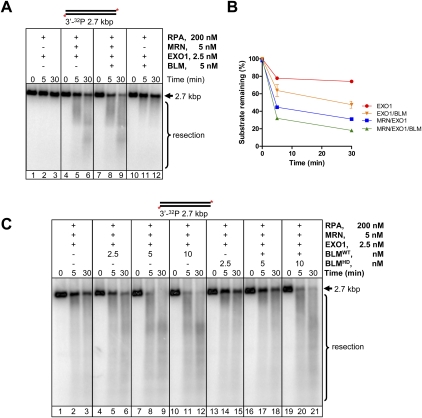
Abstract
Repair of dsDNA breaks requires processing to produce 3′-terminated ssDNA. We biochemically reconstituted DNA end resection using purified human proteins: Bloom helicase (BLM); DNA2 helicase/nuclease; Exonuclease 1 (EXO1); the complex comprising MRE11, RAD50, and NBS1 (MRN); and Replication protein A (RPA). Resection occurs via two routes. In one, BLM and DNA2 physically and specifically interact to resect DNA in a process that is ATP-dependent and requires BLM helicase and DNA2 nuclease functions. RPA is essential for both DNA unwinding by BLM and enforcing 5′ → 3′ resection polarity by DNA2. MRN accelerates processing by recruiting BLM to the end. In the other, EXO1 resects the DNA and is stimulated by BLM, MRN, and RPA. BLM increases the affinity of EXO1 for ends, and MRN recruits and enhances the processivity of EXO1. Our results establish two of the core machineries that initiate recombinational DNA repair in human cells.
Keywords: homologous recombination, DNA break repair, DNA end resection, BLM helicase, DNA2 nuclease, EXO1 nuclease
dsDNA breaks (DSBs) are generated by exogenous agents such as ionizing radiation and mutagenic chemicals. In addition, they arise endogenously from oxidative damage and replication fork collapse, and as programmed events during meiosis and immunoglobulin diversity generation (Wyman and Kanaar 2006). Regardless of their source, DNA breaks can lead to cell death if not repaired. In eukaryotes, two distinct pathways function to repair DSBs: nonhomologous end-joining and homologous recombination. Whereas the former mediates direct religation of broken ends, the latter uses the information from the undamaged homolog or sister chromatid to repair the break.
The homology-directed repair of DSBs is a multistep process requiring the activities of numerous proteins that act in a concerted fashion to re-establish the broken connection (Wyman and Kanaar 2006). Resection is one of the earliest steps of recombinational DNA repair, and is facilitated by nucleases and/or helicases that generate ssDNA. In Escherichia coli, DNA end resection is mediated by either RecBCD enzyme (Dillingham and Kowalczykowski 2008) or RecQ (a 3′ → 5′ helicase) and RecJ (a 5′ → 3′ nuclease) (Lovett and Kolodner 1989; Harmon and Kowalczykowski 1998; Handa et al. 2009). In vivo studies from Saccharomyces cerevisiae have identified two distinct pathways that act redundantly to generate long (>1-kb) resection products (Gravel et al. 2008; Mimitou and Symington 2008; Zhu et al. 2008): One pathway requires Exonuclease 1 (Exo1), a 5′ → 3′ dsDNA-specific nuclease originally identified in Schizosaccharomyces pombe (Szankasi and Smith 1992), and the other requires Sgs1 (a RecQ homolog) (Gangloff et al. 1994) and Dna2 (an ssDNA helicase/nuclease) (Budd et al. 2000). In addition, a third pathway with limited resection capability (less than a few hundred nucleotides), although sufficient for gene conversion, requires the activities of the Mre11–Rad50–Xrs2 complex (MRX) and Sae2 (Mimitou and Symington 2008; Zhu et al. 2008). MRX and Sae2 are not essential for the processing of breaks that have free ends (Llorente and Symington 2004), but are essential when the DNA breaks have covalent modifications or bulky adducts attached to the ends (Moreau et al. 1999; Gravel et al. 2008; Hartsuiker et al. 2009). The functions of the human homologs of yeast MRX and Sae2 (MRN and CtIP) are also essential for processing blocked DNA ends (Sartori et al. 2007; Buis et al. 2008). Recently, the Sgs1/Dna2 pathway has been biochemically reconstituted in vitro, verifying the in vivo findings, revealing an essential requirement for Replication protein A (RPA), and establishing a stimulatory function for MRX (Cejka et al. 2010a; Niu et al. 2010).
The identities of the DNA helicases and nucleases that are involved in DSB processing in humans are not yet as clearly defined as in yeast. This ambiguity exists, in part, because there are many candidate proteins. Although there are five RecQ homologs (Bloom helicase [BLM], Werner helicase/nuclease [WRN], RECQ1, RECQ4, and RECQ5) (Chu and Hickson 2009), convincing evidence implicates BLM in resection (Gravel et al. 2008; Nimonkar et al. 2008). The human ortholog of yeast Exo1 is EXO1 (or HEX1) (Schmutte et al. 1998; Tishkoff et al. 1998; Wilson et al. 1998). In human cells, depletion of either BLM or EXO1 using siRNA has no effect on camptothecin sensitivity, but codepletion results in increased sensitivity to camptothecin and impairment of RPA repair focus formation, indicative of a failure to produce ssDNA at the break (Gravel et al. 2008). Biochemical results indicate that EXO1 can act alone, but is stimulated by BLM to mediate DNA end resection without the need for the helicase activity of BLM (Nimonkar et al. 2008). Taken together, these findings suggest that BLM may function in two pathways: a stimulatory but nonessential role in conjunction with EXO1, and an essential but EXO1-independent role in combination with another nuclease. Although the identity of the nuclease that functions with BLM is unknown, on the basis of the yeast paradigm wherein Dna2 acts with the BLM ortholog, Sgs1 (Zhu et al. 2008), it can be hypothesized that the human homolog of Dna2, DNA2, may be the candidate nuclease. Human DNA2 displays a weak helicase activity, possesses a bipolar nuclease activity that can endonucleolytically degrade ssDNA with either a free 3′ or 5′ end, and plays an important role in DNA replication (Kim et al. 2006; Masuda-Sasa et al. 2006). Furthermore, recent work established that depletion of DNA2 results in chromosomal aberrations that are manifest as aneuploidy and the generation of internuclear chromatin bridges (Duxin et al. 2009). Although not much is known regarding the role of DNA2 in human DNA repair, it has been identified as a major DSB processing activity in Xenopus egg extracts (Liao et al. 2008; Wawrousek et al. 2010).
Thus, although recent genetic and biochemical studies have provided much information regarding the proteins involved in DSB repair in yeast, the process of DNA end resection in humans remains unclear. Here, using purified human proteins to reconstitute key elements of DNA resection in vitro, we show that BLM and DNA2 interact functionally and physically to form a core complex that resects DNA in the 5′ → 3′ direction in the presence of human RPA, and that MRN can enhance BLM- and DNA2-mediated resection. In addition, we demonstrate that MRN and RPA, as well as BLM (Nimonkar et al. 2008), stimulate the nuclease activity of EXO1. Our results define distinct roles for these proteins in DSB processing, and provide a mechanistic framework for understanding their role in recombinational DNA repair.
Results
BLM and DNA2 resect dsDNA
In vivo analyses in S. cerevisiae revealed that Sgs1 and Dna2 act together to mediate DNA end resection (Zhu et al. 2008). In vitro, the two proteins interact to form a core complex that promotes ATP-dependent processing of DNA ends (Cejka et al. 2010a; Niu et al. 2010). To determine whether the human orthologs of these two proteins perform a similar function, we tested the ability of BLM and DNA2 to resect dsDNA. We used three different substrates: a 2.7-kb plasmid-length DNA generated by linearization of pUC19, a 324-base-pair (bp) dsDNA fragment generated by PCR, and a 50-bp dsDNA fragment generated by annealing complementary oligonucleotides.
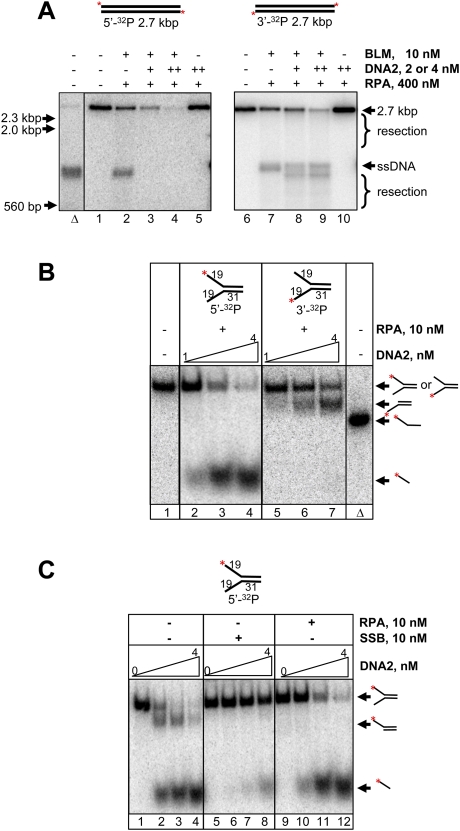
We first examined resection of the plasmid-length dsDNA that was labeled at the 5′ or 3′ end with 32P. Figure 1A shows that BLM could unwind the linear 2.7-kb substrate to produce ssDNA (lanes 2,7). Unwinding required RPA (Supplemental Fig. S1A,B; Brosh et al. 2000) and was less efficient at higher Mg2+ concentrations (Supplemental Fig. S1C,D), behaviors that are also manifest by the bacterial and yeast orthologs (Harmon and Kowalczykowski 2001; Cejka and Kowalczykowski 2010). Addition of DNA2 to reactions containing BLM and RPA led to disappearance of the 5′-end-labeled substrate (Fig. 1A, lanes 3,4). In the case of the 3′-end-labeled substrate, processing by BLM and DNA2 generated resection intermediates that appeared both just below the substrate and below the ssDNA unwinding product (Fig. 1A, lanes 8,9), and the extent of resection increased with time (Supplemental Fig. S1E, lanes 5–8). In the absence of BLM, DNA2 and RPA showed no activity on this substrate (Fig. 1A, lanes 5,10; Supplemental Fig. S1E, lane 9). We also examined processing of the shorter 5′- or 3′-end-labeled 324-bp duplex DNA, and similar results were obtained (Supplemental Fig. S1F). These data show that BLM and DNA2 can resect dsDNA ends, and collectively suggest that resection occurs on one strand, with a 5′ → 3′ polarity.
Figure 1.
BLM and DNA2 resect dsDNA. (A) Resection of plasmid-length dsDNA. Nuclease reactions were performed using 5′- or 3′-end-labeled 2.7-kb DNA; reactions contained 400 nM RPA and 2 mM MgCl2. (Lane Δ) Heat-denatured substrate. (Lanes 1,6) Substrate. (Lanes 2,7) BLM. (Lanes 3,8) BLM and 2 nM DNA2. (Lanes 4,9) BLM and 4 nM DNA2. (Lanes 5,10) DNA2 (4 nM). Single plus sign (+) and double plus signs (++) refer to 2 nM and 4 nM DNA2, respectively. The positions of the intact substrate (2.7 kb), unwound substrate (ssDNA), resection products, and molecular size markers are indicated. (B) DNA2 cleaves forked DNA with 5′ → 3′ polarity. Nuclease reactions with varying DNA2 concentrations (1, 2, and 4 nM) were performed using 5′- or 3′-end-labeled 50-bp forked DNA (1.5 nM ends, 75 nM nucleotides). Reactions contained 10 nM RPA and 5 mM MgCl2. (Lane 1) Substrate. (Lanes 2–4) Reactions with 5′-end-labeled fork. (Lanes 5–7) Reactions with 3′-end-labeled fork. (Lane Δ) Heat-denatured substrate. (C) RPA modulates DNA2-mediated cleavage of forked DNA. Nuclease reactions with DNA2 (0, 1, 2, and 4 nM) were performed using 5′-end-labeled 50-bp forked DNA in the absence or presence of indicated ssDNA-binding protein (RPA or E. coli SSB). All reactions contained 5 mM MgCl2. (Lanes 1–4) DNA2 alone. (Lanes 5–8) DNA2 and SSB. (Lanes 9–12) DNA2 and RPA. The positions of the intact fork, unwound substrate, and cleavage products are indicated schematically.
RPA enforces 5′ → 3′ resection by BLM and DNA2
To establish the direction of resection by DNA2, we examined cleavage of a forked DNA substrate labeled at 5′ or 3′ ends (Fig. 1B). Because the fork simulates partially unwound DNA, BLM was omitted from the analysis. With the 5′-end-labeled fork, a faster-migrating species consistent with cleavage of the labeled strand was observed (Fig. 1B, lanes 2–4). Correspondingly, with the 3′-end-labeled substrate, a product consistent with cleavage of the unlabeled strand was seen (Fig. 1B, lanes 5–7); unwinding of the 31-bp duplex region was not detected. We also analyzed the cleavage products by denaturing polyacrylamide gel electrophoresis (Supplemental Fig. S2A). The data show that, whereas the 5′-end-labeled strand was cleaved (Supplemental Fig. S2A, lane 2), the 3′-end-labeled strand stayed intact (Supplemental Fig. S2A, lane 3). The ssDNA product was ∼20 nucleotides (nt), indicating that DNA2 endonucleolytically cleaves the ssDNA arm near the junction with dsDNA, consistent with published data (Masuda-Sasa et al. 2006). These results show that DNA2 resects DNA with a strict 5′ → 3′ polarity in our assays.
Although the polarity of degradation by DNA2 displayed the bias expected from physical analysis of resection in vivo, it was nonetheless surprising given that DNA2 can degrade both 5′- and 3′-ssDNA tails of forked DNA (Masuda-Sasa et al. 2006). However, the nucleolytic behavior of DNA2, both human and yeast, is modulated by the cognate RPA (Bae et al. 2001; Masuda-Sasa et al. 2006, 2008; Cejka et al. 2010a; Niu et al. 2010). Consequently, we examined the role of RPA in resection by DNA2. In the absence of RPA, both ssDNA tails of the forked DNA substrate were cleaved by DNA2 (Fig. 1C, lanes 1–4); however, in the presence of RPA, DNA2 selectively promoted cleavage of the 5′ tail (Fig. 1C, lanes 9–12). E. coli SSB failed to have the same effects and, on the contrary, was inhibitory to DNA2-mediated cleavage (Fig. 1C, lanes 5–8). To further confirm the observed degradative bias, we tested DNA substrates containing 5′ or 3′ flaps that had been used previously to characterize DNA2. Supplemental Figure S2B shows that DNA2 can cleave 5′ as well as 3′ flaps in the absence of RPA (lanes 4,10). In its presence, however, RPA blocks degradation of the 3′-ssDNA flap (Supplemental Fig. S2B, lanes 11–13) but stimulates degradation of the 5′-ssDNA flap (Supplemental Fig. S2B, lanes 5–7). We therefore conclude that, even though DNA2 has the intrinsic capacity to degrade both 5′- and 3′-terminated ssDNA, RPA enforces a degradation bias that results in a unique polarity of nucleolytic action in the 5′ → 3′ direction.
The helicase activity of BLM and the nuclease activity of DNA2 are essential for resection
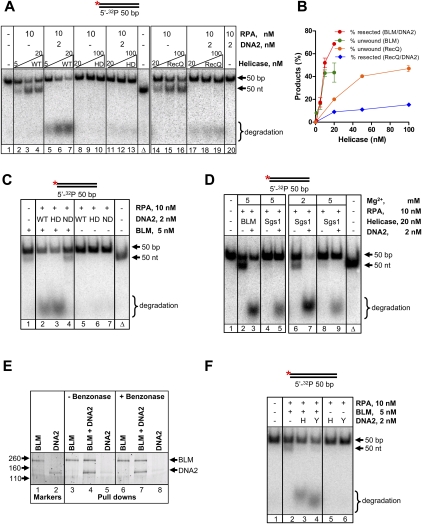
We next examined the contributions of BLM and DNA2 helicase activities in the resection process. We used a 50-bp dsDNA substrate to more accurately quantify DNA unwinding and resection. In the presence of RPA, BLM unwound this DNA (Fig. 2A, lanes 2–4), but DNA2 alone neither unwound nor degraded it (Fig. 2A, lane 20). However, DNA2 degraded this substrate when BLM was present to unwind the DNA (Fig. 2A, lanes 5–7; Supplemental Fig. S3A), consistent with the results obtained with the plasmid DNA (Fig. 1A). Omitting RPA decreased substrate unwinding and processing by BLM and DNA2 (Supplemental Fig. S3B, lane 3 vs. lane 7).
Figure 2.
BLM and DNA2 interact specifically to resect dsDNA. (A) BLM-mediated unwinding is essential for resection by BLM and DNA2. Nuclease reactions were performed using a 5′-end-labeled 50-bp DNA fragment. Where indicated, wild-type BLM (WT) was substituted with helicase-dead K695R mutant (HD) or E. coli RecQ. Reactions contained 10 nM RPA and 5 mM MgCl2. (Lane 1) Substrate. (Lanes 2–4,5–7) BLM (wild type) (5, 10, and 20 nM) in the absence or presence of DNA2, respectively. (Lanes 8–10,11–13) BLM (helicase-dead) (20, 50, and 100 nM) in the absence or presence of DNA2, respectively. (Lane Δ) Heat-denatured substrate. (Lanes 14–16,17–19) RecQ (20, 50, and 100 nM) in the absence or presence of DNA2, respectively. (Lane 20) DNA2 alone. (B) The percentage of dsDNA unwound or resected, from experiments as shown in A, plotted as a function of helicase concentration. The percentage resected or unwound was obtained by expressing the amount of DNA degraded or unwound, respectively, as a percentage of total signal, and is plotted as “products.” Error bars indicate standard deviation from three to five independent experiments and are smaller than the symbols when not evident. (C) Resection requires the nuclease but not helicase activity of DNA2. Reactions were performed as described in A, with the exception that, where indicated, DNA2 was substituted with helicase-dead (HD) or nuclease-dead (ND) DNA2. (Lane 1) BLM. (Lanes 2–4) BLM with wild-type (WT), K671E (HD), or D294A (ND) DNA2, respectively. (Lanes 5–7) Wild type (WT), helicase-dead (HD), and nuclease-dead (ND) DNA2, respectively. (Lane Δ) Heat-denatured substrate. (D) Sgs1 can substitute for BLM at low [Mg2+] (2 mM). Where indicated, BLM was replaced with Sgs1.The concentration of Mg2+ was as indicated in the figure. (Lane 1) Substrate. (Lanes 2,3) Reactions with BLM (absence or presence of DNA2). (Lanes 4–9) Reactions with Sgs1 (absence or presence of DNA2). (Lane Δ) Heat-denatured substrate. (E) BLM and DNA2 interact directly. Pull-down experiments were performed with BLM and DNA2 in three sets: BLM alone, BLM and DNA2, and DNA2 alone. The bound fractions were analyzed by gel electrophoresis followed by Sypro Orange staining. (Lanes 1,2) Approximately 250 ng of BLM and DNA2, respectively. (Lanes 3–5) Pull-downs with BLM alone, BLM and DNA2, and DNA2 alone, respectively. (Lanes 6–8) Same as lanes 3–5, but in the presence of 12.5 U of benzonase. The positions of size markers (in kilodaltons), BLM, and DNA2 are indicated. (F) Yeast Dna2 functions with BLM in DNA end resection. Reactions were performed as described in A, except that, when indicated, DNA2 was substituted with Dna2 (human RPA throughout). (Lane 1) Substrate. (Lane 2) BLM. (Lane 3) BLM–DNA2. (Lane 4) BLM–Dna2. (Lane 5) DNA2. (Lane 6) Dna2. DNA2 and Dna2 are indicated as “H” and “Y,” respectively.
To unambiguously determine the function of BLM helicase, we replaced wild-type BLM with its helicase-dead counterpart (K695R) (Bugreev et al. 2007). As reported, BLM (K695R) failed to unwind DNA (Fig. 2A, lanes 8–10). Importantly, BLM (K695R) also failed to stimulate degradation of dsDNA by DNA2 (Fig. 2A, lanes 11–13), even when the mutant helicase was present at a fivefold higher concentration than wild-type BLM. These data indicate that DNA unwinding is essential for DNA2-mediated resection of dsDNA ends.
To define the functions of the nuclease and helicase activities of DNA2 in resection, we substituted DNA2 with its nuclease-dead (D294A) or helicase-dead (K671E) counterpart. Whereas the nuclease-dead mutant (D294A) failed to resect DNA in the presence of BLM (Fig. 2C, lane 4), the helicase-dead mutant (K671E) did (Fig. 2C, lane 3), establishing that resection is due to the intrinsic nuclease activity of DNA2.
Stimulation of DNA end resection by DNA2 is specific to BLM, and results from a direct interaction between the two proteins
To determine whether the resection by BLM and DNA2 is specific to these two human proteins, we substituted other helicases. We first examined E. coli RecQ. DNA unwinding by RecQ is sensitive to the free Mg+2 concentration (Harmon and Kowalczykowski 1998) and, under the conditions here, an approximately fivefold higher protein concentration was required for unwinding compared with BLM (Fig. 2A [cf. lanes 2–4 and 14–16], 2B). Importantly, DNA2 did not efficiently degrade dsDNA when RecQ replaced BLM (Fig. 2A, lanes 17–19); in fact, DNA2 inhibited the helicase activity of RecQ (Fig. 2A, lanes 14–19).
We next substituted BLM with other RecQ homologs: human RecQ4, RecQ5, or WRN, or S. cerevisiae Sgs1. Previously, we demonstrated that the human helicases were functional (Nimonkar et al. 2008). However, at these concentrations, none of the other human helicases could significantly unwind blunt-ended DNA (Supplemental Fig. S3C,D) as reported previously (Mohaghegh et al. 2001; Garcia et al. 2004; Rossi et al. 2010). As a consequence, none supported DNA degradation by DNA2 (Supplemental Fig. S3C,D), even though some degradation by WRN nuclease was evident.
Finally, when Sgs1 was directly substituted for BLM, unwinding of the 50-bp DNA was initially not detectable (Fig. 2D, lane 4), and addition of DNA2 resulted in much less resection than the BLM–DNA2 reactions (Fig. 2D, cf. lanes 5 and 3). Sgs1 is a vigorous helicase in the presence of RPA (yeast or human) (Supplemental Fig. S3E), but is less active at the elevated Mg2+ concentration (5 mM) used here (Cejka et al. 2010a; Cejka and Kowalczykowski 2010). When examined at a lower concentration of Mg2+ in the presence of human RPA, both unwinding by Sgs1 and resection by Sgs1 and human DNA2 was evident (Fig. 2D, lanes 6,7, respectively). Thus, even though bacterial RecQ is unable to stimulate resection by DNA2, yeast Sgs1 can.
The relatively specific stimulation of DNA2 by BLM and its closely related ortholog, Sgs1, suggested the existence of a physical interaction between the two proteins. To test this possibility, pull-down experiments with the purified proteins were performed. We used Ni-NTA magnetic beads, exploiting the presence of a C-terminal His6 tag on BLM. The data show that DNA2 is pulled down only when BLM was bound to the beads (Fig. 2E, lanes 3–5), demonstrating that the two proteins interact physically. We verified that the association between the two proteins is direct, and not mediated via DNA, by confirming the pull-down results in the presence of benzonase, a nuclease that degrades all forms of DNA and RNA (Fig. 2E, lanes 6–8). This direct association between BLM and DNA2 is evolutionarily conserved because yeast Sgs1 and Dna2 also interact physically in a DNA-independent manner (Cejka et al. 2010a).
In support of these in vitro findings, BLM was shown previously to interact with yeast Dna2 by coimmunoprecipitation in vivo and to suppress the DNA damage and temperature sensitivity of dna2 cells (Imamura and Campbell 2003). In agreement, we saw that yeast Dna2 can resect DNA in conjunction with BLM in vitro (Fig. 2F, lane 4), demonstrating biochemical cooperation between the two noncognate pairs. The ability of both BLM and Sgs1 to interact with both DNA2 orthologs suggests a conserved interaction interface. The direct interaction between the two proteins suggests that they act in a concerted fashion wherein unwinding by BLM and cleavage by DNA2 are coupled.
MRN stimulates resection by BLM–DNA2–RPA by recruiting BLM to DNA ends
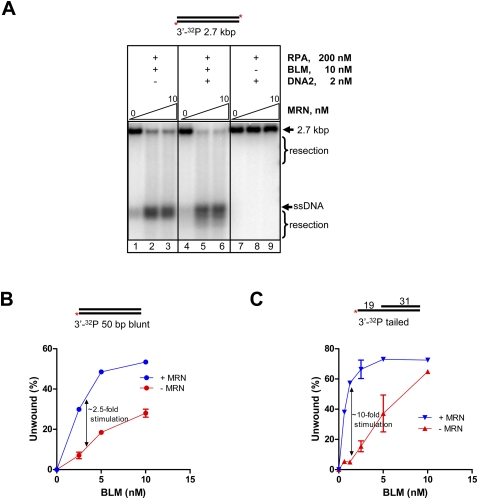
The MRN/MRX complex is one of the earliest proteins to bind dsDNA breaks (Stracker et al. 2004), and both genetic and biochemical analyses from yeast show that MRX recruits other nucleases to the DNA ends (Llorente and Symington 2004; Mimitou and Symington 2008; Zhu et al. 2008; Cejka et al. 2010a; Niu et al. 2010). Consequently, we tested the effect of MRN on the resection of 3′-end-labeled dsDNA (2.7 kb) by BLM and DNA2; to detect any possible stimulatory effects, the concentration of RPA was halved to a suboptimal concentration to limit BLM helicase activity. MRN by itself had neither helicase nor nuclease activity (Supplemental Fig. S4A, lane 2). However, MRN greatly stimulated DNA unwinding by BLM (Fig. 3A, lanes 1–3); the stimulation of unwinding was dependent on MRN concentration and was about fourfold at saturating MRN (∼5 nM) (Supplemental Fig. S4B). MRN also substantially stimulated DNA resection by BLM and DNA2 (Fig. 3A, lanes 4–6), as measured by disappearance of intact DNA and formation of products smaller than the unwound ssDNA (Fig. 3A, cf. lanes 2,3 and 5,6). Finally, MRN had no effect on DNA2 alone (Fig. 3A, lanes 7–9), eliminating the possibility that resection is an additive consequence of these two nucleases. These data showed that MRN exerts its stimulatory effect on resection by BLM and DNA2 by primarily enhancing the ability of BLM to unwind DNA.
Figure 3.
MRN stimulates processing of dsDNA by BLM–DNA2–RPA. (A) Resection by BLM–DNA2 as a function of MRN concentration. Nuclease reactions were performed at varying MRN concentrations (0, 5, and 10 nM) using 3′-end-labeled 2.7-kb DNA. Reactions contained 200 nM RPA and 2 mM MgCl2. (Lanes 1–3) BLM and MRN. (Lanes 4–6) BLM, DNA2, and MRN. (Lanes 7–9) DNA2 and MRN. (B) MRN stimulates BLM-mediated unwinding of blunt-end DNA. The percentage of dsDNA unwound from experiments as shown in Supplemental Figure S4C, plotted as a function of BLM concentration. The percentage unwound was obtained by expressing the amount of ssDNA as a percentage of the total signal. Error bars indicate standard deviation from three to five independent experiments and are smaller than the symbols when not evident. (C) MRN stimulates BLM-mediated unwinding of 3′-tailed DNA. The percentage of 3′-tailed DNA unwound from experiments as shown in Supplemental Figure S4D, plotted as a function of BLM concentration. The percentage unwound was obtained by expressing the amount of ssDNA as a percentage of the total signal. Error bars indicate standard deviation from three to five independent experiments and are smaller than the symbols when not evident. In B and C, this linear regime for DNA unwound as a function of BLM concentration was used to calculate the slope (DNA unwound/[BLM]); this value was determined in both the absence and presence of MRN, and was used to define the fold stimulation by MRN.
Yeast Mre11 interacts with and recruits Sgs1 to DNA ends (Chiolo et al. 2005; Cejka et al. 2010a; Niu et al. 2010). To investigate whether MRN functions by recruiting BLM to the ends, we compared the ability of BLM to unwind the 50-bp DNA (3′-end-labeled) in the absence and presence of MRN. Figure 3B and Supplemental Figure S4C show that DNA unwinding in the linear range of BLM concentration was ∼2.5-fold higher in the presence of MRN. MRN reduced the amount of BLM required to obtain maximum unwinding, which implied that MRN increased the affinity of BLM for DNA ends, thereby suggesting a recruitment function for MRN. MRN also enhanced BLM-mediated unwinding of a 3′-tailed substrate by ∼10-fold (in the linear region, at the lower BLM concentrations) (Fig. 3C; Supplemental Fig. S4D) showing that MRN also stimulated BLM binding to partially resected DNA.
MRN stimulates resection by EXO1
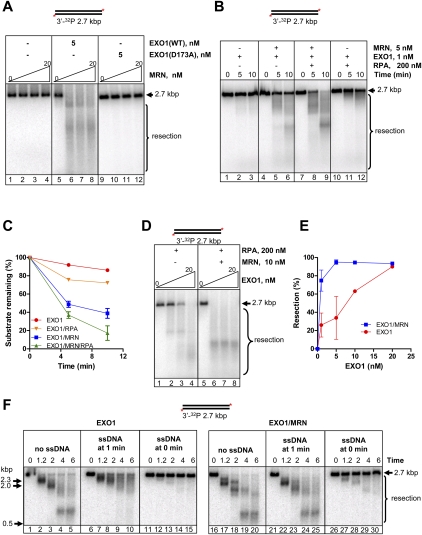
Having established a stimulatory role for MRN in resection mediated by BLM and DNA2, we next examined the influence of MRN on the nuclease activity of EXO1 using 3′-end-labeled 2.7-kb dsDNA. EXO1 alone could degrade the DNA, albeit inefficiently (Fig. 4A, lane 5) (Nimonkar et al. 2008). MRN alone had no nuclease activity (Fig. 4A, lanes 2–4), but it stimulated digestion by EXO1 to generate a smear of faster-migrating resection products (Fig. 4A, lanes 6–8). To verify that the intrinsic nuclease activity of EXO1 is responsible for resection in the MRN-stimulated resection reaction, we examined the effect MRN on EXO1 (D173A), a mutated form of EXO1 that lacks nuclease activity (Dzantiev et al. 2004). As expected, EXO1 (D173A) showed no activity (Fig. 4A, lane 9), and addition of MRN did not result in any resection (Fig. 4A, lanes 10–12).
Figure 4.
MRN stimulates resection by EXO1. Nuclease reactions with EXO1 and MRN were performed using 3′-end-labeled 2.7-kb DNA. All reactions contained 5 mM MgCl2. RPA (200 nM) was included when indicated. (A) Resection by EXO1 (wild type) and EXO1 (D173A) as a function of MRN concentration (0, 5, 10, and 20 nM). (Lanes 1–4) MRN alone. (Lanes 5–8) MRN and EXO1 (wild type). (Lanes 9–12) MRN and EXO1 (D173A). (B) Kinetics of resection by EXO1 in the presence of RPA and MRN. (Lanes 1–3) EXO1. (Lanes 4–6) EXO1 and MRN. (Lanes 7–9) EXO1, MRN, and RPA. (Lanes 10–12) EXO1 and RPA. (C) The percentage of intact DNA from experiments as shown in B, plotted as a function of time. The percentage intact was obtained relative to the 0-min time point for each set. Error bars indicate standard deviation from three to five independent experiments and are smaller than the symbols when not evident. (D) Resection as a function of EXO1 concentration (0, 5, 10, and 20 nM) in the absence or presence of MRN. Reactions contained 200 nM RPA. Incubation time was 10 min. (Lanes 1–3) EXO1. (Lanes 4–6) EXO1 and MRN. (E) The percentage of intact DNA from experiments as shown in D, plotted as a function of EXO1 concentration. The percentage intact was obtained relative to 0 nM EXO1 reaction for each set. Error bars indicate standard deviation from three to five independent experiments and are smaller than the symbols when not evident. (F) MRN increases processivity of EXO1. Inhibitor challenge experiments were performed as described in the Supplemental Material. (Lanes 1–5,16–20) EXO1 and EXO1–MRN reaction in the absence of challenger DNA, respectively. (Lanes 6–10,21–25) EXO1 and EXO1–MRN reaction with challenger DNA added at 1 min, respectively. (Lanes 11–15,26–30) EXO1 and EXO1–MRN reaction with challenger DNA added at 0 min, respectively. The concentrations of EXO1, MRN, and φX174 ssDNA were 10 nM, 10 nM, and 32 μM (nucleotides), respectively.
RPA modestly stimulated (approximately twofold) the rate of resection by EXO1 (Fig. 4B [lanes 10–12], C) and, in the presence of RPA, MRN further stimulated EXO1 by approximately threefold (Fig. 4B [lanes 7–9], C). MRN also stimulated EXO1 nucleolytic activity by approximately fivefold when RPA was absent (Fig. 4B [lanes 4–6], C). MRN lowered the concentration of EXO1 required for resection (Fig. 4D,E), and the observation that substrate utilization at lower EXO1 concentrations is greatly increased by MRN suggests, minimally, that MRN recruits EXO1 to DNA ends. Finally, resection by MRN and EXO1 proceeded in the absence of ATP (Supplemental Fig. S4E). Interestingly, yeast MRX could effectively replace MRN in the resection reaction (Supplemental Fig. S4F), revealing that stimulation of EXO1 by MRN is not species-specific. This observation suggests that either MRN and EXO1 do not interact directly or the interface is conserved, as is the case for DNA2 and BLM orthologs. Recent studies concluded that yeast MRX stimulates Exo1 without directly associating with it (Nicolette et al. 2010), and that human CtIP but not MRE11 interacts with EXO1 (Eid et al. 2010). Finally, activation of MRN nuclease by substituting or adding Mn2+ (Paull and Gellert 1998) did not generate resection products (Supplemental Fig. S5) due to the inability of one or more resection enzymes to function in the presence of Mn2+. Collectively, our data show that DNA resection by these proteins relies on the nucleolytic activity intrinsic to EXO1, and that both MRN and RPA are stimulators of EXO1-dependent resection.
MRN increases the processivity of EXO1-mediated resection
The results in Figure 4, A–E, suggested that MRN could stimulate EXO1 by recruiting it to DNA ends, which agrees with a conclusion established for the yeast orthologs (Mimitou and Symington 2010; Nicolette et al. 2010; Shim et al. 2010). However, it was evident from our results that the extent of degradation was also altered by MRN, suggesting that perhaps MRN increased the processivity of EXO1. To examine this possibility, the processivity of EXO1was measured using a procedure established previously to demonstrate the effect of MutSα on EXO1 (Genschel and Modrich 2003). In this approach, an ongoing EXO1 reaction is challenged with excess unlabeled competitor ssDNA, which binds any EXO1 that dissociates from the labeled substrate. Since the experiment relies on the ability of ssDNA to sequester free protein, RPA was omitted. Figure 4F shows that addition of the challenge ssDNA prior to initiation abolished DNA degradation by EXO1 in the absence as well as the presence of MRN, verifying that the ssDNA is an effective inhibitor of EXO1 activity (lanes 11–15,26–30). Addition of the challenge DNA 1 min after formation of the protein–DNA complexes halted further degradation by EXO1 alone (Fig. 4F, lanes 6–10), but did not significantly inhibit EXO1 activity when MRN was present (Fig. 4F, lanes 21,25). Also evident was the fact that substrate utilization but not the rate of resection is greater when MRN is present, confirming that MRN recruits EXO1 to the ends. Importantly, these data establish that MRN also enhances the intrinsic processivity of EXO1, evidently by remaining associated with the EXO1–DNA complex.
BLM increases the affinity of EXO1 for DNA ends but not its processivity
We showed previously that BLM also interacts with EXO1 to enhance resection of dsDNA (Nimonkar et al. 2008). In that study, we suggested that BLM acts by recruiting EXO1 to DNA ends. To confirm this hypothesis and eliminate the added possibility that BLM was also enhancing the processivity of EXO1, we again performed the DNA challenge experiments using the gapped circular dsDNA substrate that had been used previously (Supplemental Fig. S6; Genschel and Modrich 2003). BLM was devoid of nuclease activity (Supplemental Fig. S6B), and increased substrate use by EXO1 (Supplemental Fig. S6, cf. A and C, lanes 1–5). Addition of the challenge ssDNA prior to initiation abolished EXO1 activity in the absence as well as the presence of BLM (Supplemental Fig. S6, cf. A and C, lanes 6–10). Addition of challenge DNA 1 min after initiation blocked further degradation by EXO1 (Supplemental Fig. S6A,C, lanes 11–15). Regardless of the presence of BLM, the size distribution of the degradation product was unaltered, showing that the processivity of EXO1 was unchanged by its interaction with BLM. In the presence of BLM, however, a smaller amount of intact substrate remained (Supplemental Fig. S6, cf. A and C, cf. lanes 11–15), consistent with our previous interpretation of recruitment of EXO1 by BLM. We therefore conclude that BLM influences EXO1 by increasing its affinity for DNA ends.
Having established that MRN enhances the processivity of EXO1 (Fig. 4) and BLM increases the affinity of EXO1 to DNA ends (Supplemental Fig. S6), we next addressed whether MRN stimulated resection by EXO1 in the presence of BLM. Because the helicase activity of BLM is dispensable for stimulation of EXO1 (Nimonkar et al. 2008), we initially omitted the ATP to eliminate complications due to BLM-dependent unwinding of the substrate. Furthermore, we established that ATP is also not required for the stimulation by MRN (Supplemental Fig. S4E). Figure 5A shows that either protein stimulated DNA resection by EXO1 (lanes 4–6,10–12). In the presence of both proteins, resection was greater (Fig. 5A, lanes 7–9), showing that both can contribute to the increased activity of EXO1. Quantification of the intact substrate remaining (Fig. 5B) shows that stimulation by BLM (∼1.6-fold at 5 min) and MRN (∼2.5-fold at 5 min) is reinforcing (approximately threefold at 5 min) but not synergistic. Finally, in the presence of ATP (Fig. 5C), resection proceeds somewhat more rapidly (Fig. 5, cf. C [lanes 10–12] and A [lanes 7–9]), and the replacement of BLM with the helicase-deficient mutant still results in stimulation, although the level is reduced to a level and product pattern that is more comparable with wild-type BLM in the absence of ATP (Fig. 5, cf. C [lanes 16–18] and A [lanes 7–9]) than with BLM in the presence of ATP (Fig. 5C, lanes 10–12).
Figure 5.
Stimulation of EXO1 by MRN and BLM. Nuclease reactions were performed using 3′-end-labeled 2.7-kb DNA. All reactions contained 200 nM RPA and 5 mM MgCl2. (A) Kinetics of resection in the absence of ATP. (Lanes 1–3) EXO1. (Lanes 4–6) MRN and EXO1. (Lanes 7–9) MRN, EXO1, and BLM. (Lanes 10–12) EXO1 and BLM. (B) The percentage of intact DNA from experiments as shown in A, plotted as a function of time. The percentage intact was obtained relative to the 0-min time point for each set. Error bars indicate standard deviation from three to five independent experiments and are smaller than the symbols when not evident. (C) Kinetics of resection in the presence of ATP and also with the helicase-dead BLM mutant. (Lanes 1–3) EXO1. (Lanes 4–12) MRN, EXO1, and BLM (wild-type). (Lanes 13–21) MRN, EXO1, and BLM (helicase-dead).
Discussion
In this study, we biochemically reconstituted DNA end resection using human BLM, DNA2, EXO1, MRE11, RAD50, NBS1, and RPA. We established that two specific combinations of these proteins comprise components of alternate biochemical machineries for DNA end resection (Fig. 6). The first consists of BLM, DNA2, MRN, and RPA, and resects DNA in a coordinated ATP-dependent manner: BLM plays the role of helicase, DNA2 resects the unwound DNA, and RPA enforces endonucleolytic cleavage in the 5′ → 3′ direction. The MRN complex enhances the efficacy of this reaction by recruiting BLM to the DNA end, thereby increasing the effectiveness of its helicase function. This complex can unwind and resect DNA for at least several thousand base pairs. The second consists of EXO1, BLM, MRN, and RPA. EXO1 serves as the 5′ → 3′ resection nuclease, BLM stimulates EXO1 by increasing its binding to DNA, and MRN both recruits EXO1 to DNA and increases its processivity. Although EXO1 can resect DNA ends by itself, BLM and MRN together make EXO1 a more efficient nuclease that can also resect thousands of nucleotides from DNA ends.
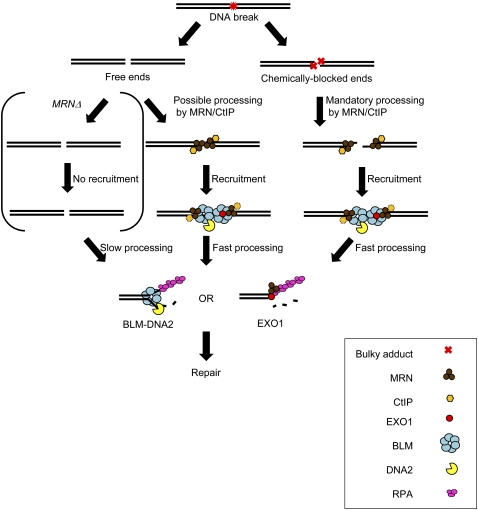
Figure 6.
Model depicting DNA end resection pathways during DSB repair. DSBs caused by exogenous or endogenous sources (represented by the red symbol) can be free or chemically blocked. The blocked ends are indicated by red crosses. DSBs are bound by MRN (brown trimer), which can recruit CtIP (orange hexagon) to form one resection complex. Resection by MRN and CtIP is required to cleave the chemically blocked ends and can resect several hundred nucleotides, but is not essential for resection of the free DNA ends. However, processing in the physical absence of MRN is kinetically delayed (“slow processing”). MRN also functions to recruit the BLM–DNA2 (BLM indicated by blue hexamer; DNA2 indicated by yellow pac molecule) or EXO1–BLM (EXO1 indicated by red sphere) to the ends (“fast processing”). Subsequently, extensive processing (>1 kb) can proceed by either the BLM–DNA2–RPA (RPA indicated by pink trimer) or the EXO1–BLM–RPA machinery.
Resection by BLM, DNA2, RPA, and MRN
Genetic, physical, and biochemical analyses in S. cerevisiae demonstrated a role for yeast Sgs1 and Dna2 in DNA end resection (Gravel et al. 2008; Mimitou and Symington 2008; Zhu et al. 2008; Cejka et al. 2010a; Niu et al. 2010). Here we established that the human orthologs BLM and DNA2 resect DNA in a similar but not entirely identical manner. Our biochemical findings show that BLM and DNA2 interact functionally, physically, and specifically to resect DNA. The ATP-dependent unwinding of dsDNA by BLM is required for DNA2-dependent resection, but the helicase function of DNA2 is dispensable; however, the nuclease activity of DNA2 is essential for resection. Despite being a bipolar nuclease (Budd et al. 2000; Masuda-Sasa et al. 2006), our data show that, in presence of RPA, DNA2 resects only in the 5′ → 3′ direction. We demonstrate that the 5′ → 3′ resection polarity is established by the attenuation of 3′ → 5′ nuclease activity of DNA2 in the presence of RPA and, at the same time, stimulation of the 5′ → 3′ resection (Masuda-Sasa et al. 2006). These key findings parallel those observed for the yeast proteins (Masuda-Sasa et al. 2008; Cejka et al. 2010a; Niu et al. 2010).
Our biochemical results also demonstrate that MRN stimulates BLM- and DNA2-dependent resection by a mechanism that does not require the nucleolytic capacity of MRN. MRN stimulates by increasing the affinity of BLM for DNA ends by 2.5-fold to 10-fold (Fig. 3B,C), indicating a recruitment function for MRN. Consistent with this interpretation, coimmunoprecipitation experiments showed that Xenopus DNA2 interacts with the NBS1 subunit of MRN (Wawrousek et al. 2010). This conclusion is also in agreement with studies in yeast that demonstrated that the nuclease activity of Mre11 is not essential for the 5′ → 3′ resection of HO endonuclease-induced DNA breaks (Llorente and Symington 2004). In the physical absence of MRX, resection is kinetically delayed (Mimitou and Symington 2008; Zhu et al. 2008), whereas in the nuclease-deficient strain, there is no delay (Llorente and Symington 2004). Furthermore, the yeast Dna2 nuclease can substitute for MRX nuclease activity, but not for MRX protein, in DSB repair (Budd and Campbell 2009). Also consistent with the recruitment concept, MRX interacts with Sgs1 (Chiolo et al. 2005). Recently, biochemical reconstitution of yeast Sgs1- and Dna2-dependent resection established that MRX stimulates Sgs1 helicase by recruiting it to DNA ends (Cejka et al. 2010a; Niu et al. 2010). Finally, biochemical reconstitution of resection using the Pyrococcus furiosus Mre11, Rad50, HerA, and NurA proteins also established that the nuclease function of Mre11 is dispensable (Hopkins and Paull 2008). In vertebrates, the nuclease activity of murine MRE11 is essential for the repair of breaks that are created by ionizing radiation or I-SceI (Buis et al. 2008). Ionizing radiation can chemically alter DNA ends, and I-SceI stays bound to one end after DNA cleavage (Perrin et al. 1993), consistent with the view that MRN is required only when ends are chemically modified or altered so as to preclude resection by EXO1 or DNA2. We therefore conclude that, when DNA breaks are free of such impediments, MRN minimally serves as the platform for recruitment of other components (BLM and EXO1) of the resection machinery to the DNA end. MRN and CtIP may have an additional role that involves limited resection to produce resected DNA with a short 3′-ssDNA overhang (as in yeast, where only a few hundred nucleotides are resected by MRX) (Mimitou and Symington 2008; Zhu et al. 2008); such a resection could be minimally sufficient for DNA repair, or the tailed DNA could serve as an even better substrate for recruitment of BLM–DNA2 by MRN to the break site (Fig. 3C).
Resection by EXO1, BLM, RPA, and MRN
EXO1 is capable of degrading one strand of dsDNA in the 5′–3′ direction (Lee and Wilson 1999). However, several studies have shown that that the activity of EXO1 can be increased by other proteins. For example, the mismatch repair protein MUTSα increases the processivity of EXO1 (Genschel and Modrich 2003). Previously, we demonstrated that BLM also potentiates the activity of EXO1 (Nimonkar et al. 2008). The salient feature of the BLM–EXO1 interaction is its ATP independence, which eliminates the requirement for BLM-catalyzed unwinding or translocation. This ability of BLM to modulate EXO1 distinguishes it from what is known about Sgs1 (Mimitou and Symington 2008; Zhu et al. 2008). Our DNA challenge experiments confirm our prior observation that BLM enhances the affinity of EXO1 to a DNA end (Nimonkar et al. 2008) but does not affect the processivity of EXO1.
Our biochemical findings demonstrate that EXO1 is also stimulated by MRN. MRN increases the binding of EXO1 for DNA by ∼10-fold (Fig. 4E). In addition to recruiting EXO1 to DNA ends, MRN also increases the processivity of EXO1-mediated resection. This indicates that MRN presumably exerts its influence away from the break. Because the DNA challenge experiments also preclude reassociation of MRN with the EXO1 that is bound to the dsDNA being resected, this finding further implies that MRN is associated with EXO1 as it resects DNA. Our work thereby provides biochemical evidence for a stimulatory role of MRN during DNA break repair in conjunction with BLM and EXO1, and is in agreement with recent studies of DNA resection in yeast (Cejka et al. 2010a; Mimitou and Symington 2010; Nicolette et al. 2010; Niu et al. 2010; Shim et al. 2010).
The role of BLM
Individuals afflicted with Bloom syndrome display many clinical symptoms, including a predisposition of early cancer development (German 1993), and Bloom syndrome cells manifest a high frequency of chromosomal exchanges (Luo et al. 2000). The viability of individuals with Bloom syndrome demonstrates that functional BLM protein is not necessarily essential. The BLM protein, like its orthologs, functions in many steps of DNA break repair (Harmon and Kowalczykowski 1998; Chu and Hickson 2009; Cejka et al. 2010a,b). Here we showed that, although BLM is an essential component of the DNA2-dependent pathway of resection, it is one of two stimulatory components of the alternative EXO1-dependent pathway. Thus, resection in blm−/− cells could bypass resection by DNA2 and employ EXO1 for DSB repair, as is the case for yeast cells defective for Sgs1 (Mimitou and Symington 2008; Zhu et al. 2008). However, blm−/− cells display elevated levels of recombination, as measured by increased chromosomal crossing over (Luo et al. 2000). This phenotype is a consequence of two other activities of BLM helicase and its orthologs: disruption of DNA pairing intermediates (Harmon and Kowalczykowski 1998; Bugreev et al. 2007) and the dissolution of double Holliday junctions (Wu and Hickson 2003; Cejka et al. 2010b). The disruption of pairing complexes results in the elimination of recombination intermediates that can mature into Holliday junctions, and the dissolution activity results in recombinational DNA repair without crossing over. Thus, loss of BLM function would result in elevated crossover recombination. Finally, BLM functions with Topoisomerase IIIα (Chu and Hickson 2009); deletion of BLM would result in loss of Topoisomerase IIIα function, and could explain the lethality of BLM disruption and certain blm alleles (Chester et al. 1998; Luo et al. 2000).
The role of MRN
The MRN complex is one of the first proteins to bind DSBs (Stracker et al. 2004). It plays multiple roles in the DNA damage response, where it functions as a sensor for DSBs and also signals the existence of the break to downstream factors. In addition, MRX–CtIP has direct functions in the repair of the DSB (Sartori et al. 2007). In vivo, DSBs can be “clean,” where the break site is not chemically altered (e.g., from collapse of a replication fork at a nick, or artificially generated by cleavage HO endonuclease), or “dirty,” where the break site is modified by a damaged nucleotide or a covalent adduct (e.g., due to ionizing radiation or SPO11-dependent cleavage). MRN recruits CtIP, and the two constitute a resection machine that can perform limited processing. Recently, CtIP has been shown to interact with and affect the activity of EXO1 (Eid et al. 2010). Although CtIP reduced EXO1 activity on nicked DNA, its preincubation with blunt-end DNA increased processing by EXO1; thus, perhaps the MRN–CtIP complex might act synergistically to recruit EXO1 to DSBs. Several lines of evidence, already outlined above, established that the nuclease activity MRN/MRX is not mandatory if the broken DNA ends are chemically normal. However, the resection activity of MRN and CtIP is especially critical when the ends are blocked with covalent adducts (Sartori et al. 2007; Gravel et al. 2008). Regardless of the nature of the ends, MRN functions as an important recruiting factor to target DNA helicases and resection nucleases to the ends, a function that is conserved in S. cerevisiae as well (Mimitou and Symington 2010; Shim et al. 2010).
Homologous pairing of resected DNA
Analyses in budding yeast indicate that resection of 100–200 bp is sufficient for efficient gene conversion (Mimitou and Symington 2008; Zhu et al. 2008). Nevertheless, long-range resection must play an important role during DNA repair, since sequences that are 2–3 kb away from the break site are preferred for homology search and repair (Inbar and Kupiec 1999). In addition, given that resection in vivo occurs at ∼4 kb per hour, the delay of more than an hour between induction of DNA breaks and the occurrence of strand invasion and DNA synthesis indicates that extended resection can occur before homologous pairing (White and Haber 1990; Zhu et al. 2008). Such extensive resection also ensures repair with homologs that are not identical. In bacteria, the primary resection machine for DSB repair, RecBCD enzyme, resects as much as 30 kb of DNA (Dillingham and Kowalczykowski 2008). This seemingly excessive resection is useful when the DNA break is opposite a heterology in the DNA pairing donor; in such cases, as much as 10 kb of heterologous DNA can be resected to reveal homology that is downstream from the break (Myers et al. 1995). The extended resection observed in our reconstituted system is consistent with a role for resection over kilobase distances in DSB repair in human cells.
While the choice of processing pathway may be dictated by the nature of the break, the net result is the same: generation of ssDNA. The ssDNA tail then serves as the substrate for DNA strand exchange proteins that promote homologous pairing. Given that RAD51, the human strand exchange protein, interacts directly with BLM (Wu et al. 2001), the presence of BLM in both machineries may not be a coincidence, since BLM may function to target RAD51 to the nascent ssDNA substrate. Indeed, we showed previously that BLM–EXO1-resected DNA is used by RAD51 for pairing (Nimonkar et al. 2008). The in vitro reconstitution of DNA end resection using purified proteins described in this study significantly advances our understanding of end resection during initiation of recombinational DNA repair in humans, and opens avenues to study the connection between steps that precede resection in DSB repair and those that follow, such as DNA pairing and resolution.
Materials and methods
Nuclease and helicase assays
Reaction were conducted using 1.5 nM end DNA (2.7 kb [4 μM nucleotides]), 324 bp (0.5 μM nucleotides), or 50 bp (75 nM nucleotides) in standard buffer (20 mM Na-HEPES at pH 7.5, 2 mM ATP, 0.1 mM DTT, 100 μg/mL BSA, 0.05% Triton-X100 , MgCl2 concentration as indicated) for 30 min at 37°C. For experiments with the short substrates (forked as well as dsDNA), the MgCl2 concentration was maintained at 5 mM to avoid melting of the substrate. The concentrations of proteins are noted in each figure or legend. Variations in buffer components and incubation times are also similarly noted. Reactions were assembled on ice and initiated by transferring to 37°C. Reactions were stopped by addition of 2 μg/μL proteinase K, 50 mM EDTA, and 1% SDS (final concentrations) and incubating for 30 min. In the case of a 50-bp substrate, a 20-fold molar excess of the identical but unlabeled oligonucleotide was included in the termination buffer to prevent spontaneous reannealing of the unwound substrate strands. Products were analyzed by 1% native agarose gel (reactions with linear pUC19), 6% native polyacrylamide (reactions with a 324-bp fragment), or 12% native polyacrylamide gel electrophoresis (reactions with 50-nt dsDNA). All electrophoresis were performed using TAE (40 mM Tris-acetateat pH 8.0, 1 mM EDTA). Gels were dried on DE81 paper (Whatman), and visualized and quantified by storage phosphor analysis with a Molecular Dynamics Storm 860 (GE) using ImageQuant version 5.2. Data were analyzed using GraphPad Prism version 5.
EXO1 processivity assays
The assays to test the processivity of EXO1 were performed as described previously (Genschel and Modrich 2003) and are described in detail in the Supplemental Material.
Pull-down assays
DNA2 and BLM (1 μg each) were incubated in standard buffer lacking ATP, MgCl2, and BSA and containing 175 mM NaCl and 50 mM imidazole for 30 min at 37°C. Ni-NTA magnetic beads (Qiagen) were added to a concentration of 1% and incubated for 30 min at 37°C with constant mixing. The beads were isolated using a magnet and washed (once with 50 μL and twice with 100 μL) with standard buffer containing 100 mM imidazole. The beads were resuspended in loading buffer. Fractions were analyzed by 10% SDS–polyacrylamide gel electrophoresis and Sypro Orange staining (Molecular Probes).
Acknowledgments
We thank S.C.K. laboratory members for comments. We are grateful to Dr. David Chen (University of Texas Southwestern) for WRN, Dr. Ian Hickson (Oxford University) for RecQ5, Dr. Alex Mazin (Drexel University) for BLM helicase mutant (K695R), Dr. Patrick Sung (Yale University) for RecQ4, Petr Cejka (this laboratory) for Sgs1 and yDna2, Behzad Rad (this laboratory) for RecQ and SSB, Katsumi Morimatsu (this laboratory) for RecJ, and Zeynep Ozsoy (former laboratory member) for RPA. This work was supported by NIH grant GM-62653 to S.C.K., NCI program project SBDR 5PO1CA092548 and Netherlands Organization for Scientific Research-Chemical Sciences (NWO-CW) Vici award to C.W., Army Research Office grant ARO 09-1-0041 and NIH grant GM-78666 to J.L.C, and NIH grant GM-45190 to P.M. P.M. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.2003811.
Supplemental material is available for this article.
References
- Bae SH, Bae KH, Kim JA, Seo YS 2001. RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature 412: 456–461 [DOI] [PubMed] [Google Scholar]
- Brosh RM Jr, Li JL, Kenny MK, Karow JK, Cooper MP, Kureekattil RP, Hickson ID, Bohr VA 2000. Replication protein A physically interacts with the Bloom's syndrome protein and stimulates its helicase activity. J Biol Chem 275: 23500–23508 [DOI] [PubMed] [Google Scholar]
- Budd ME, Campbell JL 2009. Interplay of Mre11 nuclease with Dna2 plus Sgs1 in Rad51-dependent recombinational repair. PLoS ONE 4: e4267 doi: 10.1371/journal/pone.0004267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd ME, Choe W, Campbell JL 2000. The nuclease activity of the yeast DNA2 protein, which is related to the RecB-like nucleases, is essential in vivo. J Biol Chem 275: 16518–16529 [DOI] [PubMed] [Google Scholar]
- Bugreev DV, Yu X, Egelman EH, Mazin AV 2007. Novel pro- and anti-recombination activities of the Bloom's syndrome helicase. Genes Dev 21: 3085–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buis J, Wu Y, Deng Y, Leddon J, Westfield G, Eckersdorff M, Sekiguchi JM, Chang S, Ferguson DO 2008. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell 135: 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P, Kowalczykowski SC 2010. The full-length Saccharomyces cerevisiae Sgs1 protein is a vigorous DNA helicase that preferentially unwinds Holliday junctions. J Biol Chem 285: 8290–8301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P, Cannavo E, Polaczek P, Masuda-Sasa T, Pokharel S, Campbell JL, Kowalczykowski SC 2010a. DNA end resection by Dna2–Sgs1–RPA and its stimulation by Top3–Rmi1 and Mre11–Rad50–Xrs2. Nature 467: 112–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P, Plank JL, Bachrati CZ, Hickson ID, Kowalczykowski SC 2010b. Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1-Top3. Nat Struct Mol Biol 17: 1377–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chester N, Kuo F, Kozak C, O'Hara CD, Leder P 1998. Stage-specific apoptosis, developmental delay, and embryonic lethality in mice homozygous for a targeted disruption in the murine Bloom's syndrome gene. Genes Dev 12: 3382–3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiolo I, Carotenuto W, Maffioletti G, Petrini JH, Foiani M, Liberi G 2005. Srs2 and Sgs1 DNA helicases associate with Mre11 in different subcomplexes following checkpoint activation and CDK1-mediated Srs2 phosphorylation. Mol Cell Biol 25: 5738–5751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu WK, Hickson ID 2009. RecQ helicases: Multifunctional genome caretakers. Nat Rev Cancer 9: 644–654 [DOI] [PubMed] [Google Scholar]
- Dillingham MS, Kowalczykowski SC 2008. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol Mol Biol Rev 72: 642–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duxin JP, Dao B, Martinsson P, Rajala N, Guittat L, Campbell JL, Spelbrink JN, Stewart SA 2009. Human dna2 is a nuclear and mitochondrial DNA maintenance protein. Mol Cell Biol 29: 4274–4282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzantiev L, Constantin N, Genschel J, Iyer RR, Burgers PM, Modrich P 2004. A defined human system that supports bidirectional mismatch-provoked excision. Mol Cell 15: 31–41 [DOI] [PubMed] [Google Scholar]
- Eid W, Steger M, El-Shemerly M, Ferretti LP, Pena-Diaz J, Konig C, Valtorta E, Sartori AA, Ferrari S 2010. DNA end resection by CtIP and exonuclease 1 prevents genomic instability. EMBO Rep 11: 962–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S, McDonald JP, Bendixen C, Arthur L, Rothstein R 1994. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: A potential eukaryotic reverse gyrase. Mol Cell Biol 14: 8391–8398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia PL, Liu Y, Jiricny J, West SC, Janscak P 2004. Human RECQ5β, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J 23: 2882–2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschel J, Modrich P 2003. Mechanism of 5′-directed excision in human mismatch repair. Mol Cell 12: 1077–1086 [DOI] [PubMed] [Google Scholar]
- German J 1993. Bloom syndrome: A mendelian prototype of somatic mutational disease. Medicine 72: 393–406 [PubMed] [Google Scholar]
- Gravel S, Chapman JR, Magill C, Jackson SP 2008. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev 22: 2767–2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa N, Morimatsu K, Lovett ST, Kowalczykowski SC 2009. Reconstitution of initial steps of dsDNA break repair by the RecF pathway of E. coli. Genes Dev 23: 1234–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon FG, Kowalczykowski SC 1998. RecQ helicase, in concert with RecA and SSB proteins, initiates and disrupts DNA recombination. Genes Dev 12: 1134–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon FG, Kowalczykowski SC 2001. Biochemical characterization of the DNA helicase activity of the Escherichia coli RecQ helicase. J Biol Chem 276: 232–243 [DOI] [PubMed] [Google Scholar]
- Hartsuiker E, Neale MJ, Carr AM 2009. Distinct requirements for the Rad32(Mre11) nuclease and Ctp1(CtIP) in the removal of covalently bound topoisomerase I and II from DNA. Mol Cell 33: 117–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins BB, Paull TT 2008. The P. furiosus Mre11/Rad50 complex promotes 5′ strand resection at a DNA double-strand break. Cell 135: 250–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura O, Campbell JL 2003. The human Bloom syndrome gene suppresses the DNA replication and repair defects of yeast dna2 mutants. Proc Natl Acad Sci 100: 8193–8198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inbar O, Kupiec M 1999. Homology search and choice of homologous partner during mitotic recombination. Mol Cell Biol 19: 4134–4142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Kim HD, Ryu GH, Kim DH, Hurwitz J, Seo YS 2006. Isolation of human Dna2 endonuclease and characterization of its enzymatic properties. Nucleic Acids Res 34: 1854–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BI, Wilson DM III 1999. The RAD2 domain of human exonuclease 1 exhibits 5′ to 3′ exonuclease and flap structure-specific endonuclease activities. J Biol Chem 274: 37763–37769 [DOI] [PubMed] [Google Scholar]
- Liao S, Toczylowski T, Yan H 2008. Identification of the Xenopus DNA2 protein as a major nuclease for the 5′ → 3′ strand-specific processing of DNA ends. Nucleic Acids Res 36: 6091–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente B, Symington LS 2004. The Mre11 nuclease is not required for 5′ to 3′ resection at multiple HO-induced double-strand breaks. Mol Cell Biol 24: 9682–9694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett ST, Kolodner RD 1989. Identification and purification of a single-stranded-DNA-specific exonuclease encoded by the recJ gene of Escherichia coli. Proc Natl Acad Sci 86: 2627–2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G, Santoro IM, McDaniel LD, Nishijima I, Mills M, Youssoufian H, Vogel H, Schultz RA, Bradley A 2000. Cancer predisposition caused by elevated mitotic recombination in Bloom mice. Nat Genet 26: 424–429 [DOI] [PubMed] [Google Scholar]
- Masuda-Sasa T, Imamura O, Campbell JL 2006. Biochemical analysis of human Dna2. Nucleic Acids Res 34: 1865–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda-Sasa T, Polaczek P, Peng XP, Chen L, Campbell JL 2008. Processing of G4 DNA by Dna2 helicase/nuclease and replication protein A (RPA) provides insights into the mechanism of Dna2/RPA substrate recognition. J Biol Chem 283: 24359–24373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS 2008. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455: 770–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS 2010. Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J 29: 3358–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohaghegh P, Karow JK, Brosh RM Jr, Bohr VA, Hickson ID 2001. The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res 29: 2843–2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau S, Ferguson JR, Symington LS 1999. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol Cell Biol 19: 556–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers RS, Stahl MM, Stahl FW 1995. Chi recombination activity in phage λ decays as a function of genetic distance. Genetics 141: 805–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolette ML, Lee K, Guo Z, Rani M, Chow JM, Lee SE, Paull TT 2010. Mre11–Rad50–Xrs2 and Sae2 promote 5′ strand resection of DNA double-strand breaks. Nat Struct Mol Biol 17: 1478–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar AV, Ozsoy AZ, Genschel J, Modrich P, Kowalczykowski SC 2008. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc Natl Acad Sci 105: 16906–16911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H, Chung WH, Zhu Z, Kwon Y, Zhao W, Chi P, Prakash R, Seong C, Liu D, Lu L, et al. 2010. Mechanism of the ATP-dependent DNA end resection machinery from Saccharomyces cerevisiae. Nature 467: 108–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT, Gellert M 1998. The 3′ to 5′ exonuclease activity of Mre11 facilitates repair of DNA double-strand breaks. Mol Cell 1: 969–979 [DOI] [PubMed] [Google Scholar]
- Perrin A, Buckle M, Dujon B 1993. Asymmetrical recognition and activity of the I-SceI endonuclease on its site and on intron–exon junctions. EMBO J 12: 2939–2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi ML, Ghosh AK, Kulikowicz T, Croteau DL, Bohr VA 2010. Conserved helicase domain of human RecQ4 is required for strand annealing-independent DNA unwinding. DNA Repair (Amst) 9: 796–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP 2007. Human CtIP promotes DNA end resection. Nature 450: 509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutte C, Marinescu RC, Sadoff MM, Guerrette S, Overhauser J, Fishel R 1998. Human exonuclease I interacts with the mismatch repair protein hMSH2. Cancer Res 58: 4537–4542 [PubMed] [Google Scholar]
- Shim EY, Chung WH, Nicolette ML, Zhang Y, Davis M, Zhu Z, Paull TT, Ira G, Lee SE 2010. Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J 29: 3370–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracker TH, Theunissen JW, Morales M, Petrini JH 2004. The Mre11 complex and the metabolism of chromosome breaks: The importance of communicating and holding things together. DNA Repair (Amst) 3: 845–854 [DOI] [PubMed] [Google Scholar]
- Szankasi P, Smith GR 1992. A DNA exonuclease induced during meiosis of Schizosaccharomyces pombe. J Biol Chem 267: 3014–3023 [PubMed] [Google Scholar]
- Tishkoff DX, Amin NS, Viars CS, Arden KC, Kolodner RD 1998. Identification of a human gene encoding a homologue of Saccharomyces cerevisiae EXO1, an exonuclease implicated in mismatch repair and recombination. Cancer Res 58: 5027–5031 [PubMed] [Google Scholar]
- Wawrousek KE, Fortini BK, Polaczek P, Chen L, Liu Q, Dunphy WG, Campbell JL 2010. Xenopus DNA2 is a helicase/nuclease that is found in complexes with replication proteins And-1/Ctf4 and Mcm10 and DSB response proteins Nbs1 and ATM. Cell Cycle 9: 1156–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CI, Haber JE 1990. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J 9: 663–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DM III, Carney JP, Coleman MA, Adamson AW, Christensen M, Lamerdin JE 1998. Hex1: A new human Rad2 nuclease family member with homology to yeast exonuclease 1. Nucleic Acids Res 26: 3762–3768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Hickson ID 2003. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426: 870–874 [DOI] [PubMed] [Google Scholar]
- Wu L, Davies SL, Levitt NC, Hickson ID 2001. Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J Biol Chem 276: 19375–19381 [DOI] [PubMed] [Google Scholar]
- Wyman C, Kanaar R 2006. DNA double-strand break repair: All's well that ends well. Annu Rev Genet 40: 363–383 [DOI] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G 2008. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134: 981–994 [DOI] [PMC free article] [PubMed] [Google Scholar]