Abstract
The cJun N-terminal kinase (JNK) signal transduction pathway is implicated in the regulation of neuronal function. JNK is encoded by three genes that play partially redundant roles. Here we report the creation of mice with targeted ablation of all three Jnk genes in neurons. Compound JNK-deficient neurons are dependent on autophagy for survival. This autophagic response is caused by FoxO-induced expression of Bnip3 that displaces the autophagic effector Beclin-1 from inactive Bcl-XL complexes. These data identify JNK as a potent negative regulator of FoxO-dependent autophagy in neurons.
Keywords: autophagy, Beclin 1, Bnip3, JNK, Neurons
The cJun N-terminal kinases (JNKs) are encoded by three genes (Davis 2000). Two of these genes (Jnk1 and Jnk2) are expressed ubiquitously, while the Jnk3 gene is selectively expressed in neurons (Gupta et al. 1996). Compound mutation of these Jnk genes causes early embryonic lethality in mice (Kuan et al. 1999; Sabapathy et al. 1999). Consequently, studies of JNK deficiency in neurons have focused on an analysis of mice with partial loss of JNK (Davis 2000; Weston and Davis 2007). These studies have demonstrated isoform-specific functions of JNK in neurons (Brecht et al. 2005).
It is established that JNK plays an important role in the regulation of microtubule stability in neurons. JNK-induced phosphorylation of microtubule-associated proteins—including Doublecortin (Gdalyahu et al. 2004), MAP1B (Chang et al. 2003; Barnat et al. 2010), MAP2 (Chang et al. 2003), the stathmin protein family of microtubule-destabilizing proteins (Tararuk et al. 2006), and Tau (Yoshida et al. 2004)—may influence microtubule function. This action of JNK is important for neurite formation. Thus, JNK contributes to bone morphogenic protein-stimulated dendrite formation (Podkowa et al. 2010), the structure of dendritic architecture (Coffey et al. 2000; Bjorkblom et al. 2005), axodendritic length (Tararuk et al. 2006), and axonal regeneration (Barnat et al. 2010). Moreover, JNK can regulate kinesin-mediated fast axonal transport on microtubules (Morfini et al. 2006, 2009) and contributes to the regulation of synaptic plasticity (Chen et al. 2005; Zhu et al. 2005; Li et al. 2007; Thomas et al. 2008). Together, these data demonstrate that JNK plays a key role in the physiological regulation of neuronal activity (Waetzig et al. 2006).
The JNK signaling pathway has also been implicated in stress-induced apoptosis (Kuan et al. 1999; Tournier et al. 2000), including neuronal death in models of excitotoxicity (Yang et al. 1997) and stroke (Kuan et al. 2003; Pirianov et al. 2007). This JNK-induced apoptotic response is mediated, in part, by the expression and/or phosphorylation of members of the Bcl2-related protein family (Weston and Davis 2007; Hubner et al. 2008; Morel et al. 2009; Hubner et al. 2010). These data indicate that JNK plays a critical role during the injury response associated with neurodegeneration and stroke.
The dual role of JNK in mediating both physiological responses (e.g., neurite development) and pathological responses (e.g., neuronal injury) requires that the actions of JNK are context-specific (Waetzig and Herdegen 2005). These effects of JNK may be mediated by compartmentalization of specific pools of JNK in different subcellular locations or within different signaling complexes (Coffey et al. 2000). JNK may also cooperate with other signal transduction pathways to generate context-specific responses (Lamb et al. 2003). However, the fundamental role of JNK in neurons and the mechanisms that account for these divergent biological responses to JNK signaling remain poorly understood.
Studies of mice with deficiency of one Jnk gene have provided a foundation for current knowledge of the role of JNK in neurons. However, partial loss of JNK expression represents a limitation of these studies because of redundant functions of JNK isoforms (Tournier et al. 2000; Jaeschke et al. 2006). Creation of a model of compound JNK deficiency is important because compound JNK deficiency represents a more relevant model for understanding the effects of pharmacological JNK inhibition than deficiency of a single JNK isoform. JNK inhibitors have been identified that may be useful for the treatment of neurodegenerative diseases and stroke (Borsello et al. 2003; Hirt et al. 2004; Repici et al. 2007; Carboni et al. 2008; Esneault et al. 2008; Wiegler et al. 2008; Probst et al. 2011). A model of neuronal compound JNK deficiency is required to test whether the actions of these drugs are mediated by loss of JNK function. Moreover, an experimental model of compound JNK deficiency in neurons would provide insight into the physiological role of JNK in wild-type neurons.
The purpose of this study was to examine the properties of neurons with simultaneous ablation of the Jnk1, Jnk2, and Jnk3 genes. We report the creation and characterization of mice with triple deficiency of neuronal JNK isoforms in vivo and in primary cultures in vitro.
Results
Establishment of neurons with compound JNK deficiency in vitro
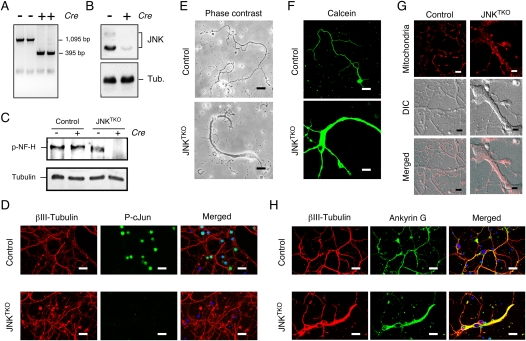
To examine the function of JNK in neurons, we prepared primary cerebellar granule neurons (CGNs) from mice with conditional Jnk alleles. Cre-mediated deletion of conditional Jnk resulted in neurons that lack expression of JNK (Fig. 1A,B) and exhibit defects in the phosphorylation of the JNK substrates cJun (Davis 2000) and neurofilament heavy chain (Fig. 1C,D; Brownlees et al. 2000). These triple Jnk knockout (JNKTKO) neurons exhibited altered morphology, including hypertrophy (Figs. 1E–G; Supplemental Fig. S1). Immunofluorescence analysis using an antibody to Tau (data not shown) and Ankyin G demonstrated the presence of hypertrophic axons (Fig. 1H).
Figure 1.
Establishment of JNK-deficient neurons. Wild-type (control) and Jnk1LoxP/LoxP Jnk2−/− Jnk3−/− CGNs were infected with Ad-cre at 3 d of culture in vitro (DIV) and then examined at 10 DIV. (A) Genotype analysis of JNKTKO neurons. The floxed Jnk1 and deleted Jnk1 alleles are detected as 1095-base-pair (bp) and 395-bp PCR products, respectively. (B) Extracts prepared from JNKTKO neurons were examined by immunoblot analysis using antibodies to JNK and α-Tubulin. (C) Control and JNKTKO neurons were examined at 10 DIV by immunoblot analysis using antibodies to phospho-neurofilament H and α-Tubulin. (D) Control and JNKTKO neurons were examined by immunofluorescence microscopy by staining with DAPI and antibodies to βIII tubulin and phospho-Ser63 cJun. Bar, 20 μm. (E) Control and JNKTKO neurons were examined by phase-contrast microscopy. Bar, 75 μm. (F) Control and JNKTKO neurons were stained with calcein-am ester and examined by fluorescence microscopy. Bar, 65 μm. (G) Wild-type (control) and JNKTKO neurons were stained with Mitotracker Red at 10 DIV and imaged by differential interference contrast (DIC) and fluorescence microscopy. Bar, 8 μm. (H) Control and JNKTKO neurons were examined by immunofluorescence microscopy by staining with DAPI and antibodies to βIII tubulin and Ankyrin G. Bar, 20 μm.
The JNK signaling pathway is implicated in microtubule stabilization and the regulation of axodendritic morphology (Coffey et al. 2000; Chang et al. 2003; Bjorkblom et al. 2005; Tararuk et al. 2006; Barnat et al. 2010). JNK inhibition may therefore increase microtubule instability and cause neurite retraction. Indeed, the JNKTKO neuronal hypertrophy was associated with a reduction in the number of dendrites (Fig. 1H; Supplemental Fig. S1). To test whether JNKTKO neurons exhibited increased microtubule instability, we examined the presence of stable microtubules containing detyrosinated Tubulin by immunofluorescence analysis (Schulze et al. 1987; Webster et al. 1987; Khawaja et al. 1988). Contrary to expectations, no decrease in microtubules with detyrosinated Tubulin was detected in JNKTKO neurons compared with control neurons (Supplemental Fig. S2). Together, these data confirm that JNK regulates neuronal morphology, but the mechanism may be only partially accounted for by altered microtubule stability.
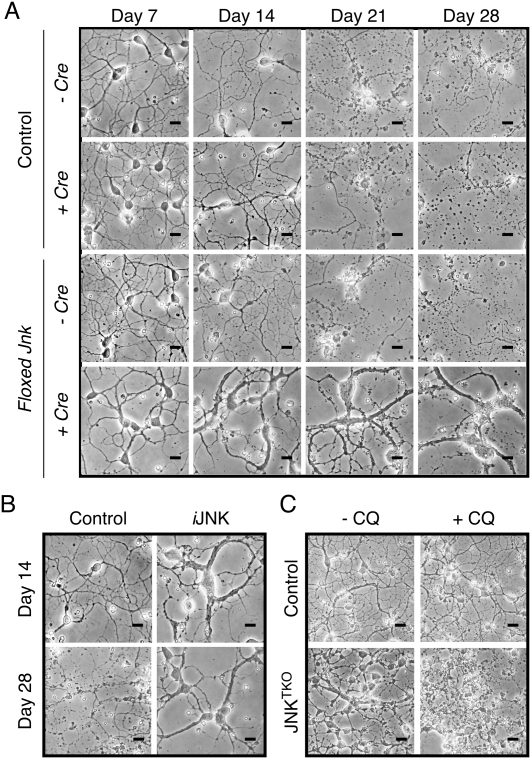
Comparison of control and JNKTKO neurons demonstrated that JNK deficiency caused a marked increase in life span during culture in vitro (Fig. 2A; Supplemental Fig. S3A). To confirm that the loss of JNK activity increased life span, we employed a chemical genetic strategy using neurons prepared from mice with germline point mutations that confer sensitivity of JNK to the predesigned small molecule drug 1NM-PP1 (Jaeschke et al. 2006). This chemical genetic analysis confirmed that JNK inhibition caused both hypertrophy and increased neuronal viability in vitro (Fig. 2B; Supplemental Fig. S3B).
Figure 2.
Increased life span of JNK-deficient neurons during culture in vitro. (A) Wild-type (control) and JNKTKO CGNs infected without or with Ad-cre at 3 DIV were examined by phase-contrast microscopy at 7–28 DIV. Bar, 45 μm. (B) Jnk1LoxP/LoxP Jnk2M108G/M108G Jnk3−/− CGNs were untreated (control) or infected with Ad-cre at 3 DIV and with the drug 1-NM-PP1 (1 μM) at 7 DIV (iJNK). The CGNs were examined using phase-contrast microscopy. Bar, 45 μm. (C) Wild-type (control) and JNKTKO CGNs infected with Ad-cre at 3 DIV were incubated without and with 1 μM chloroquine (CQ) at 11 DIV and then examined by phase-contrast microscopy at 12 DIV. Bar, 65 μm. Quantitative analysis of neuronal viability is presented in Supplemental Figure S3.
A defect in transport might contribute to the axonal hypertrophy of JNKTKO neurons (Griffin and Watson 1988). Indeed, it is established that JNK acts as a negative regulator of kinesin-mediated fast axonal transport (Morfini et al. 2006, 2009). These data suggest that JNKTKO neurons may exhibit altered kinesin-mediated transport. We found an accumulation of mitochondria (Fig.1G), synaptic vesicles (Supplemental Figs. S4, S5), and lysosomes (Supplemental Fig. S6) in JNKTKO neurons. Live-cell imaging of mitochondria demonstrated the presence of fast transport in wild-type neurons, but mitochondria were immobile in JNKTKO neurons (Supplemental Fig. S7). This loss of transport in JNKTKO neurons contrasts with expectations that JNK deficiency might increase transport (Morfini et al. 2006, 2009). It is established that fast transport of mitochondria is mediated by the conventional kinesin KIF5b (Tanaka et al. 1998). However, no decrease in Kif5b expression was detected in JNKTKO CGNs (Supplemental Fig. S8). A more general defect in trafficking may therefore account for the mislocalization of organelles in JNKTKO neurons.
Neuronal JNK deficiency causes increased autophagy in vitro
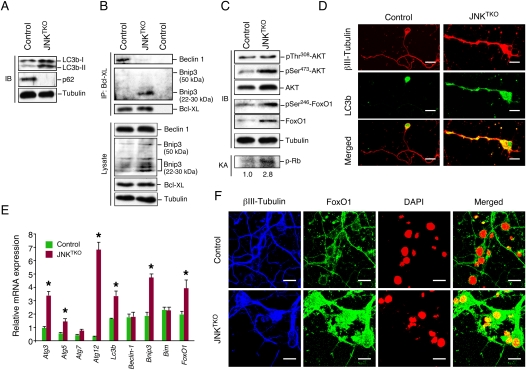
Live-cell imaging indicated that the morphology of mitochondria in JNKTKO neurons was different than control neurons (Fig. 1G). Electron microscopy confirmed that JNKTKO mitochondria were larger than control mitochondria (Supplemental Fig. S9). Numerous double-membrane structures, morphologically similar to autophagosomes, were detected in JNKTKO neurons, but not in control neurons. The presence of large numbers of autophagosomes in JNKTKO neurons suggests that these cells may exhibit increased autophagy. Indeed, biochemical analysis demonstrated that an increased amount of the autophagic effector protein Atg8/LC3b was processed by conjugation of phosphatidylethanolamine to the C terminus of the LC3b-I form to create LC3b-II, which is tightly associated with the autophagosomal membrane (Kabeya et al. 2004; Sou et al. 2008) in JNKTKO neurons compared with control neurons (Fig. 3A). Atg8/LC3b expression was increased in JNKTKO neurons (Fig. 3A,E), and Atg8/LC3b was redistributed from a location primarily in the soma of control neurons to the neurites of JNKTKO neurons (Fig. 3D). The Atg8/LC3b immunofluoresence detected in JNKTKO neurons was punctate (Supplemental Fig. S10), consistent with localization to autophagosomal membranes. Moreover, the p62/SQSTM1 protein, which directly binds the autophagic effector Atg8/LC3 (Pankiv et al. 2007), was detected in wild-type neurons but not in JNKTKO neurons (Fig. 3A).
Figure 3.
JNK deficiency in neurons causes increased autophagy. (A) Wild-type (control) and JNKTKO CGNs infected with Ad-cre at 3 DIV were harvested at 10 DIV to prepare protein extracts that were examined using antibodies to LC3b, p62/SQSTM1, and α-Tubulin. (B) Extracts prepared from control and JNKTKO CGNs were examined by immunoblot analysis by probing with antibodies to Bcl-XL, Bnip3, Beclin-1, and α-Tubulin. Coimmunoprecipitation assays were performed by immunoblot analysis of Bcl-XL immunoprecipitates. (C) Extracts prepared from control and JNKTKO CGNs were examined by immunoblot (IB) analysis by probing with antibodies to AKT, pSer308-AKT, pSer473-AKT, FoxO1, pSer246-FoxO1, and α-Tubulin. CDK2 activity was measured in an immunecomplex kinase assay (KA) using Rb as the substrate. The relative CDK2 activity is indicated below. (D) Control and JNKTKO CGNs were stained with βIII-Tubulin and LC3b antibodies and examined by fluorescence microscopy. Bar, 10 μm. (E) Gene expression in CGNs was examined by quantitative RT–PCR analysis of mRNA and normalized to the amount of Gapdh mRNA in each sample (mean ± SD; n = 3). Statistically significant differences are indicated. (*) P < 0.05. (F) Control and JNKTKO CGNs were stained with DAPI and antibodies to FoxO1 and βIII-Tubulin. The neurons were examined by fluorescence microscopy. The merged image represents colocalization of FoxO1 with DAPI. Bar, 10 μm.
The loss of p62/SQSTM1 suggests that autophagic flux is increased in JNKTKO neurons compared with control neurons (Klionsky et al. 2008). To confirm this conclusion, we examined the effect of lysosomal inhibition on the conversion of LC3b-I to LC3b-II. If the autophagic flux is increased, blocking autophagy should lead to increased accumulation of LC3b-II. Consistent with an increase in autophagic flux, we found that inhibition of autophagy caused a greater increase in LC3b-II in JNKTKO neurons compared with control neurons (Supplemental Fig. S11). Together, these data demonstrate the presence of an active autophagic response in JNKTKO neurons.
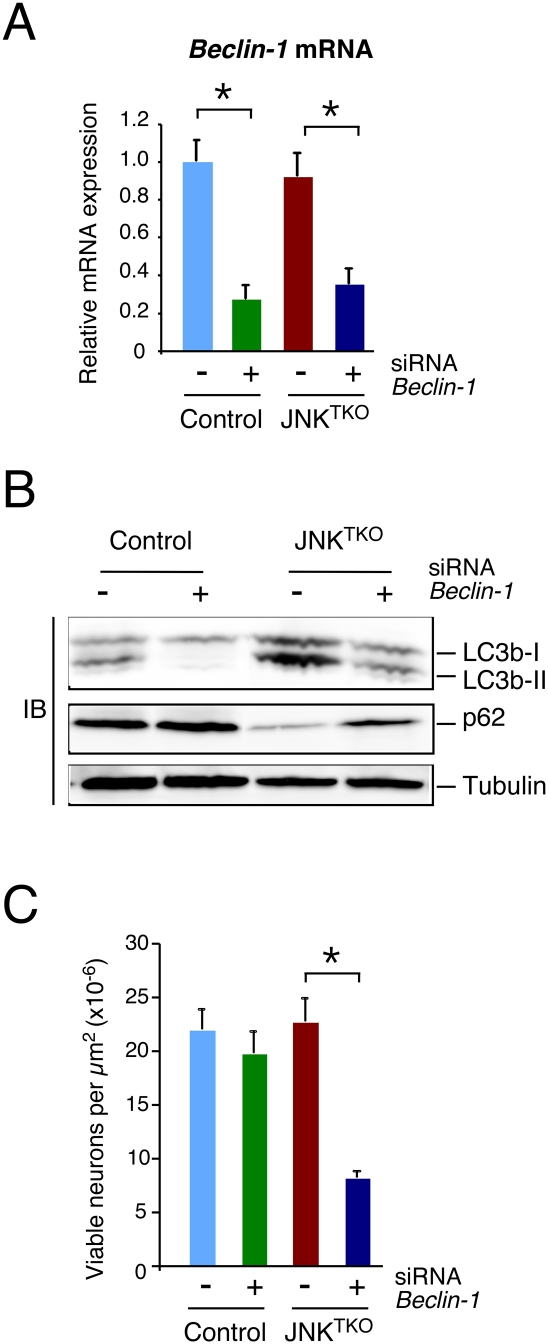
Autophagy may contribute to the increased survival of JNKTKO neurons (Hara et al. 2006; Komatsu et al. 2006). Indeed, studies using a pharmacological inhibitor demonstrated that autophagy was required for the increased life span of JNKTKO neurons compared with control neurons (Fig. 2C; Supplemental Fig. S3). Moreover, RNAi-mediated knockdown of the autophagic effector Beclin-1 caused decreased survival of JNKTKO neurons, but not control neurons (Fig. 4). Together, these data demonstrate that the survival of JNKTKO neurons depends on autophagy.
Figure 4.
Effect of RNAi-mediated knockdown of Beclin-1 on autophagy and survival of JNKTKO neurons. (A) Wild-type (control) and Jnk1LoxP/LoxP Jnk2−/− Jnk3−/− (JNKTKO) neurons infected with Ad-cre at 3 DIV were transfected at 7 DIV with Beclin-1 siRNA or control siRNA. The expression of Beclin-1 mRNA was examined at 11 DIV by quantitative RT–PCR analysis of mRNA and normalized to the amount of Gapdh mRNA in each sample (mean ± SD; n = 3). Statistically significant differences are indicated. (*) P < 0.05. (B) Control and JNKTKO neurons transfected with scrambled sequence or Beclin-1 siRNA were examined at 11 DIV by immunoblot analysis with antibodies to LC3b, p62/SQSTM1, and α-Tubulin. (C) The survival of RNAi transfected control and JNKTKO neurons at 11 DIV was quantitated (mean ± SD; n = 20). Statistically significant differences are indicated. (*) P < 0.05.
TORC1 does not mediate the effects of JNK deficiency on neuronal autophagy
The mTOR protein kinase complex TORC1 is a potent negative regulator of autophagy (Guertin and Sabatini 2007). Decreased TORC1 activity in JNK-deficient neurons may therefore account for the observed increase in autophagy. To test TORC1 function, we examined the phosphorylation of the TORC1 substrate pSer389 p70S6K. We found that JNK deficiency did not alter the phosphorylation of this TORC1 substrate in neurons (Supplemental Fig. S12). These data demonstrate that JNK deficiency regulates autophagy by a TORC1-independent mechanism.
Increased autophagy in JNK-deficient neurons is mediated by a FoxO1/Bnip3/Beclin-1 pathway
The finding that JNK deficiency in neurons triggers an autophagic response (Fig. 3) was unexpected, because studies of nonneuronal cells have implicated JNK in the induction of autophagy (Yu et al. 2004; Ogata et al. 2006; Wei et al. 2008) or as an effector of autophagy-associated cell death (Yu et al. 2004; Shimizu et al. 2010). Indeed, we found that autophagy caused by serum withdrawal was compromised in compound mutant fibroblasts that lack JNK expression (Supplemental Fig. S13). This finding markedly contrasts with the effect of compound JNK deficiency in neurons to induce spontaneous autophagy (Fig. 3). These data indicate that the role of JNK in autophagy suppression may be restricted to neurons.
To test whether the autophagic mediator Beclin-1 may be relevant to autophagy caused by JNK deficiency in neurons, we examined the effect of RNAi-mediated knockdown of Beclin-1 expression. Knockdown of Beclin-1 suppressed biochemical markers of autophagy in JNKTKO neurons, including increased LC3b-II and decreased p62/SQSTM1 (Fig. 4). These data demonstrate that Beclin-1 may mediate the effects of JNK deficiency to cause increased autophagy in neurons.
It is established that the JNK-regulated interaction of Bcl2 with the BH3 domain of Beclin-1 may contribute to autophagy (Wei et al. 2008). We therefore examined the interaction of Beclin-1 with Bcl2 family proteins in neurons. No coimmunoprecipitation of Beclin-1 with Bcl2 was detected in control neurons. However, Beclin-1 was found to coimmunoprecipitate with Bcl-XL in control neurons, but this interaction was markedly suppressed in JNKTKO neurons (Fig. 3B). The BH3 domain-binding activity of Bcl-XL is negatively regulated by phosphorylation of Bcl-XL on Ser62 (Upreti et al. 2008), but no increase in Bcl-XL phosphorylation was detected in JNKTKO neurons by immunoblot analysis with a phospho-specific antibody (data not shown). An alternative mechanism must therefore mediate the dissociation of Beclin-1. Release of Beclin-1 from Bcl-XL complexes could be mediated by competition with another BH3 domain protein. Indeed, we found that JNKTKO neurons expressed increased amounts of Bnip3, a BH3-only member of the Bcl2 protein family (Fig. 3B). Coimmunoprecipitation analysis demonstrated that the release of Beclin-1 from Bcl-XL complexes was associated with increased interaction of Bcl-XL with Bnip3 (Fig. 3B).
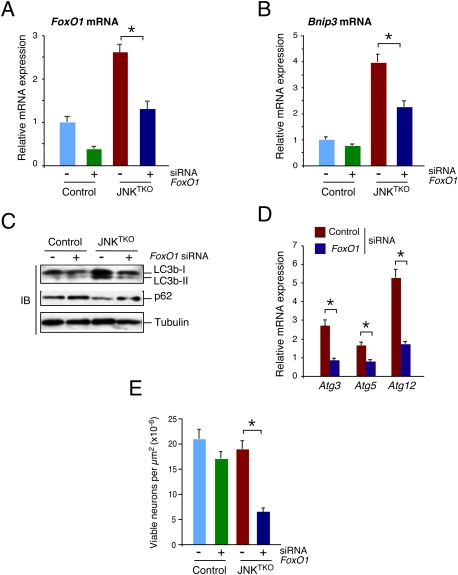
The Bnip3 gene is known to be a target of FoxO transcription factors that also increase the expression of the autophagy-related genes Atg8/Lc3b and Atg12 (Salih and Brunet 2008). The increased expression of these genes in JNKTKO neurons (Fig. 3A,B,D,E) suggests that JNK deficiency leads to FoxO activation. Indeed, gene expression analysis demonstrated increased FoxO1 mRNA and protein expression in JNKTKO neurons (Fig. 3C–E). To test whether FoxO1 contributes to the increased autophagy detected in JNKTKO neurons, we examined the effect of RNAi-mediated knockdown of FoxO1. Knockdown of FoxO1 in JNKTKO neurons caused decreased expression of Bnip3 and Atg genes, suppressed the increase in LC3b-II and the decrease in p62/SQSTM1, and caused decreased neuronal survival (Fig. 5). These data demonstrate that FoxO1 is required for the increased autophagy and survival of JNKTKO neurons.
Figure 5.
Effect of RNAi-mediated knockdown of FoxO1 on autophagy and survival of JNKTKO neurons. (A,B) Wild-type (control) and Jnk1LoxP/LoxP Jnk2−/− Jnk3−/− (JNKTKO) neurons infected with Ad-cre at 3 DIV were transfected at 7 DIV with FoxO1 siRNA or control siRNA. The expression of FoxO1 mRNA (A) and Bnip3 mRNA (B) was examined at 11 DIV by quantitative RT–PCR analysis of mRNA and normalized to the amount of Gapdh mRNA in each sample (mean ± SD; n = 3). Statistically significant differences are indicated. (*) P < 0.05. (C) Control and JNKTKO neurons transfected with scrambled sequence or FoxO1 siRNA were examined at 11 DIV by immunoblot analysis with antibodies to LC3b, p62/SQSTM1, and α-Tubulin. (D) RNAi transfected JNKTKO neurons were examined at 11 DIV by quantitative RT–PCR analysis of Atg3, Atg5, and Atg12 mRNA and normalized to the amount of Gapdh mRNA in each sample (mean ± SD; n = 3). Statistically significant differences are indicated. (*) P < 0.05. (E) The survival of RNAi transfected control and JNKTKO neurons at 11 DIV was quantitated (mean ± SD; n = 20). Statistically significant differences are indicated. (*) P < 0.05.
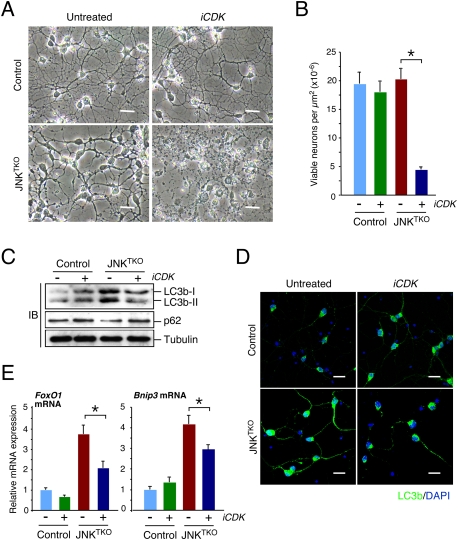
Cytoplasmic sequestration is a major mechanism of FoxO1 regulation by signal transduction pathways, including AKT (Salih and Brunet 2008). We found a small increase AKT phosphorylation on Thr308 and Ser473 in JNKTKO neurons (Fig. 3C), indicating that AKT activity may be moderately increased in JNKTKO neurons compared with control neurons. Nevertheless, we found increased nuclear localization of FoxO1 in JNKTKO neurons compared with control neurons (Fig. 3F). This nuclear redistribution of FoxO1 in JNKTKO neurons was associated with increased phosphorylation of FoxO1 on Ser246 (Fig. 3C), a site that dominantly induces nuclear accumulation of FoxO1 and is phosphorylated by cyclin-dependent protein kinases (CDKs) (Yuan et al. 2008). Abortive cell cycle re-entry has been observed during neurodegenerative processes (Kim and Bonni 2008), including stroke (Kuan et al. 2004). Indeed, we found that CDK2 was activated in JNKTKO neurons compared with control neurons (Fig. 3C). To test whether increased CDK activity contributes to the phenotype of JNKTKO neurons, we examined the effect of CDK inhibition on control and JNKTKO neurons. We found that CDK inhibition suppressed the increase in Bnip3 and FoxO1 expression detected in JNKTKO neurons (Fig. 6A). Moreover, CDK inhibition suppressed the autophagy-related increase in LC3b-II, decrease in p62/SQSTM1, and survival of JNKTKO neurons compared with control neurons (Fig. 6B–E). These data confirm a role for CDK activity in the induction of autophagy and survival by a FoxO1/Bnip3/Beclin-1 pathway in JNK-deficient neurons.
Figure 6.
CDK activity is required for the viability of JNKTKO neurons. (A,B) Wild-type (control) and Jnk1f/f Jnk2−/− Jnk3−/− (JNKTKO) neurons infected with Ad-cre at 3 DIV were treated without or with the CDK inhibitor roscovitine (5 μM; iCDK) at 10 DIV. (A) The neurons were examined by phase-contrast microscopy at 11 DIV. Bar, 45 μm. (B) The number of viable neurons was examined at 11 DIV (mean ± SD; n = 20). Statistically significant differences are indicated. (*) P < 0.05. (C) Control and JNKTKO neurons were examined after treatment on 10 DIV with roscovitine (iCDK) for 8 h by immunoblot analysis using antibodies to LC3b, p62SQTM1, and α-Tubulin. (D) Control and JNKTKO neurons were examined by immunofluorescence analysis after treatment with roscovitine (iCDK) for 8 h using an antibody to LC3b. DNA was stained with DAPI. Bar, 20 μm. (E) Control and JNKTKO neurons were examined after treatment with roscovitine (iCDK) for 8 h by quantitative RT–PCR analysis of FoxO1 and Bnip3 mRNA and normalized to the amount of Gapdh mRNA in each sample (mean ± SD; n = 3). Statistically significant differences are indicated. (*) P < 0.05.
Mice with compound JNK deficiency in neurons in vivo
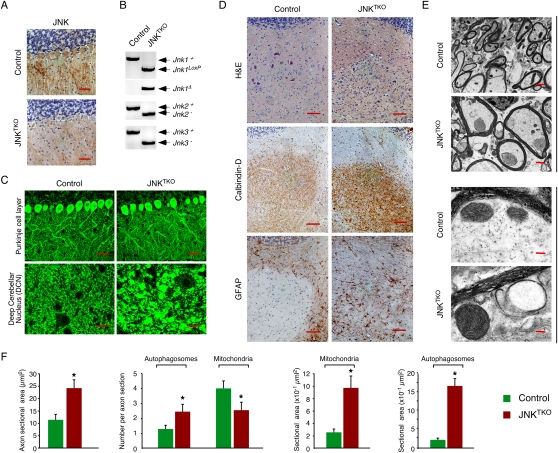
We tested the effect of transgenic expression of Cre recombinase in the brain of mice with floxed Jnk on neuronal function in vivo. Initial studies using Nestin-Cre mice demonstrated that triple JNK deficiency in neuronal progenitor cells caused early embryonic death (data not shown). Similarly, expression of Cre recombinase in a more limited region of the brain (telencephalon) using Foxg1-Cre transgenic mice also caused early embryonic death (data not shown). The early death of these JNKTKO mice precluded analysis of the effects of triple JNK deficiency on the brain. We therefore examined the effect of Cre expression in a subset of neurons that are nonessential for mouse survival. A mouse strain with Cre recombinase inserted in the Pcp2 gene expresses Cre recombinase in cerebellar Purkinje cells (Barski et al. 2000). This Pcp2-Cre strain enabled the creation of viable mice with triple neuronal deficiency of JNK1, JNK2, and JNK3 (Fig. 7). Purkinje cell defects represent one cause of cerebellar ataxia (Grusser-Cornehls and Baurle 2001), but ataxia was not detected in mice with compound JNK-deficient Purkinje cells that were examined (Figs. 7, 8). This observation indicates that Purkinje cells can function without the JNK signaling pathway.
Figure 7.
Compound deficiency of JNK in neurons in vivo. Young adult (8-wk-old) Pcp2-Cre mice (control) and Pcp2-Cre Jnk1LoxP/LoxP Jnk2−/− Jnk3−/− mice (JNKTKO) that express Cre recombinase selectively in cerebellar Purkinje cells were examined. (A) Sections of the Purkinje cell layer of control and JNKTKO mice were examined by immunohistochemical staining with antibodies to JNK1/2. Bar, 100 μm. (B) Cerebellar DNA was examined by PCR analysis to detect Jnk1+ (1550-bp), Jnk1LoxP (1095-bp), Jnk1Δ (395-bp), Jnk2+ (400-bp), Jnk2− (270-bp), Jnk3+ (430-bp), and Jnk3− (250-bp) alleles. (C) Sections of the Purkinje cell layer and DCN of control and JNKTKO mice were examined by immunofluorescence staining with an antibody to Calbindin D-28k. Bar, 40 μm. (D) Serial sections of the DCN of control and JNKTKO mice were examined by staining with H&E and by immunohistochemical staining antibodies to Calbindin D-28k and GFAP. Bar, 100 μm. (E) The myelinated axons in the DCN of control and JNKTKO mice were examined by transmission electron microscopy. Bars: top panels, 2 μm; bottom panels 0.125μm. (F) The axon area and the number and area of autophagosomes and mitochondria in the myelinated axons of control and JNKTKO mice were measured. The data are presented as mean ± SEM of 20 axons of three different mice per group. Statistically significant differences between control and JNKTKO mice are indicated. (*) P < 0.05.
Figure 8.
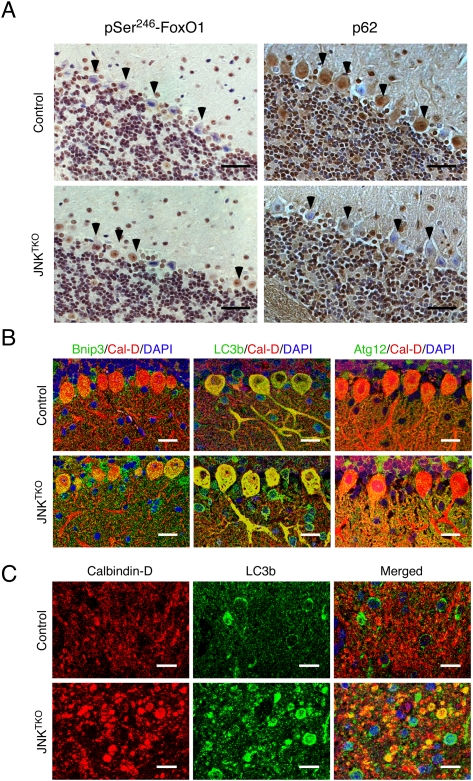
Compound JNK deficiency in Purkinje cells causes increased autophagy. Young adult (8-wk-old) Pcp2-Cre mice (control) and Pcp2-Cre Jnk1LoxP/LoxP Jnk2−/− Jnk3−/− mice (JNKTKO) that express Cre recombinase selectively in cerebellar Purkinje cells were examined. (A) Sections of the Purkinje cell layer of Pcp2-Cre mice (control) and Pcp2-Cre Jnk1LoxP/LoxP Jnk2−/− Jnk3−/− mice (JNKTKO) were examined by immunohistochemical staining with antibodies to pSer246-FoxO1 and p62/SQSTM1. Bar, 100 μm. (B) Sections of the Purkinje cell layer of control and JNKTKO mice were stained with antibodies (to Bnip3, LC3b, Atg12, and Calbidin-D28k) and examined by fluorescence microscopy. Bar, 20 μm. (C) Sections of the DCN of control and JNKTKO mice were stained with antibodies (to Calbindin D-28k and LC3b) and examined by fluorescence microscopy. Bar, 10 μm.
Immunocytochemistry analysis demonstrated the loss of JNK protein in the Purkinje cell layer of the cerebellum (Fig. 7A), and genotype analysis of cerebellar DNA led to the identification of loss-of-function alleles of Jnk1, Jnk2, and Jnk3 (Fig. 7B). The JNKTKO Purkinje cells exhibited reduced dendritic arborization (Supplemental Fig. S14). Immunofluorescence analysis using an antibody to Calbindin D-28k indicated the presence of hypertrophic Purkinje cell axons in deep cerebellar nuclei (DCN) (Fig. 7C). These hypertrophic axons were also identified in sections of the JNKTKO DCN stained with H&E (Fig. 7D), by immunohistochemical staining with an antibody to Calbindin D-28k (Fig. 7D), and staining using the Golgi reagent (Supplemental Fig. S14). Staining with an antibody to GFAP demonstrated that the axonal hypertrophy was associated with reactive gliosis (Fig. 7D). Electron microscopy confirmed the hypertrophy of myelinated Purkinje cell axons in the DCN of JNKTKO mice (Fig. 7E; Supplemental Fig. S15). Quantitative image analysis demonstrated that the cross-sectional area of Purkinje cell axons was significantly larger in the DCN of JNKTKO mice compared with control mice (Fig. 7F). Fewer axonal mitochondria and increased numbers of autophagosomes were detected in JNKTKO mice compared with control mice (Fig. 7F). In contrast, the size of both autophagosomes and mitochondria were increased in JNKTKO mice compared with control mice (Fig. 7F).
Neuronal JNK deficiency causes increased autophagy in vivo
The observation that compound JNK deficiency causes increased autophagy in primary cultures of neurons in vitro (Fig. 3) suggests that JNK may suppress neuronal autophagy in vivo. To test this hypothesis, we examined autophagy in mice with triple deficiency of JNK1, JNK2, and JNK3 in Purkinje cells (Fig. 8). Electron microscopy demonstrated that autophagy was influenced by compound JNK deficiency because the size of axonal autophagosomes in the DCN was significantly increased compared with control mice (Fig. 7F). However, the altered size of autophagosomes could be caused by either an increase or a decrease in neuronal autophagy. We therefore examined the amount of p62/SQSTM1 protein (which directly binds the autophagic effector Atg8/LC3) (Pankiv et al. 2007) in Purkinje cells by immunohistochemistry. The p62/SQSTM1 protein was detected in the Purkinje cell soma of control mice, but not in mice with compound deficiency of JNK in Purkinje cells (Fig. 8A). This loss of p62/SQSTM1 suggests that autophagic flux is increased in JNKTKO neurons compared with control neurons (Klionsky et al. 2008). The increased autophagy was associated with nuclear phosphorylation of the transcription factor FoxO1 on the activating site Ser246 (Fig. 8A) and increased expression of Bnip3 and Atg12 (Fig. 8B). The amount of LC3b in the Purkinje cell soma was moderately increased in compound JNK-deficient Purkinje cells (Fig. 8B), but a large increase in LC3b was detected in Purkinje cell axons within the DCN (Fig. 8C). Together, these data indicate that the FoxO1–Bnip3 pathway that induces autophagy is activated in compound JNK-deficient Purkinje cells in vivo.
Discussion
Studies of nonneuronal cells have implicated JNK in the induction of autophagy (Yu et al. 2004; Ogata et al. 2006; Wei et al. 2008). Indeed, we confirmed the conclusion that JNK can contribute to increased autophagy by examining primary mouse embryonic fibroblasts (MEFs) with compound JNK deficiency (Supplemental Fig. S13). The mechanism of JNK-induced autophagy may be mediated by phosphorylation of Bcl2 by JNK and the subsequent release of the autophagic effector Beclin-1 (Wei et al. 2008). The sites of JNK phosphorylation on Bcl2 (Yamamoto et al. 1999) are conserved in the related protein Bcl-XL (Kharbanda et al. 2000; Upreti et al. 2008). This conservation suggests that phosphorylation of Bcl2 and Bcl-XL is functionally important. Phosphorylation of Bcl2 and Bcl-XL by JNK (Yamamoto et al. 1999; Kharbanda et al. 2000) and other protein kinases (Tournier et al. 2001; Terrano et al. 2010) may represent an important mechanism of autophagy regulation (Wei et al. 2008). Indeed, the properties of JNK as a stress-responsive kinase provide an elegant mechanism for coupling stress exposure to the induction of autophagy (Ogata et al. 2006).
The JNK signaling pathway suppresses neuronal autophagy
Studies of nonneuronal cells demonstrate that JNK is markedly activated from a low basal state when cells are exposed to stress (Davis 2000). However, JNK is regulated very differently in neurons. JNK1 remains constitutively activated under basal conditions, while JNK2 and JNK3 exhibit low basal activity and are stress-responsive (Coffey et al. 2000, 2002). The proautophagy role of JNK in nonneuronal cells has been reported to be mediated by JNK1 (Wei et al. 2008). It is therefore intriguing that JNK1 is constitutively activated in neurons. Based on studies of nonneuronal cells (Wei et al. 2008), the constitutive activation of JNK1 in neurons should cause autophagy. A mechanism must therefore exist to prevent autophagy activation by constitutively activated JNK1 in neurons. Although the mechanism is unclear, these considerations indicate that neurons are refractory to the proautophagy JNK1 signaling pathway that has been identified in nonneuronal cells (Wei et al. 2008).
Our analysis of compound JNK-deficient neurons demonstrates that JNK regulates neuronal autophagy. In contrast to the proautophagy role of JNK nonneuronal cells, neuronal JNK acts to suppress autophagy. Loss of neuronal JNK function causes engagement of a transcriptional program that leads to increased expression of autophagy-related genes and the induction of an autophagic response (Fig. 3). One consequence of autophagy induction caused by JNK deficiency is improved neuronal survival (Figs. 2; Supplemental Fig. S3).
JNK can act as a molecular switch that regulates FoxO-induced autophagy and apoptosis
FoxO transcription factors are implicated in the induction of both cell death (apoptosis) and cell survival (autophagy) responses (Salih and Brunet 2008). The results of this study identify JNK as a signaling molecule that may contribute to the coordination of these divergent responses to FoxO transcription factor activation.
FoxO activation in neurons leads to the expression of the target gene Bim, a proapoptotic BH3-only protein, and causes cell death (Gilley et al. 2003). JNK activation in neurons promotes expression of Bim, most likely because JNK-dependent AP-1 activity is required for Bim expression (Whitfield et al. 2001). Moreover, JNK phosphorylates Bim on an activating site (Hubner et al. 2008), and also causes the release of Bim from complexes with the anti-apoptotic Bcl2 family protein Mcl-1 (Morel et al. 2009). Together, these processes initiate JNK-dependent apoptosis. JNK inhibition can therefore prevent neuronal cell death. Indeed, small molecule inhibitors of JNK cause neuroprotection in models of neurodegenerative disease (Borsello et al. 2003; Hirt et al. 2004; Repici et al. 2007; Carboni et al. 2008; Esneault et al. 2008; Wiegler et al. 2008; Probst et al. 2011).
Activation of FoxO transcription factors can also cause increased expression of autophagy-related genes, including Atg8/Lc3b, Atg12, and Bnip3 (Salih and Brunet 2008). While JNK cooperates with FoxO to increase proapoptotic Bim expression (Whitfield et al. 2001), JNK deficiency prevents induction of Bim expression (Fig. 3E) and promotes a survival response that is mediated by increased FoxO-dependent expression of the autophagy-related target genes Atg8/Lc3b, Atg12, and Bnip3 (Figs. 3E, 5B–D). Indeed, inhibition of autophagy in JNK-deficient neurons causes rapid death (Figs. 2C, 4C). This neuronal survival response is relevant to stroke models in which neuronal death is mediated by a JNK-dependent mechanism (Kuan et al. 2003; Pirianov et al. 2007).
Together, these data demonstrate that cross-talk between the FoxO and JNK signaling pathways leads to neuronal death. In contrast, loss of JNK promotes FoxO-induced survival mediated by increased autophagy. JNK therefore acts as a molecular switch that defines the physiological consequence of FoxO activation in neurons.
Conclusions
JNK is implicated in the induction of autophagy in nonneuronal cells. However, JNK1 is constitutively activated in neurons, and these cells are refractory to JNK-induced autophagy. Instead, JNK acts to suppress autophagy in neurons by inhibiting FoxO-induced expression of autophagy-related genes (e.g., Atg8/Lc3b, Atg12, and Bnip3) and increasing the expression of proapoptotic genes (e.g., Bim). JNK inhibition causes neuroprotection that is mediated by loss of proapoptotic gene expression and increased autophagy.
Materials and methods
Mice
We described Jnk1−/− mice (Dong et al. 1998), Jnk1LoxP/LoxP mice (Das et al. 2007), Jnk2−/− mice (Yang et al. 1998), Jnk2M108G/M108G mice (Jaeschke et al. 2006), and Jnk3−/− mice (Yang et al. 1997). B6.129-Tg(Pcp2-cre)2Mpin/J mice (Barski et al. 2000), B6.Cg-Tg(Nes-cre)1Kln/J mice (Tronche et al. 1999), and B6.129P2(Cg)-Foxg1tm1(cre)Skm/J mice (Eagleson et al. 2007) were obtained from The Jackson Laboratories. These mice were backcrossed to the C57BL/6J strain (Jackson Laboratories) and were housed in a facility accredited by the American Association for Laboratory Animal Care. The animal studies were approved by the Institutional Animal Care and Use Committee of the University of Massachusetts Medical School.
Genotype analysis
Genomic DNA was examined by PCR analysis using primers to identify wild-type and Jnk1− (Dong et al. 1998), Jnk1LoxP (Das et al. 2007), Jnk1Δ (Das et al. 2007), Jnk2− (Yang et al. 1998), Jnk2M108G (Jaeschke et al. 2006), Jnk3− (Yang et al. 1997), and Cre+ (Das et al. 2007) alleles.
Tissue culture
Wild-type and Jnk1−/− Jnk2−/− MEFs (Tournier et al. 2000) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum (Invitrogen). Primary cultures of CGNs were prepared from postnatal day 6 mice (Kennedy et al. 2007). The CGNs were cultured 2 d in vitro with neurobasal medium containing B27 supplements, 1% glutamine, 1% penicillin/streptomycin, 25 mM glucose, and 25 mM KCl; seeded in poly-D-lysine/laminin-coated chamber slides (Becton Dickenson) or dishes (MatTek); and then infected with adenovirus-Cre (Ad5CMVCre; ∼100 multiplicity of infection) (Gene Transfer Vector Core, University of Iowa) each day for 3 d. RNAi transfection studies were performed using the PepMute siRNA transfection reagent (SignaGen Laboratories) with 20 nM siRNA (NM_019584 or NM_019739, Dharmacon RNA Technologies) at 7 d of culture in vivo (DIV) and again at 8 DIV. Some cultures were treated with 1 μM 1-naphthylmethyl-4-amino-1-tert-butyl-3-(p-methylphenyl)pyrazolo[3,4-d]pyrimidine (1NM-PP1; Calbiochem), 1 μM chloroquine (Sigma), or 5 μM roscovitine (LC Laboratories). Neurons were also stained with calcein-am ester (Calbiochem) and imaged by confocal fluorescence microscopy with a Leica SP2 instrument (Kennedy et al. 2007).
RNA analysis
The expression of mRNA was examined by quantitative PCR analysis using a 7500 Fast Real-Time PCR machine (Applied Biosystems). TaqMan assays were used to quantitate Atg3 (Mm00471287_m1), Atg5 (Mm00504340_m1), Atg7 (Mm00512209_m1), Atg8/Lc3b (Mm00782868_m1), Atg12 (Mm00503201_m1), Beclin-1 (Mm01265461_m1), Bim (Mm01975020_s1), Bnip3 (Mm01275601_g1), FoxO1 (Mm00490672_M1), Gapdh (4352339E), Kif3a (Mm00492876_m1), Kif5a (Mm00515258_m1), Kif5b (Mm00515276_m1), and Kif5c (Mm00515265_m1) (Applied Biosystems). The relative mRNA expression was normalized by measurement of the amount of Gapdh mRNA in each sample using TaqMan assays (Applied Biosystems).
Immunoblot analysis
Cell extracts were prepared using Triton lysis buffer (20 mM Tris at pH 7.4, 1% Triton X-100, 10% glycerol, 137 mM NaCl, 2 mM EDTA, 25 mM β-glycerophosphate, 1 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride, 10 μg/mL aprotinin and leupeptin). Extracts (20–50 μg of protein) were examined by protein immunoblot analysis by probing with antibodies to LC3b (Novus Biologicals); p62/SQSTM1; CDK-2 (Santa Cruz Biotechnology); AKT, pSer308-AKT, pSer473-AKT, Beclin-1, and Bcl-XL; pThr389-S6Kand S6K (Cell Signaling); JNK1/2 (BD Biosciences Pharmingen); phospho-neurofilament H (SMI-31R, Covance); phospho-serine, Bnip3, and FoxO1 (Abcam); and α-Tubulin (Sigma). The antibody to pSer246-FoxO1 was provided by Dr Azad Bonni (Yuan et al. 2008). Immunecomplexes were detected by enhanced chemiluminescence (NEN). Immunoblot analysis of immunoprecipitates was performed using the One-Step Complete Immunoprecipitation-Western kit (Genescript Corp.).
Protein kinase assays
CDK2 activity was measured in an in vitro kinase assay using Rb-C fusion protein (Cell Signaling) as the substrate, and was quantitated using a PhosphorImager (Molecular Dynamics).
Immunofluorescence analysis
Primary CGNs were fixed by incubation with 4% paraformaldehyde for 1 h at room temperature and were permeabilized by incubation with 90% methanol containing 5% acetic acid for 5 min at −20°C. The slides were then blocked with 1% skim milk in phosphate-buffered saline (PBS) for 1 h at room temperature and incubated with antibodies to phospho-Ser63-cJun (Cell Signaling), detyrosinated Tubulin, Synaptophysin, and Tau (Chemicon); Ankyrin G and Lamp-1 (Santa Cruz Biotechnology); Snap25 and FoxO1 (Abcam); LC3b (Novus Biologicals); and βIII-Tubulin (Covance) in PBS supplemented with 1% skim milk overnight at 4°C. Secondary antibodies were conjugated with Alexa Fluor 488 or 546 (Molecular Probes) for 1 h at room temperature. CGNs were loaded with 100 nM MitoTracker Red (Molecular Probes) for 15 min at 37°C. All washed slides were mounted with VectaShield mounting medium with DAPI (Vector Laboratories) and were examined with a Leica SP2 laser-scanning confocal fluorescence microscope.
Time-lapse fluorescence microscopy
The CGNs were cultured 12 d in vitro in poly-D-lysine/laminin-coated 35-mm glass-bottom microwell dishes (MatTek) and incubated with 100 nM MitoTracker Green (Molecular Probes) for 3 min. Time-lapse fluorescence microscopy of CGN cells was performed using a Nikon TE2000-E2 microscope with a Yokogawa CSU10b spinning-disk confocal scan head and custom laser launch, acoustical optical tunable filter (NEOS), and relay optics (Solamere Technology Group). Multiwavelength confocal Z-series were acquired with a Nikon 60× Plan Apo oil objective (NA = 1.4) and a QImaging Rolera MGi camera using the digitizer with electron multiplication gain. Metamorph software controlled the microscope hardware and image acquisition. The frames were collected every 3 secs with an exposure time of 100 msec.
Electron microscopy
Cells and tissue were fixed with 1.25% glutaraldehyde for 30 min at room temperature and with 2.5% gluteraldehyde in cacodylate buffer for 14 h at 4°C. The cells were then post-fixed with 1% (w/v) osmium tetraoxide in PBS, dehydrated, and embedded in Lx 112/Araldite 502 epoxy resin. Ultrathin sections were mounted on copper support grids in serial order, contrasted with lead citrate and uranyl acetate, and examined on a Philips CM 10 transmission electron microscope (Gangwani et al. 2005). Quantitation of electron micrographs was performed by image analysis using the program AxioVision release 4.5 (Zeiss).
Immunohistochemical and immunofluorescence analysis of tissue sections
Perfusion fixation of mice was performed using PBS supplemented with 4% (w/v) paraformaldehyde. Fixed tissues (24 h at 4°C) were processed and embedded in paraffin, and 4-μm sections were prepared. These sections were stained with antibodies to JNK1/2 (BD Biosciences Pharmingen), p62/SQSTM1 (Abnova), or pSer246-FOXO1 (Yuan et al. 2008) using indirect immunoperoxidase detection (Xu et al. 1998). Sections were also stained by immunofluorescence after paraffin removal using antigen retrieval with antigen unmasking solution (Vector Laboratories) and microwave irradiation. The sections were subsequently blocked with 0.4% Triton X-100, 10% goat serum, 150 mM NaCl, and 10 mM Tris-HCl (pH 7.4). Sections were incubated with antibodies to Calbindin D-28k (Sigma), Bnip3 and Atg12 (Cell Signaling), or LC3b (Novus Biologicals) for 12 h at 4°C and washed. Immunecomplexes were detected by incubation with secondary antibodies conjugated to Alexa Fluor 488 or 546 (Molecular Probes) for 1 h at 25°C. The slides were washed and mounted with VectaShield mounting medium with DAPI (Vector Laboratories) and examined with a Leica SP2 laser-scanning confocal fluorescence microscope. Frozen sections (100 μm) of the cerebellum were processed using the Rapid Golgi stain kit (FD NeuroTechnologies).
Statistical analysis
Differences between groups were examined for statistical significance using the Student's test or analysis of variance (ANOVA) with the Fisher's test.
Acknowledgments
We thank Azad Bonni for providing the pSer246-FoxO1 antibody; Tammy Barrett, Vicky Benoit, and Jian-Hua Liu for expert technical assistance; and Kathy Gemme for administrative assistance. These studies were supported by a grant from the National Institutes of Health (NS054948). Core facilities at the University of Massachusetts used by these studies were supported by the NIDDK Diabetes and Endocrinology Research Center (DK32520). R.J.D. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1984311.
Supplemental material is available for this article.
References
- Barnat M, Enslen H, Propst F, Davis RJ, Soares S, Nothias F 2010. Distinct roles of c-Jun N-terminal kinase isoforms in neurite initiation and elongation during axonal regeneration. J Neurosci 30: 7804–7816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski JJ, Dethleffsen K, Meyer M 2000. Cre recombinase expression in cerebellar Purkinje cells. Genesis 28: 93–98 [PubMed] [Google Scholar]
- Bjorkblom B, Ostman N, Hongisto V, Komarovski V, Filen JJ, Nyman TA, Kallunki T, Courtney MJ, Coffey ET 2005. Constitutively active cytoplasmic c-Jun N-terminal kinase 1 is a dominant regulator of dendritic architecture: Role of microtubule-associated protein 2 as an effector. J Neurosci 25: 6350–6361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsello T, Clarke PG, Hirt L, Vercelli A, Repici M, Schorderet DF, Bogousslavsky J, Bonny C 2003. A peptide inhibitor of c-Jun N-terminal kinase protects against excitotoxicity and cerebral ischemia. Nat Med 9: 1180–1186 [DOI] [PubMed] [Google Scholar]
- Brecht S, Kirchhof R, Chromik A, Willesen M, Nicolaus T, Raivich G, Wessig J, Waetzig V, Goetz M, Claussen M, et al. 2005. Specific pathophysiological functions of JNK isoforms in the brain. Eur J Neurosci 21: 363–377 [DOI] [PubMed] [Google Scholar]
- Brownlees J, Yates A, Bajaj NP, Davis D, Anderton BH, Leigh PN, Shaw CE, Miller CC 2000. Phosphorylation of neurofilament heavy chain side-arms by stress activated protein kinase-1b/Jun N-terminal kinase-3. J Cell Sci 113: 401–407 [DOI] [PubMed] [Google Scholar]
- Carboni S, Boschert U, Gaillard P, Gotteland JP, Gillon JY, Vitte PA 2008. AS601245, a c-Jun NH2-terminal kinase (JNK) inhibitor, reduces axon/dendrite damage and cognitive deficits after global cerebral ischaemia in gerbils. Br J Pharmacol 153: 157–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Jones Y, Ellisman MH, Goldstein LS, Karin M 2003. JNK1 is required for maintenance of neuronal microtubules and controls phosphorylation of microtubule-associated proteins. Dev Cell 4: 521–533 [DOI] [PubMed] [Google Scholar]
- Chen JT, Lu DH, Chia CP, Ruan DY, Sabapathy K, Xiao ZC 2005. Impaired long-term potentiation in c-Jun N-terminal kinase 2-deficient mice. J Neurochem 93: 463–473 [DOI] [PubMed] [Google Scholar]
- Coffey ET, Hongisto V, Dickens M, Davis RJ, Courtney MJ 2000. Dual roles for c-Jun N-terminal kinase in developmental and stress responses in cerebellar granule neurons. J Neurosci 20: 7602–7613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey ET, Smiciene G, Hongisto V, Cao J, Brecht S, Herdegen T, Courtney MJ 2002. c-Jun N-terminal protein kinase (JNK) 2/3 is specifically activated by stress, mediating c-Jun activation, in the presence of constitutive JNK1 activity in cerebellar neurons. J Neurosci 22: 4335–4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das M, Jiang F, Sluss HK, Zhang C, Shokat KM, Flavell RA, Davis RJ 2007. Suppression of p53-dependent senescence by the JNK signal transduction pathway. Proc Natl Acad Sci 104: 15759–15764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RJ 2000. Signal transduction by the JNK group of MAP kinases. Cell 103: 239–252 [DOI] [PubMed] [Google Scholar]
- Dong C, Yang DD, Wysk M, Whitmarsh AJ, Davis RJ, Flavell RA 1998. Defective T cell differentiation in the absence of Jnk1. Science 282: 2092–2095 [DOI] [PubMed] [Google Scholar]
- Eagleson KL, Schlueter McFadyen-Ketchum LJ, Ahrens ET, Mills PH, Does MD, Nickols J, Levitt P 2007. Disruption of Foxg1 expression by knock-in of cre recombinase: Effects on the development of the mouse telencephalon. Neuroscience 148: 385–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esneault E, Castagne V, Moser P, Bonny C, Bernaudin M 2008. D-JNKi, a peptide inhibitor of c-Jun N-terminal kinase, promotes functional recovery after transient focal cerebral ischemia in rats. Neuroscience 152: 308–320 [DOI] [PubMed] [Google Scholar]
- Gangwani L, Flavell RA, Davis RJ 2005. ZPR1 is essential for survival and is required for localization of the survival motor neurons (SMN) protein to Cajal bodies. Mol Cell Biol 25: 2744–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gdalyahu A, Ghosh I, Levy T, Sapir T, Sapoznik S, Fishler Y, Azoulai D, Reiner O 2004. DCX, a new mediator of the JNK pathway. EMBO J 23: 823–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilley J, Coffer PJ, Ham J 2003. FOXO transcription factors directly activate bim gene expression and promote apoptosis in sympathetic neurons. J Cell Biol 162: 613–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin JW, Watson DF 1988. Axonal transport in neurological disease. Ann Neurol 23: 3–13 [DOI] [PubMed] [Google Scholar]
- Grusser-Cornehls U, Baurle J 2001. Mutant mice as a model for cerebellar ataxia. Prog Neurobiol 63: 489–540 [DOI] [PubMed] [Google Scholar]
- Guertin DA, Sabatini DM 2007. Defining the role of mTOR in cancer. Cancer Cell 12: 9–22 [DOI] [PubMed] [Google Scholar]
- Gupta S, Barrett T, Whitmarsh AJ, Cavanagh J, Sluss HK, Derijard B, Davis RJ 1996. Selective interaction of JNK protein kinase isoforms with transcription factors. EMBO J 15: 2760–2770 [PMC free article] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, et al. 2006. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441: 885–889 [DOI] [PubMed] [Google Scholar]
- Hirt L, Badaut J, Thevenet J, Granziera C, Regli L, Maurer F, Bonny C, Bogousslavsky J 2004. D-JNKI1, a cell-penetrating c-Jun-N-terminal kinase inhibitor, protects against cell death in severe cerebral ischemia. Stroke 35: 1738–1743 [DOI] [PubMed] [Google Scholar]
- Hubner A, Barrett T, Flavell RA, Davis RJ 2008. Multisite phosphorylation regulates Bim stability and apoptotic activity. Mol Cell 30: 415–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner A, Cavanagh-Kyros J, Rincon M, Flavell RA, Davis RJ 2010. Functional cooperation of the proapoptotic Bcl2 family proteins Bmf and Bim in vivo. Mol Cell Biol 30: 98–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeschke A, Karasarides M, Ventura JJ, Ehrhardt A, Zhang C, Flavell RA, Shokat KM, Davis RJ 2006. JNK2 is a positive regulator of the cJun transcription factor. Mol Cell 23: 899–911 [DOI] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T 2004. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci 117: 2805–2812 [DOI] [PubMed] [Google Scholar]
- Kennedy NJ, Martin G, Ehrhardt AG, Cavanagh-Kyros J, Kuan CY, Rakic P, Flavell RA, Treistman SN, Davis RJ 2007. Requirement of JIP scaffold proteins for NMDA-mediated signal transduction. Genes Dev 21: 2336–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharbanda S, Saxena S, Yoshida K, Pandey P, Kaneki M, Wang Q, Cheng K, Chen YN, Campbell A, Sudha T, et al. 2000. Translocation of SAPK/JNK to mitochondria and interaction with Bcl-x(L) in response to DNA damage. J Biol Chem 275: 322–327 [DOI] [PubMed] [Google Scholar]
- Khawaja S, Gundersen GG, Bulinski JC 1988. Enhanced stability of microtubules enriched in detyrosinated tubulin is not a direct function of detyrosination level. J Cell Biol 106: 141–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim AH, Bonni A 2008. Cdk1-FOXO1: A mitotic signal takes center stage in post-mitotic neurons. Cell Cycle 7: 3819–3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, et al. 2008. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 4: 151–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, et al. 2006. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441: 880–884 [DOI] [PubMed] [Google Scholar]
- Kuan CY, Yang DD, Samanta Roy DR, Davis RJ, Rakic P, Flavell RA 1999. The Jnk1 and Jnk2 protein kinases are required for regional specific apoptosis during early brain development. Neuron 22: 667–676 [DOI] [PubMed] [Google Scholar]
- Kuan CY, Whitmarsh AJ, Yang DD, Liao G, Schloemer AJ, Dong C, Bao J, Banasiak KJ, Haddad GG, Flavell RA, et al. 2003. A critical role of neural-specific JNK3 for ischemic apoptosis. Proc Natl Acad Sci 100: 15184–15189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuan CY, Schloemer AJ, Lu A, Burns KA, Weng WL, Williams MT, Strauss KI, Vorhees CV, Flavell RA, Davis RJ, et al. 2004. Hypoxia-ischemia induces DNA synthesis without cell proliferation in dying neurons in adult rodent brain. J Neurosci 24: 10763–10772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb JA, Ventura JJ, Hess P, Flavell RA, Davis RJ 2003. JunD mediates survival signaling by the JNK signal transduction pathway. Mol Cell 11: 1479–1489 [DOI] [PubMed] [Google Scholar]
- Li XM, Li CC, Yu SS, Chen JT, Sabapathy K, Ruan DY 2007. JNK1 contributes to metabotropic glutamate receptor-dependent long-term depression and short-term synaptic plasticity in the mice area hippocampal CA1. Eur J Neurosci 25: 391–396 [DOI] [PubMed] [Google Scholar]
- Morel C, Carlson SM, White FM, Davis RJ 2009. Mcl-1 integrates the opposing actions of signaling pathways that mediate survival and apoptosis. Mol Cell Biol 29: 3845–3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morfini G, Pigino G, Szebenyi G, You Y, Pollema S, Brady ST 2006. JNK mediates pathogenic effects of polyglutamine-expanded androgen receptor on fast axonal transport. Nat Neurosci 9: 907–916 [DOI] [PubMed] [Google Scholar]
- Morfini GA, You YM, Pollema SL, Kaminska A, Liu K, Yoshioka K, Bjorkblom B, Coffey ET, Bagnato C, Han D, et al. 2009. Pathogenic huntingtin inhibits fast axonal transport by activating JNK3 and phosphorylating kinesin. Nat Neurosci 12: 864–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K, et al. 2006. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol 26: 9220–9231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282: 24131–24145 [DOI] [PubMed] [Google Scholar]
- Pirianov G, Brywe KG, Mallard C, Edwards AD, Flavell RA, Hagberg H, Mehmet H 2007. Deletion of the c-Jun N-terminal kinase 3 gene protects neonatal mice against cerebral hypoxic–ischaemic injury. J Cereb Blood Flow Metab 27: 1022–1032 [DOI] [PubMed] [Google Scholar]
- Podkowa M, Zhao X, Chow CW, Coffey ET, Davis RJ, Attisano L 2010. Microtubule stabilization by bone morphogenetic protein receptor-mediated scaffolding of c-Jun N-terminal kinase promotes dendrite formation. Mol Cell Biol 30: 2241–2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst GD, Bowers S, Sealy JM, Truong AP, Hom RK, Galemmo RA Jr, Konradi AW, Sham HL, Quincy DA, Pan H, et al. 2011. Highly selective c-Jun N-terminal kinase (JNK) 2 and 3 inhibitors with in vitro CNS-like pharmacokinetic properties prevent neurodegeneration. Bioorg Med Chem Lett 21: 315–319 [DOI] [PubMed] [Google Scholar]
- Repici M, Centeno C, Tomasi S, Forloni G, Bonny C, Vercelli A, Borsello T 2007. Time-course of c-Jun N-terminal kinase activation after cerebral ischemia and effect of D-JNKI1 on c-Jun and caspase-3 activation. Neuroscience 150: 40–49 [DOI] [PubMed] [Google Scholar]
- Sabapathy K, Jochum W, Hochedlinger K, Chang L, Karin M, Wagner EF 1999. Defective neural tube morphogenesis and altered apoptosis in the absence of both JNK1 and JNK2. Mech Dev 89: 115–124 [DOI] [PubMed] [Google Scholar]
- Salih DA, Brunet A 2008. FoxO transcription factors in the maintenance of cellular homeostasis during aging. Curr Opin Cell Biol 20: 126–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze E, Asai DJ, Bulinski JC, Kirschner M 1987. Posttranslational modification and microtubule stability. J Cell Biol 105: 2167–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S, Konishi A, Nishida Y, Mizuta T, Nishina H, Yamamoto A, Tsujimoto Y 2010. Involvement of JNK in the regulation of autophagic cell death. Oncogene 29: 2070–2082 [DOI] [PubMed] [Google Scholar]
- Sou YS, Waguri S, Iwata JI, Ueno T, Fujimura T, Hara T, Sawada N, Yamada A, Mizushima N, Uchiyama Y, et al. 2008. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol Biol Cell 19: 4762–4775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Kanai Y, Okada Y, Nonaka S, Takeda S, Harada A, Hirokawa N 1998. Targeted disruption of mouse conventional kinesin heavy chain, kif5B, results in abnormal perinuclear clustering of mitochondria. Cell 93: 1147–1158 [DOI] [PubMed] [Google Scholar]
- Tararuk T, Ostman N, Li W, Bjorkblom B, Padzik A, Zdrojewska J, Hongisto V, Herdegen T, Konopka W, Courtney MJ, et al. 2006. JNK1 phosphorylation of SCG10 determines microtubule dynamics and axodendritic length. J Cell Biol 173: 265–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrano DT, Upreti M, Chambers TC 2010. Cyclin-dependent kinase 1-mediated Bcl-xL/Bcl-2 phosphorylation acts as a functional link coupling mitotic arrest and apoptosis. Mol Cell Biol 30: 640–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas GM, Lin DT, Nuriya M, Huganir RL 2008. Rapid and bi-directional regulation of AMPA receptor phosphorylation and trafficking by JNK. EMBO J 27: 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournier C, Hess P, Yang DD, Xu J, Turner TK, Nimnual A, Bar-Sagi D, Jones SN, Flavell RA, Davis RJ 2000. Requirement of JNK for stress-induced activation of the cytochrome c-mediated death pathway. Science 288: 870–874 [DOI] [PubMed] [Google Scholar]
- Tournier C, Dong C, Turner TK, Jones SN, Flavell RA, Davis RJ 2001. MKK7 is an essential component of the JNK signal transduction pathway activated by proinflammatory cytokines. Genes & Dev 15: 1419–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronche F, Kellendonk C, Kretz O, Gass P, Anlag K, Orban PC, Bock R, Klein R, Schutz G 1999. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet 23: 99–103 [DOI] [PubMed] [Google Scholar]
- Upreti M, Galitovskaya EN, Chu R, Tackett AJ, Terrano DT, Granell S, Chambers TC 2008. Identification of the major phosphorylation site in Bcl-xL induced by microtubule inhibitors and analysis of its functional significance. J Biol Chem 283: 35517–35525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waetzig V, Herdegen T 2005. Context-specific inhibition of JNKs: Overcoming the dilemma of protection and damage. Trends Pharmacol Sci 26: 455–461 [DOI] [PubMed] [Google Scholar]
- Waetzig V, Zhao Y, Herdegen T 2006. The bright side of JNKs-multitalented mediators in neuronal sprouting, brain development and nerve fiber regeneration. Prog Neurobiol 80: 84–97 [DOI] [PubMed] [Google Scholar]
- Webster DR, Gundersen GG, Bulinski JC, Borisy GG 1987. Assembly and turnover of detyrosinated tubulin in vivo. J Cell Biol 105: 265–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Pattingre S, Sinha S, Bassik M, Levine B 2008. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell 30: 678–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston CR, Davis RJ 2007. The JNK signal transduction pathway. Curr Opin Cell Biol 19: 142–149 [DOI] [PubMed] [Google Scholar]
- Whitfield J, Neame SJ, Paquet L, Bernard O, Ham J 2001. Dominant-negative c-Jun promotes neuronal survival by reducing BIM expression and inhibiting mitochondrial cytochrome c release. Neuron 29: 629–643 [DOI] [PubMed] [Google Scholar]
- Wiegler K, Bonny C, Coquoz D, Hirt L 2008. The JNK inhibitor XG-102 protects from ischemic damage with delayed intravenous administration also in the presence of recombinant tissue plasminogen activator. Cerebrovasc Dis 26: 360–366 [DOI] [PubMed] [Google Scholar]
- Xu P, Hashimoto S, Miyazaki H, Asabe K, Shiraishi S, Sueishi K 1998. Morphometric analysis of the immunohistochemical expression of Clara cell 10-kDa protein and surfactant apoproteins A and B in the developing bronchi and bronchioles of human fetuses and neonates. Virchows Arch 432: 17–25 [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Ichijo H, Korsmeyer SJ 1999. BCL-2 is phosphorylated and inactivated by an ASK1/Jun N-terminal protein kinase pathway normally activated at G(2)/M. Mol Cell Biol 19: 8469–8478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DD, Kuan CY, Whitmarsh AJ, Rincon M, Zheng TS, Davis RJ, Rakic P, Flavell RA 1997. Absence of excitotoxicity-induced apoptosis in the hippocampus of mice lacking the Jnk3 gene. Nature 389: 865–870 [DOI] [PubMed] [Google Scholar]
- Yang DD, Conze D, Whitmarsh AJ, Barrett T, Davis RJ, Rincon M, Flavell RA 1998. Differentiation of CD4+ T cells to Th1 cells requires MAP kinase JNK2. Immunity 9: 575–585 [DOI] [PubMed] [Google Scholar]
- Yoshida H, Hastie CJ, McLauchlan H, Cohen P, Goedert M 2004. Phosphorylation of microtubule-associated protein tau by isoforms of c-Jun N-terminal kinase (JNK). J Neurochem 90: 352–358 [DOI] [PubMed] [Google Scholar]
- Yu L, Alva A, Su H, Dutt P, Freundt E, Welsh S, Baehrecke EH, Lenardo MJ 2004. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science 304: 1500–1502 [DOI] [PubMed] [Google Scholar]
- Yuan Z, Becker EB, Merlo P, Yamada T, DiBacco S, Konishi Y, Schaefer EM, Bonni A 2008. Activation of FOXO1 by Cdk1 in cycling cells and postmitotic neurons. Science 319: 1665–1668 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Pak D, Qin Y, McCormack SG, Kim MJ, Baumgart JP, Velamoor V, Auberson YP, Osten P, van Aelst L, et al. 2005. Rap2-JNK removes synaptic AMPA receptors during depotentiation. Neuron 46: 905–916 [DOI] [PubMed] [Google Scholar]