstruction using a maximum likelihood algorithm demonstrated that no dramatic capsid transformation occurred upon DNA packaging. Virions tended to form tightly packed clusters or quasi-crystalline arrays while procapsids mostly scattered outside or on the edges of the clusters. The study revealed vivid images of STIV assembly, maturation and particle distribution in cell.
Keywords: Electron cryotomography (ECT), Sulfolobus turreted icosahedral virus (STIV), in vivo virus assembly, Sulfolobus, inner-membrane containing viruses
Introduction
To gain mechanistic understanding of virus assembly, it is important to follow those events in the context of the host cell. Electron cryotomography (ECT) is an emerging technique that allows such processes to be imaged in 3-D with "macromolecular" (4–6 nm) resolution (Grunewald and Cyrklaff, 2006; Koster et al., 1997; Lucic et al., 2005). Briefly, cells are preserved in close-to-native states by plunge-freezing (Dubochet et al., 1988; Iancu et al., 2006), a tilt series of projection images are recorded while the sample is kept frozen, and then a 3-D reconstruction of the object is calculated by back-projection or other algorithms (Jensen and Briegel, 2007; Lucic et al., 2005).
Here we applied ECT to the intact archaea Sulfolobus solfataricus cells infected with Sulfolobus turreted icosahedral viruses (STIV). STIV belongs to the PRD1-Adeno lineage of dsDNA viruses as demonstrated by the structural similarity of the major capsid protein and the overall capsid architecture in this lineage (Khayat et al., 2005). The icosahedral capsid is 74 nm in diameter, has a pseudo T=31d quasi-equivalent surface lattice and an inner membrane that encloses a 17.3 kbp circular dsDNA genome. At the twelve five-fold vertices are turret-like appendages extending 13 nm above the capsid shell (Rice et al., 2004). Mass spectrometry studies of purified virions suggest that the inner membrane contains several viral gene products and shows the presence of acidic tetraether lipids corresponding to an enrichment of a subpopulation of lipid found in the host (Maaty et al., 2006).
The STIV replication cycle can be followed using a near synchronous, single cycle infection of S. solfataricus (strain P2) which grows optimally at 80°C and pH ~3 (Ortmann et al., 2006). S. solfataricus has an oval shape about 1 µm in diameter. Exterior to the cytoplasmic membrane is a surface protein layer (S-layer) that is formed by a single glycosylated protein (Engelhardt and Peters, 1998; Grimm et al., 1998). Microarray analysis and transmission EM (TEM) of thin sections demonstrated that the transcription of the STIV genome peaks at around 24 hr post-infection (hpi) and cells containing assembled viral particles become dominate at around 32 hpi (Brumfield et al., 2009; Ortmann et al., 2006). TEM and scanning EM (SEM) analysis of cells infected by STIV show pyramid-like structural protrusions from the cell surfaces (Brumfield et al., 2009). These protrusions appear to be the sites where infected cells lyse at the late infection stage in order to release virions (Brumfield et al., 2009). Some viral particles shown in TEM did not appear to contain genomes and were proposed to be procapsids. Although possible artifacts, caused by negative stain sample preparation, could not be ruled out, the results suggested the existence of a procapsid prior to the formation of a genome containing virion (Brumfield et al., 2009).
We plunge-froze S. solfataricus at different time points post-infection. The whole cell tomograms showed STIV particles in different states of assembly. A maximum likelihood (ML) algorithm was developed to automatically identify, classify and reconstruct two classes of viral particles within the cell. The reconstructions were used as models to identify and orient particles in other tomograms based on a template matching approach (Frangakis et al., 2002). This procedure made it feasible to robustly determine STIV particle positions and orientations in large numbers of tomograms and revealed vivid images of virus maturation and their distribution in the cell over the time course of infection.
Results
The Ultrastructure of STIV Infected Sulfolobus
To capture viral particle assembly in vivo and characterize related cellular changes, cells were plunge-frozen at various stages post-infection in the original culture medium to preserve the ultrastructures as they existed at the moment of vitrification. The pyramid-like protrusions on cell surfaces are a characteristic feature of STIV infected cells. Tilt series of 2d projections were acquired and back-projected to reconstruct 3-D volumes of whole cells. A 20 nm slice of a reconstructed cell at 32 hpi is shown in Fig 1A. While Sulfolobus cells are quite thick (0.5–1 µm) for ECT, nevertheless the outer S-layer and inner cytoplasmic membrane of the cell were clearly defined, constituting a cell wall approximately 45 nm in width. A tomographic slice of a typical uninfected cell is shown in Fig S1.
Fig. 1. Whole cell ECT of S. solfataricus infected with STIV.
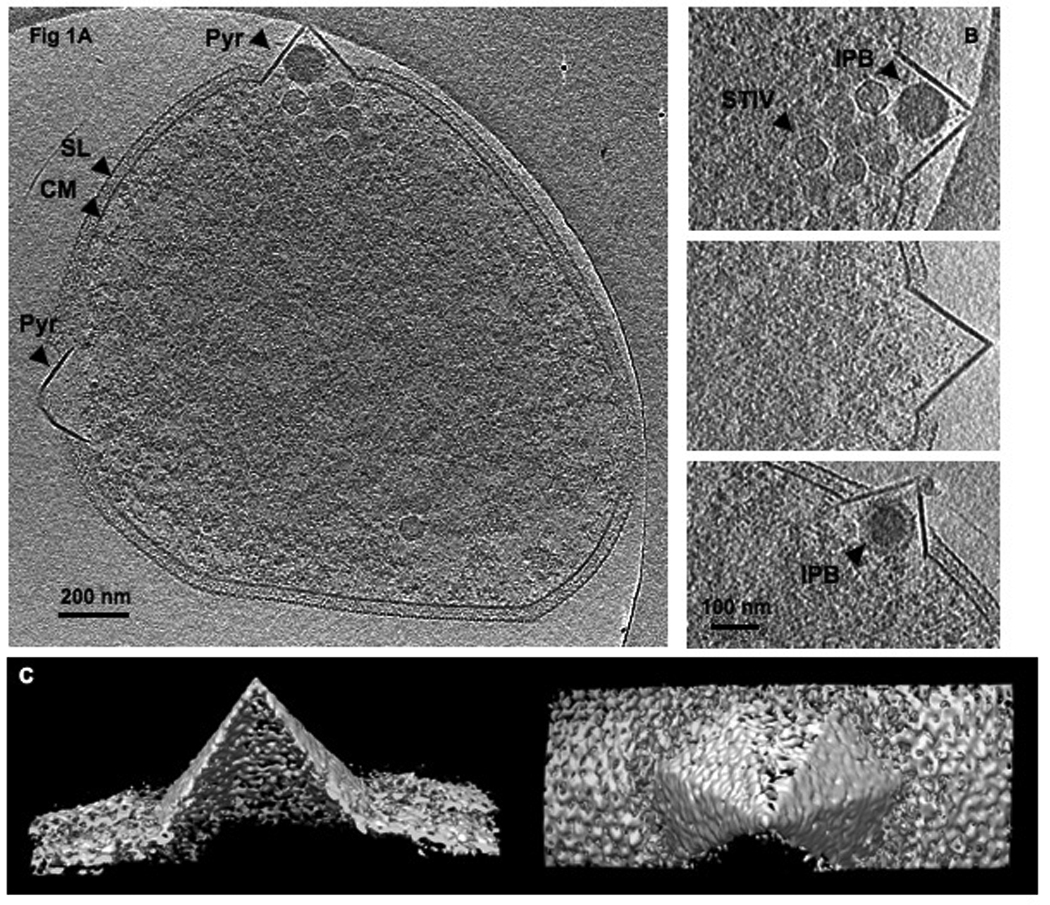

(A) A 20 nm slice of a 3-D tomogram, computationally sectioned perpendicular to the direction of beam. (B) Enlarged views of some representative pyramids. (C) Surface representations of a pyramid viewed from the side and the top of the structure. (D) Enlarged views of some pyramids at early stages of formation. (E) A model of pyramid formation. A pyramid forms by either mechanically protruding and/or enzymatically digesting through the S-layer. The S-layer structure is perturbed and finally detaches from pyramid membrane. Specific proteins and lipids are recruited as a pyramid builds. SL, S-layer; CM, cytoplasmic membrane; Pyr, pyramid like protrusion; STIV, STIV particles; IPB, intra-pyramidal body. Scale bar, 200 nm (A), 100 nm (B,D).
The pyramid protrusions were not covered by the S-layer and appeared thicker in cross-section than the cytoplasmic membrane, suggesting that the pyramids have a protein and/or lipid composition that differs from the cellular membrane (Fig 1B)(Quax et al.). Some pyramid bases seemed to extend inside the cytoplasmic membrane when viewed in cross section. 3-D isosurfaces showed that the facets of the pyramids were sharply defined (Fig 1C, note that the weak and missing facets are a well-understood artifact of the missing "wedge" of data that cannot be acquired from planar samples). In the images taken by SEM, pyramid facets collapse and the pyramids exhibit star-shaped cross-sections (Brumfield et al., 2009) which were not observed our study by ECT. The difference in the two types of imaging indicates that strong scaffolding may exist at the edges between facets while the faces themselves collapsed in the SEM due to the harsh dehydration and staining procedures. Pyramids started to form before viral particles became visible in the cytoplasm as seen in the samples at 22 hpi & 24 hpi. Fig 1D shows some pyramids in the process of protruding out of a thinning cell wall and perturbing the S-layer. Electron dense bodies with diameters of 90–110 nm were often found inside the pyramids and are referred to as intra-pyramidal bodies (IPBs) (Fig 1A,B). Electron dense bodies were also observed in random locations in uninfected cells; however, they were almost always associated with pyramids in infected cells. Some IPBs appeared to have extra layers of material wrapped around them that were less dense than the cores (Fig 1B, bottom panel). The compositions and functions of IBPs and whether they are different from those observed in the uninfected cells remain to be characterized.
Assembly and Maturation of STIV Observed in vivo
Particles with icosahedral shape and dimension similar to STIV could be clearly identified in the cytoplasm (Fig 1A). Full and empty particles were readily distinguished (Fig 2A, B). The radial density plots illustrated the differences in internal density in full and empty particles (approximately 0–15 nm radius from the centers), which corresponded to the packed DNA genome as characterized by single particle reconstruction of STIV virions (Fig S2, 3D) (Rice et al., 2004). Both particle types had an outer capsid shell (~ 37.5 nm in outer radius) and an inner lipid membrane (~ 25 nm in outer radius), comparable to what was seen in the single particle reconstruction (Fig S2). These observations confirm that the STIV life cycle involves a procapsid (empty particles) where the capsid and membrane are assembled prior to genome packaging.
Fig. 2. STIV virions, procapsids, and partially assembled particles observed in vivo.

Sub-tomographic slices displaying STIV virions (A) and procapsids (B) and partially assembled particles (C) that contain parts of capsid and membrane viewed perpendicular to the direction of the beam (x-y plane). Scale bar, 100 nm (C)
See also Figure S2
Fig. 3. The reconstructed density of the two classes of STIV particles determined by the ML algorithm.

The reconstructions of a virion (A) and a procapsid (B) determined with 123 particle-containing subvolumes of the cellular tomogram. (C) The surface representation of the single molecule reconstruction of purified virions. (D) The radial density plots of the sub-tomographic reconstructions of the virion (blue), procapsid (red) and the single molecule reconstruction of purified virions (black).
See also Figure S3.
Detailed 3-D reconstructions of the STIV virions were needed to show the changes that accompany virus maturation in vivo. Electron cryotomograms are challenging to interpret due to the missing wedge artifact and low signal-to-noise ratio. To fill in the missing data and boost the signal-to-noise ratio, particles from multiple sub-tomograms were aligned and averaged. An average 3-D reconstruction of the particles was produced with a multi-class ML algorithm from 123 sub-volumes containing one STIV particle each. The algorithm, as it was applied, allows for multiple reconstructions to be derived simultaneously (Fig 3A,B). Following a detailed analysis of these reconstructions there were only two distinct structures that emerged; empty and full. If other subtle differences in particle structure exist, the data were not of sufficient resolution to resolve them. More detailed information can be found in the methods and Fig S3. The three perpendicular cross-sections of the averaged volumes (Fig S3C) display turrets at 5-fold vertices, indicating the correct computation of orientations for each sub-volume. The resolution was estimated to be approximately 6.5 nm based on the Fourier Shell Correlation (FSC) method with a 0.5 threshold. The radial density plots for the procapsid and virion reconstructions displayed overall agreement for the capsid and membrane radii with each other and with the single particle reconstruction (Fig 3D). Comparison of the full and empty particles demonstrate that no large-scale reorganization of the capsid or membrane occur upon DNA packaging in STIV. The appearance of turret-like structures at 5-fold vertices in the reconstructions did not result from model-bias since the ML approach is ab initio. Both virions and procapsids had closely similar turret-like structures, showing that they were assembled on the particle prior to genome packaging. A three-class reconstruction was also carried out (data not shown). The resulting reconstructions display one DNA-free and two DNA-containing reconstructions. However, no significant differences in the two DNA-containing reconstructions can be concluded. A three-class reconstruction cannot identify partially packaged particles if there are any.
In addition to virions and procapsids, partially assembled shells formed by capsid and membranes were also observed (Fig 2C). These partial assemblies have curvature and spacing between capsid and membrane layers closely similar to those of fully assembled procapsids. They were not associated with the cytoplasmic membrane and clearly did not form by a budding mechanism. The data indicate that the STIV capsid and membrane co-assemble in the cytoplasm to form a procapsid, which further matures to a virion through a DNA packaging event.
Distribution and Packing of Viral Particles in vivo
In order to robustly classify viral particles and determine their locations and orientations in a large number of tomograms, without applying the computationally intensive ML process for each tomogram, a correlation-based approach was employed using the reconstructions derived by the ML method as references. More detailed information can be found in the methods and Fig S4, S5. A gallery of cells sampled at 29–32 hpi is shown in Fig 4a. Each cell has between 22–166 particles. Viral clusters or arrays were frequently present and contained the majority of particles in the cell (table I). Cells that did not have viral clusters had low particle number (22–44) and a high ratio of empty procapsids to virions (55%–73%), indicating that these cells were at an early stage of infection. Particle clusters mainly consisted of virions. Procapsids were only found on the edge of the clusters or not associated with the clusters at all. Particles in clusters were packed very tightly as illustrated in the blow-up view of a cluster (Fig 4B). The narrow distribution of distances between the centers of particles and their closest neighbors are shown in Fig S6A. The average center-to-center distance between adjacent particles is 75 nm, corresponding to the particle diameter without the turrets. This is consistent with the orientation analysis that demonstrates the turrets of nearest neighbors avoid each other in the arrays (Fig 4B, S6B). Some viral clusters had close-packed lattices with quasi-crystalline, inter-particle spacing as illustrated in Fig S6C and table SI.
Fig. 4. The analysis of viral distribution and packing.

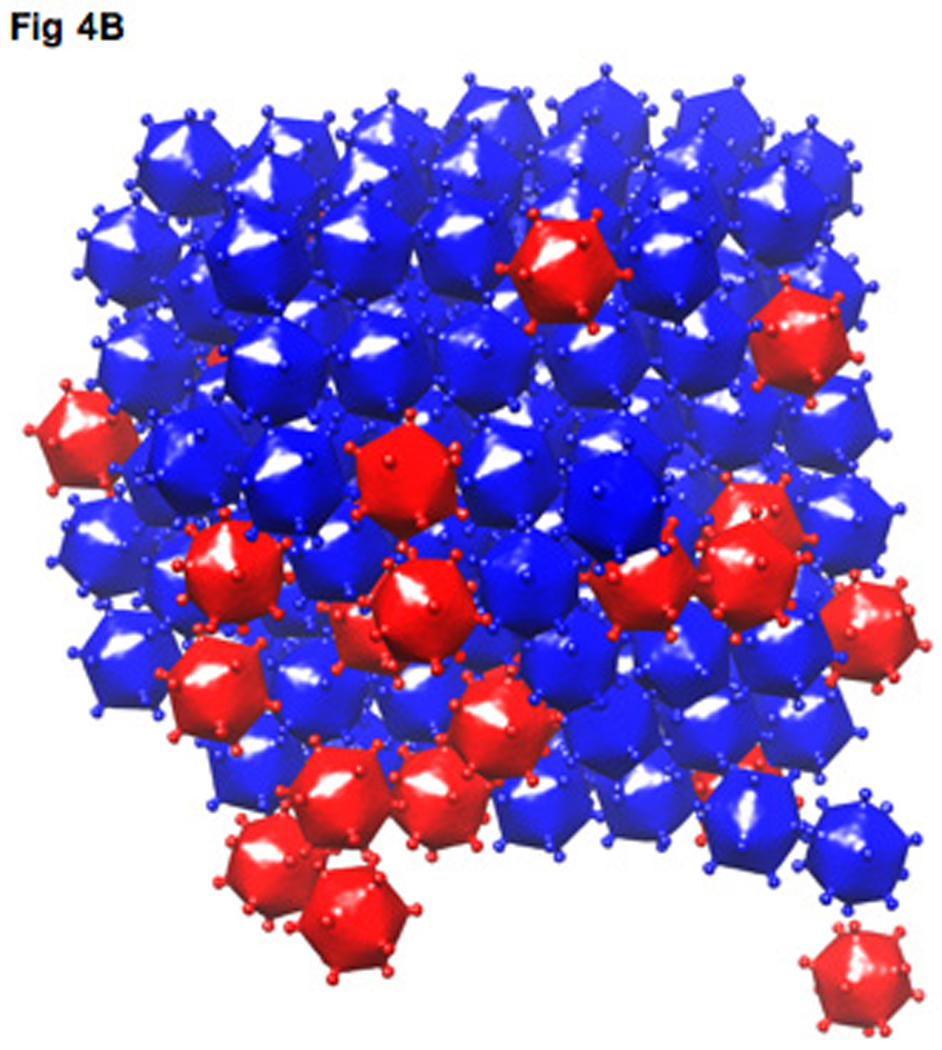
(A) A gallery of model representations of viral distributions with cell periphery outlined in gray. (B) The model representation of a quasi-crystalline packing of a viral array (virions, blue icosahedra; procapsids, red icosahedra).
See also Figure S4, S5 and S6 and Table S1.
Table I.
The analysis of viral distribution and array packing
| Total # of particles | 22–166 |
| % of virions | 27%–86% |
| % of particles in viral clusters | 67%–99% |
| % of particles in viral clustrers being virions | 74%–94% |
Discussion
STIV infection induces dramatic host morphological changes. The pyramid structures are unlike cellular protrusions typically observed in eukaryotes (Charras and Paluch, 2008; Mattila and Lappalainen, 2008). Based on what was characterized by ECT, we propose a model for pyramid formation (Fig 1E). The pyramids could form by either mechanically protruding and/or enzymatically digesting through the S-layer, perturbing the S-layer integrity and detaching it from the pyramid membrane. The thicker pyramid membrane and sharply defined facets indicate different protein and/or lipid compositions from the cytoplasmic membrane, implying the transport of specific proteins and extra lipids to the target sites to build pyramids. The compositions and functions of IPB remain to be characterized. The fact that not every pyramid has IPB and pyramids at an early stage of formation do not have IPB suggests that they may not relate to the pyramid formation or growth but likely relate to some pyramid-mediated viral release mechanism.
The control mechanisms that assemble a viral capsid with hundreds copies of repeated subunits are yet to be fully understood (Caspar, 1980; Caspar and Klug, 1962; Fane and Prevelige, 2003; Johnson and Speir, 1997). The issue is further complicated in viruses that contain inner membranes. Outstanding questions regarding these processes include: (1) How does the membrane core assemble? Does it form independently, followed by capsid assembly or do capsid proteins and membranes co-assemble? (2) Where does viral lipid originate in cells that lack membrane-containing organelles i.e. does the membrane core bud off the cytoplasmic membrane or form de novo in the cytoplasm? This ECT study of STIV provided valuable insights of in vivo assembly and explicitly addresses some of these points. The observation of partially built particles supports the hypothesis that assembly of the STIV capsid shell and the membrane are tightly coupled. The curvature and layer spacing of partial shells resembled those of fully assembled procapsids, implying defined local interactions between capsid and membrane and between capsid subunits as assembly proceeds. There is no evidence from our ECT study that partially assembled particles bud off the cytoplasmic membrane. Indeed, the mass spectrometry study concluded that the lipid compositions of viral and cellular membranes are different (Maaty et al., 2006).
The crystal structures of PRD1 and PM2, viruses in the same lineage as STIV, revealed how proteins and membranes organize in virions (Abrescia et al., 2004; Abrescia et al., 2008; Cockburn et al., 2004). About half of the membrane’s mass is attributable to proteins, which is likely to be the case for the STIV internal membrane. The crystal structures reported suggest that the membrane-associated proteins may function as tape-measures or scaffolding proteins, similar to those in viruses that lack inner membranes (Abrescia et al., 2004; Abrescia et al., 2008; Cockburn et al., 2004; Fane and Prevelige, 2003). STIV gene products may embed in the nascent lipid creating curvature and affinity for the major capsid protein. In a previous study, the c-terminal region of the STIV capsid protein was shown to interact with membrane (Khayat et al., 2005). We propose that the viral lipids derive de novo and associate through their hydrophobic nature. The association of trans-membrane proteins organizes the membrane through specific inter-subunit interactions that facilitates correct binding interactions of capsid proteins as assembly continues.
The ECT study showed that the STIV life cycle involved a procapsid, as observed in PRD1 (Martin et al., 2001), human adenovirus (Ostapchuk and Hearing, 2005), as well as dsDNA bacteriophages (Casjens and King, 1975). By comparing the reconstructions of the procapsid and the virion, it is clear that DNA packaging does not induce capsid conformational change that can be resolved at ~ 6.5 nm resolution. At the current resolution, subtle changes cannot be detected. For instance in PRD1, single particle reconstructions show a subtle reduction of separation between the capsid and membrane due to a 5% radial expansion and 10% thinning of the membrane when the genome was encapsidated (Butcher et al., 1995; Martin et al., 2001).
The five-fold vertices of icosahedral viruses are often found to be involved in receptor binding and/or genome translocation (Abrescia et al., 2004; Abrescia et al., 2008; Gowen et al., 2003; Johnson and Chiu, 2007; Moore and Prevelige, 2002). The presence of turret-like densities at the 5-fold vertices in both virions and procapsids indicates that the vertex proteins associate while procapsids assemble. It remains to be determined whether any of the 12 turrets can function in DNA packaging or if there is a specialized vertex for DNA packaging like in PRD1, Adenoviruses, Herpes virus, and dsDNA phages (Bazinet and King, 1985; Cardone et al., 2007; Chang et al., 2007; Karhu et al., 2007; Ostapchuk and Hearing, 2005).
We propose that crystalline viral arrays offer an important advantage as they allow the limited cellular space to accommodate the greatest number of viruses. In fact, virus arrays have been observed in other systems such as in papillomaviruses (Campo, 2002; Wang et al., 2009), FHV (Lanman et al., 2008), and iridovirus (Darlington et al., 1966). The high image quality of cellular ECT and the robust computational analysis allowed the STIV arrays to be analyzed in exceptional detail. Virus factories or viroplasm have been reported in eukaryotic and prokaryotic systems where proteins and newly synthesized genomes are confined within specific compartments for efficient viral replication and assembly (Bravo et al., 2005; Cook, 1999; Netherton et al., 2007). The analysis of the distribution of STIV virions and procapsids in multiple cells suggests that genome packaging leads to redistribution of particles. The viral clusters of STIV may accommodate DNA and packaging enzymes where capsid assembly and genome packaging are tightly coupled, accounting for the observation that procapsids appear only on the edge of the clusters. Alternatively, particles may assemble outside the clusters and traffic to specific sites where viral genome packaging takes place with the formation of tightly packed clusters.
STIV infected sulfolobus provided an ideal system for the ECT study of complex virus assembly and maturation as well as raising intriguing questions about virus-induced cellular changes. Future studies to elucidate greater detail regarding the proteins directing and participating in the all aspects of the virus life cycle will utilize specific gold-labeled antibodies to cellular and viral gene products as well as an available infectious clone for STIV that allows the mutation or deletion of specific viral genes.
Experimental Procedures
Sample Preparation
S. solfataricus strain 2-2-12 cultures were infected with STIV as previously described (Ortmann et al., 2006). Sulfolobus were grown up from glycerol stock in media 182 at pH 3.5, 80°C and passage once. The cultures were passed second time to media 182 at pH 2.5 and infected with STIV at MOI ~2 at OD 650 nm ~0.4. At 22–32 hr post-infection, cells were concentrated ~20 fold by centrifugation. Equal volume of cells were mixed with BSA-treated 10-nm gold colloidal beads and applied to plasma cleaned Quantifoil holey carbon films. The grids were plunge-frozen in liquid ethane using a Vitrobot (FEI Inc.) and stored in liquid nitrogen (Iancu et al., 2006).
Tomography Data Collection and Image Processing
Specimens were imaged using a FEI Polara transmission electron microscope at an accelerating voltage of 300 kV. Tilt series covering an angular range from −60° to +60° with 1° increment were acquired automatically using Leginon at 18,000× magnification, 10–14 µm underfocus, with 110–160 electrons /Å2 total dose (Suloway et al., 2005; Suloway et al., 2009). Images were recorded with a lens-coupled Gatan UltraCam 4k × 4k CCD camera binned by two so the final pixel size represented 1.26 nm on the specimen. Tilt series were aligned with gold fiducials and 3-D reconstructions were calculated with IMOD (Kremer et al., 1996). Tomograms were filtered with a 3-D median filter with 3dmod and visualized and/or segmented using 3dmod and Chimera (Kremer et al., 1996; Pettersen et al., 2004). Totally about 60 tomograms were analyzed.
Maximum Likelihood Reconstruction
The icosahedrally symmetric reconstructions were computed from the tomography volumes by a modification of the single particle cryo-EM reconstruction algorithm (Prust et al., 2009; Yin et al., 2003). First 123 sub-volumes, each containing one STIV particle, were extracted from the tomography by cross correlating with a spherically symmetric template. Then the modification of the algorithm of (Yin et al. 2003) was applied to jointly compute two 3-D icosahedrally-symmetric reconstructions, which correspond to full and empty particles. Third, the STIV particle shown in each of the sub-volumes used in the reconstruction was classified with respect to being a full or an empty particle.
The modification of the ML reconstruction algorithm (Yin et al. 2003) takes into account the following aspects of electron tomography. In (Yin et al. 2003) the cryo-EM image is a linear transformation of the 3-D distribution of electron scattering intensity and the transformation includes projection from 3-D to 2-D with unknown projection angles, translation of the location of particle center (two component vector), and the contrast transfer function. Now the transformation includes rotation of the particle in the sub-volume, translation of the location of the center of the particle in the sub-volume (three component vector), and the fact that a wedge of data in 3-D reciprocal space is missing due to the fact that the tilt angle is limited to +/−60 degrees.
To estimate the resolution, the whole data set of 123 sub-volumes was separated into two subsets. For each data set, reconstructions assuming two classes of particles are performed. Then, for each class, Fourier Shell Correlations are calculated between reconstructions from the two different subsets. Using a cut-off of 0.5, the resolutions for Class 1 and class 2 reconstructions are 6.5 nm.
Template Matching Approach
The reconstructions were used as references to determine particle classes (i.e. full vs. empty), locations, and orientations in the second cell tomogram. A correlation-based approach was used as described in the following steps: (1) A one-class icosahedrally-symmetric reconstruction was computed as described previously in this paper and spherically averaged to provide a template. (2) The template was used to locate particle centers in the tomograms by cross-correlation. (3) Sub-volumes of 65×65×65 voxels were extracted from the cell tomogram based on the center locations. (4) The icosahedrally-symmetric reconstructions obtained in the two-class reconstruction were used to generate templates to determine particle orientations. For each reconstruction, a library of 5000 templates was constructed to cover the reconstruction in different orientations described by Euler angles (α, β, γ) (all within 3 adjacent fundamental domains of the icosahedral group). The missing wedge was taking into account when generating templates. (5) Using the 10,000 templates from step (4), the sub-tomogram centered at each location determined in step (2) and extracted in step (3) is both labeled and orientated by normalized correlation. Then, based on these estimated orientations and class labels, each particle is aligned and averaged within its class with the application of the icosahedral symmetry operations. The maximum peak value of the correlation attainable over orientations at each SNR was simulated by correlating the templates and the templates plus white Gaussian noise.
Supplementary Material
Fig. 5. A model of STIV Assembly and Maturation.

The lipids and trans-membrane proteins assemble to form viral membrane as the transmembrane proteins serve as tape-measure scaffolding and facilitate correct assembly of cpasid proteins. The growing membrane and capsid form a procapsid. The genome is packaged through either a turret vertex or a specialized portal vertex. A procapsid matures to a virion without undergoing large-scale capsid transformation.
Acknowledgements
This work was supported by the National Institutes of Health (NIH) grant GM54076 to J.E.J., the NIH grant GM082545 to G.J.J. and the National Science Foundation (NSF) grants 0735297 and 0836656 to P.C.D., the NSF grant EF0802200 to M.J.Y., and gifts to Caltech from the Gordon and Betty Moore Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrescia NG, Cockburn JJ, Grimes JM, Sutton GC, Diprose JM, Butcher SJ, Fuller SD, San Martin C, Burnett RM, Stuart DI, et al. Insights into assembly from structural analysis of bacteriophage PRD1. Nature. 2004;432:68–74. doi: 10.1038/nature03056. [DOI] [PubMed] [Google Scholar]
- Abrescia NG, Grimes JM, Kivela HM, Assenberg R, Sutton GC, Butcher SJ, Bamford JK, Bamford DH, Stuart DI. Insights into virus evolution and membrane biogenesis from the structure of the marine lipid-containing bacteriophage PM2. Mol Cell. 2008;31:749–761. doi: 10.1016/j.molcel.2008.06.026. [DOI] [PubMed] [Google Scholar]
- Bazinet C, King J. The DNA translocating vertex of dsDNA bacteriophage. Annu Rev Microbiol. 1985;39:109–129. doi: 10.1146/annurev.mi.39.100185.000545. [DOI] [PubMed] [Google Scholar]
- Bravo A, Serrano-Heras G, Salas M. Compartmentalization of prokaryotic DNA replication. FEMS Microbiol Rev. 2005;29:25–47. doi: 10.1016/j.femsre.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Brumfield SK, Ortmann AC, Ruigrok V, Suci P, Douglas T, Young MJ. Particle assembly and ultrastructural features associated with replication of the lytic archaeal virus sulfolobus turreted icosahedral virus. J Virol. 2009;83:5964–5970. doi: 10.1128/JVI.02668-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher SJ, Bamford DH, Fuller SD. DNA packaging orders the membrane of bacteriophage PRD1. EMBO J. 1995;14:6078–6086. doi: 10.1002/j.1460-2075.1995.tb00298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campo MS. Animal models of papillomavirus pathogenesis. Virus Res. 2002;89:249–261. doi: 10.1016/s0168-1702(02)00193-4. [DOI] [PubMed] [Google Scholar]
- Cardone G, Winkler DC, Trus BL, Cheng N, Heuser JE, Newcomb WW, Brown JC, Steven AC. Visualization of the herpes simplex virus portal in situ by cryo-electron tomography. Virology. 2007;361:426–434. doi: 10.1016/j.virol.2006.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S, King J. Virus assembly. Annu Rev Biochem. 1975;44:555–611. doi: 10.1146/annurev.bi.44.070175.003011. [DOI] [PubMed] [Google Scholar]
- Caspar DL. Movement and self-control in protein assemblies. Quasi-equivalence revisited. Biophys J. 1980;32:103–138. doi: 10.1016/S0006-3495(80)84929-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar DL, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harb Symp Quant Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- Chang JT, Schmid MF, Rixon FJ, Chiu W. Electron cryotomography reveals the portal in the herpesvirus capsid. J Virol. 2007;81:2065–2068. doi: 10.1128/JVI.02053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charras G, Paluch E. Blebs lead the way: how to migrate without lamellipodia. Nat Rev Mol Cell Biol. 2008;9:730–736. doi: 10.1038/nrm2453. [DOI] [PubMed] [Google Scholar]
- Cockburn JJ, Abrescia NG, Grimes JM, Sutton GC, Diprose JM, Benevides JM, Thomas GJ, Jr, Bamford JK, Bamford DH, Stuart DI. Membrane structure and interactions with protein and DNA in bacteriophage PRD1. Nature. 2004;432:122–125. doi: 10.1038/nature03053. [DOI] [PubMed] [Google Scholar]
- Cook PR. The organization of replication and transcription. Science. 1999;284:1790–1795. doi: 10.1126/science.284.5421.1790. [DOI] [PubMed] [Google Scholar]
- Darlington RW, Granoff A, Breeze DC. Viruses and renal carcinoma of Rana pipiens. II. Ultrastructural studies and sequential development of virus isolated from normal and tumor tissue. Virology. 1966;29:149–156. doi: 10.1016/0042-6822(66)90204-2. [DOI] [PubMed] [Google Scholar]
- Dubochet J, Adrian M, Chang JJ, Homo JC, Lepault J, McDowall AW, Schultz P. Cryo-electron microscopy of vitrified specimens. Q Rev Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- Engelhardt H, Peters J. Structural research on surface layers: a focus on stability, surface layer homology domains, and surface layer-cell wall interactions. J Struct Biol. 1998;124:276–302. doi: 10.1006/jsbi.1998.4070. [DOI] [PubMed] [Google Scholar]
- Fane BA, Prevelige PE., Jr Mechanism of scaffolding-assisted viral assembly. Adv Protein Chem. 2003;64:259–299. doi: 10.1016/s0065-3233(03)01007-6. [DOI] [PubMed] [Google Scholar]
- Frangakis AS, Bohm J, Forster F, Nickell S, Nicastro D, Typke D, Hegerl R, Baumeister W. Identification of macromolecular complexes in cryoelectron tomograms of phantom cells. Proc Natl Acad Sci U S A. 2002;99:14153–14158. doi: 10.1073/pnas.172520299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowen B, Bamford JK, Bamford DH, Fuller SD. The tailless icosahedral membrane virus PRD1 localizes the proteins involved in genome packaging and injection at a unique vertex. J Virol. 2003;77:7863–7871. doi: 10.1128/JVI.77.14.7863-7871.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm R, Singh H, Rachel R, Typke D, Zillig W, Baumeister W. Electron tomography of ice-embedded prokaryotic cells. Biophys J. 1998;74:1031–1042. doi: 10.1016/S0006-3495(98)74028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald K, Cyrklaff M. Structure of complex viruses and virus-infected cells by electron cryo tomography. Curr Opin Microbiol. 2006;9:437–442. doi: 10.1016/j.mib.2006.06.016. [DOI] [PubMed] [Google Scholar]
- Iancu CV, Tivol WF, Schooler JB, Dias DP, Henderson GP, Murphy GE, Wright ER, Li Z, Yu Z, Briegel A, et al. Electron cryotomography sample preparation using the Vitrobot. Nat Protoc. 2006;1:2813–2819. doi: 10.1038/nprot.2006.432. [DOI] [PubMed] [Google Scholar]
- Jensen GJ, Briegel A. How electron cryotomography is opening a new window onto prokaryotic ultrastructure. Curr Opin Struct Biol. 2007;17:260–267. doi: 10.1016/j.sbi.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Johnson JE, Chiu W. DNA packaging and delivery machines in tailed bacteriophages. Curr Opin Struct Biol. 2007;17:237–243. doi: 10.1016/j.sbi.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Johnson JE, Speir JA. Quasi-equivalent viruses: a paradigm for protein assemblies. J Mol Biol. 1997;269:665–675. doi: 10.1006/jmbi.1997.1068. [DOI] [PubMed] [Google Scholar]
- Karhu NJ, Ziedaite G, Bamford DH, Bamford JK. Efficient DNA packaging of bacteriophage PRD1 requires the unique vertex protein P6. J Virol. 2007;81:2970–2979. doi: 10.1128/JVI.02211-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khayat R, Tang L, Larson ET, Lawrence CM, Young M, Johnson JE. Structure of an archaeal virus capsid protein reveals a common ancestry to eukaryotic and bacterial viruses. Proc Natl Acad Sci U S A. 2005;102:18944–18949. doi: 10.1073/pnas.0506383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster AJ, Grimm R, Typke D, Hegerl R, Stoschek A, Walz J, Baumeister W. Perspectives of molecular and cellular electron tomography. J Struct Biol. 1997;120:276–308. doi: 10.1006/jsbi.1997.3933. [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Lanman J, Crum J, Deerinck TJ, Gaietta GM, Schneemann A, Sosinsky GE, Ellisman MH, Johnson JE. Visualizing flock house virus infection in Drosophila cells with correlated fluorescence and electron microscopy. J Struct Biol. 2008;161:439–446. doi: 10.1016/j.jsb.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucic V, Forster F, Baumeister W. Structural studies by electron tomography: from cells to molecules. Annu Rev Biochem. 2005;74:833–865. doi: 10.1146/annurev.biochem.73.011303.074112. [DOI] [PubMed] [Google Scholar]
- Maaty WS, Ortmann AC, Dlakic M, Schulstad K, Hilmer JK, Liepold L, Weidenheft B, Khayat R, Douglas T, Young MJ, Bothner B. Characterization of the archaeal thermophile Sulfolobus turreted icosahedral virus validates an evolutionary link among double-stranded DNA viruses from all domains of life. J Virol. 2006;80:7625–7635. doi: 10.1128/JVI.00522-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin CS, Burnett RM, de Haas F, Heinkel R, Rutten T, Fuller SD, Butcher SJ, Bamford DH. Combined EM/X-ray imaging yields a quasi-atomic model of the adenovirus-related bacteriophage PRD1 and shows key capsid and membrane interactions. Structure. 2001;9:917–930. doi: 10.1016/s0969-2126(01)00642-6. [DOI] [PubMed] [Google Scholar]
- Mattila PK, Lappalainen P. Filopodia: molecular architecture and cellular functions. Nat Rev Mol Cell Biol. 2008;9:446–454. doi: 10.1038/nrm2406. [DOI] [PubMed] [Google Scholar]
- Moore SD, Prevelige PE., Jr DNA packaging: a new class of molecular motors. Curr Biol. 2002;12:R96–R98. doi: 10.1016/s0960-9822(02)00670-x. [DOI] [PubMed] [Google Scholar]
- Netherton C, Moffat K, Brooks E, Wileman T. A guide to viral inclusions, membrane rearrangements, factories, and viroplasm produced during virus replication. Adv Virus Res. 2007;70:101–182. doi: 10.1016/S0065-3527(07)70004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortmann AC, Wiedenheft B, Douglas T, Young M. Hot crenarchaeal viruses reveal deep evolutionary connections. Nat Rev Microbiol. 2006;4:520–528. doi: 10.1038/nrmicro1444. [DOI] [PubMed] [Google Scholar]
- Ostapchuk P, Hearing P. Control of adenovirus packaging. J Cell Biochem. 2005;96:25–35. doi: 10.1002/jcb.20523. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Prust CJ, Doerschuk PC, Lander GC, Johnson JE. Ab initio maximum likelihood reconstruction from cryo electron microscopy images of an infectious virion of the tailed bacteriophage P22 and maximum likelihood versions of Fourier Shell Correlation appropriate for measuring resolution of spherical or cylindrical objects. J Struct Biol. 2009;167:185–199. doi: 10.1016/j.jsb.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quax TE, Krupovic M, Lucas S, Forterre P, Prangishvili D. The Sulfolobus rod-shaped virus 2 encodes a prominent structural component of the unique virion release system in Archaea. Virology. 404:1–4. doi: 10.1016/j.virol.2010.04.020. [DOI] [PubMed] [Google Scholar]
- Rice G, Tang L, Stedman K, Roberto F, Spuhler J, Gillitzer E, Johnson JE, Douglas T, Young M. The structure of a thermophilic archaeal virus shows a double-stranded DNA viral capsid type that spans all domains of life. Proc Natl Acad Sci U S A. 2004;101:7716–7720. doi: 10.1073/pnas.0401773101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Suloway C, Shi J, Cheng A, Pulokas J, Carragher B, Potter CS, Zheng SQ, Agard DA, Jensen GJ. Fully automated, sequential tilt-series acquisition with Leginon. J Struct Biol. 2009;167:11–18. doi: 10.1016/j.jsb.2009.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HK, Duffy AA, Broker TR, Chow LT. Robust production and passaging of infectious HPV in squamous epithelium of primary human keratinocytes. Genes Dev. 2009;23:181–194. doi: 10.1101/gad.1735109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, Zheng Y, Doerschuk PC, Natarajan P, Johnson JE. A statistical approach to computer processing of cryo-electron microscope images: virion classification and 3-D reconstruction. J Struct Biol. 2003;144:24–50. doi: 10.1016/j.jsb.2003.09.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


