Abstract
In Saccharomyces cerevisiae cells, B-type cyclin-dependent kinases (CDKs) target two origin recognition complex (ORC) subunits, Orc2 and Orc6, to inhibit helicase loading. We show that helicase loading by ORC is inhibited by two distinct CDK-dependent mechanisms. Independent of phosphorylation, binding of CDK to an “RXL” cyclin-binding motif in Orc6 sterically reduces the initial recruitment of the Cdt1/Mcm2–7 complex to ORC. CDK phosphorylation of Orc2 and Orc6 inhibits the same step in helicase loading. This phosphorylation of Orc6 is stimulated by the RXL motif and mediates the bulk of the phosphorylation-dependent inhibition of helicase loading. Direct binding experiments show that CDK phosphorylation specifically blocks one of the two Cdt1-binding sites on Orc6. Consistent with the inactivation of one Cdt1-binding site preventing helicase loading, CDK phosphorylation of ORC causes a twofold reduction of initial Cdt1/Mcm2–7 recruitment but results in nearly complete inhibition of Mcm2–7 loading. Intriguingly, in addition to being a target of both CDK inhibitory mechanisms, the Orc6 RXL/cyclin-binding motif plays a positive role in the initial recruitment of Cdt1/Mcm2–7 to the origin, suggesting that this motif is critical for the switch between active and inhibited ORC function at the G1-to-S-phase transition.
Keywords: Clb5, DNA replication, cell cycle regulation, origin licensing, prereplicative complex (pre-RC), rereplication
DNA replication in dividing eukaryotic cells is tightly controlled to ensure that the genome is replicated exactly once per cell cycle (Sclafani and Holzen 2007). This regulation is achieved primarily through the temporal separation of two events required for replication initiation: (1) loading of the core engine of the replicative DNA helicase, the Mcm2–7 complex, at origins of replication; and (2) helicase activation and replisome assembly. Helicase loading, which is also referred to as prereplicative complex (pre-RC) formation or origin licensing, occurs exclusively during the G1 phase of the cell cycle. In contrast, helicase activation occurs only after cells enter S phase. Thus, during each cell cycle, helicase loading and activation can occur only once, ensuring that no DNA can be replicated more than once.
Helicase loading requires the coordinated action of multiple proteins to assemble a double hexamer of the Mcm2–7 complex around dsDNA (Remus and Diffley 2009). The first step in this process is the binding of the origin recognition complex (ORC) to origin DNA. During G1, ORC recruits Cdc6 and a heptameric complex between Cdt1 and Mcm2–7. In a mechanism that requires ATP hydrolysis by Cdc6 (Randell et al. 2006), these proteins assemble a head-to-head double hexamer of the Mcm2–7 complex around dsDNA (Evrin et al. 2009; Remus et al. 2009). The lack of intermediates with only a single Mcm2–7 complex around origin DNA (Remus et al. 2009) suggests that either this event involves the coordinated assembly of two Mcm2–7 complexes, or separately loaded Mcm2–7 complexes only stably bind origin DNA after they form a double hexamer.
The mechanisms that ensure that the genome is replicated only once per cell cycle vary between organisms, but primarily inhibit helicase loading outside of G1 (Arias and Walter 2007). In the yeast Saccharomyces cerevisiae, this inhibition is mediated by B-type cyclin-dependent kinase (CDK) phosphorylation of helicase loading proteins (Nguyen et al. 2001). ORC, Cdc6, and the Mcm2–7 complex are each modified by CDK in a manner that inhibits helicase loading. CDK phosphorylation of Cdc6 leads to its SCF-dependent degradation (Drury et al. 1997, 2000; Elsasser et al. 1999). In contrast, CDK phosphorylation of soluble Mcm2–7 leads to its export to the cytoplasm, although the localization of chromatin-associated Mcm2–7 is unaffected (Labib et al. 1999; Nguyen et al. 2000; Liku et al. 2005). Finally, CDK phosphorylation of two ORC subunits, Orc2 and Orc6, inhibits helicase loading through an unknown mechanism (Nguyen et al. 2001; Green et al. 2006; Tanny et al. 2006). Orc6 also includes a “RXL” cyclin-binding motif that interacts with Clb5, the cyclin subunit for the most prominent S-phase CDK (Wilmes et al. 2004). Analysis of mutations lacking the Orc6 RXL motif suggests that this interaction also contributes to the prevention of helicase loading outside of G1 (Wilmes et al. 2004; Tanny et al. 2006).
Here we investigate the mechanism of CDK inhibition of ORC function. We show that CDK can inhibit helicase loading by two mechanisms: sterically inhibiting ORC function by binding to the Orc6 RXL motif, and by phosphorylating Orc2 and Orc6. In each case, these modifications inhibit the initial recruitment of the Cdt1/Mcm2–7 complex to the origin DNA. Further studies support a model in which two Cdt1-binding sites on ORC are required for helicase loading, and reveal a central role for the Orc6 RXL motif in the CDK inhibition of this event.
Results
Three motifs contribute to ORC-dependent rereplication control
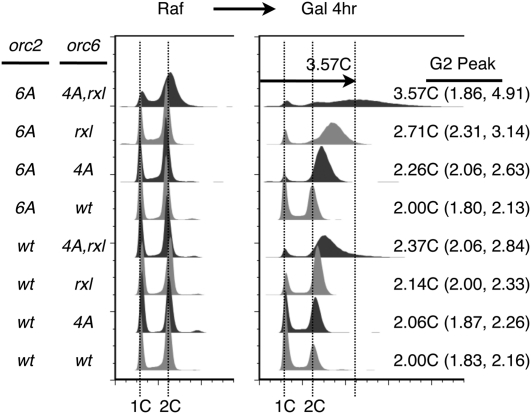
To address the contribution of different CDK targets on ORC to the prevention of rereplication, we generated yeast strains that included different combinations of mutations in the CDK target sites found in ORC: Orc2 phosphorylation sites (S16, T24, T70, T174, S188, and S206; orc2-6A), Orc6 phosphorylation sites (S106, S116, S123, and T146; orc6-4A) (Nguyen et al. 2001), and the RXL cyclin-binding motif on Orc6 (R178A and L180A; orc6-rxl) (Wilmes et al. 2004). Each strain also included mutations that bypassed CDK inhibition of Cdc6 and Mcm2–7. In the case of CDC6, we used a mutation that eliminated amino acids 1–47, which stabilizes the remainder of Cdc6 by removing a CDK phosphorylation-dependent degron that is recognized by the SCF-Cdc4 ubiquitin ligase (Drury et al. 1997; Nguyen et al. 2001). This gene was placed under the control of the GAL1 promoter to restrict its expression to cells grown in galactose (Gal-ΔntCDC6-HA). To bypass CDK regulation of Mcm2–7 localization, we used an allele of MCM7 that is fused to the SV-40 nuclear localization sequence at its C terminus (MCM7-NLS). This fusion protein maintains the Mcm2–7 complex in the nucleus even after CDK phosphorylation normally causes Mcm2–7 nuclear export. Importantly, neither of these mutations has a significant rereplication phenotype on its own, and, even when combined, they only cause substantial rereplication at only one locus (Green et al. 2006). Although a subset of these ORC mutations has been tested for rereplication previously (Wilmes et al. 2004; Archambault et al. 2005), we examined the complete set of strains to better compare how each mutation/combination of mutations affects rereplication.
The extent of rereplication correlated with the number of ORC mutations present in the strain. Prior to induction of ΔntCdc6-HA, each strain grew normally, cycling between 1C and 2C DNA content (Fig. 1). Upon induction of ΔntCdc6-HA overexpression, we observed varying levels of rereplication, as indicated by cells with >2C DNA content. To compare rereplication levels, we determined the average ploidy of the replicated DNA. Consistent with previous results (Wilmes et al. 2004; Archambault et al. 2005), we found that the strain in which all three CDK target motifs were mutated exhibited the most extensive rereplication. Intermediate levels of rereplication were observed for each pairwise combination of ORC mutations. Finally, only limited rereplication was observed for strains expressing any individual ORC mutation. Analysis of Cdc6 protein levels showed that the extent of rereplication was not correlated with the level of Cdc6 overexpression (Supplemental Fig. S1), suggesting that it was present in excess. Instead, we conclude that the mutations in ORC were responsible for the differences in rereplication, and that all three mutations contributed additively.
Figure 1.
Three motifs in Orc2 and Orc6 inhibit genomic rereplication. Cells with the indicated ORC2 and ORC6 genotypes were grown in selective medium overnight and transferred to YPRaffinose medium for 4 h before inducing with 3% galactose. All cells contained a constitutively nuclear-localized Mcm7 and a galactose-inducible, nondegradable Cdc6. The DNA contents of samples from before and after galactose induction were analyzed by flow cytometry. The extent of rereplication is expressed as the position of the peak of replicated DNA, with its 50% height positions indicated in parentheses.
CDK phosphorylation of ORC inhibits helicase loading in vitro
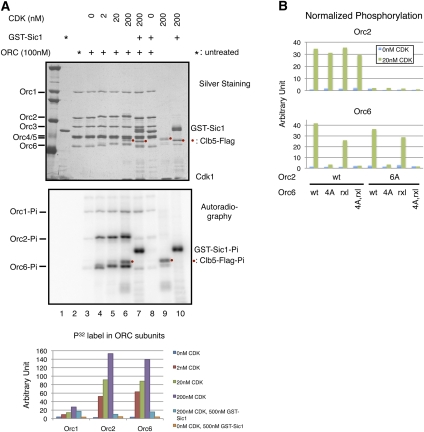
We measured phosphorylation of ORC by CDK (Clb5-Cdk1) using purified proteins (Supplemental Fig. S2A). Although very weak background phosphorylation was observed in the absence of added kinase, increasing concentrations of CDK resulted in a 20-fold to 50-fold increase in phosphorylation of Orc2 and Orc6, with weak phosphorylation of Orc1 at high CDK levels (Fig. 2A). Addition of the CDK inhibitor Sic1 eliminated phosphorylation of Orc2, Orc6, and Clb5, while Sic1 was strongly phosphorylated (Fig. 2A, cf. lanes 6,7,9,10). Thus, CDK specifically targeted Orc2, Orc6, and, to a lesser extent, Orc1.
Figure 2.
Purified CDK specifically phosphorylates Orc2 and Orc6 subunits in vitro. (A) CDK phosphorylation of wild-type ORC. Purified wild-type ORC (100 nM) was phosphorylated with purified CDK and separated by SDS-PAGE, stained with silver (top), and exposed to autoradiography (middle). The mobility shifts of phosphorylated Orc2 and Orc6 bands are shown in the silver staining. (Bottom) The incorporation of 32P-phosphate into subunits Orc1, Orc2, and Orc6 was quantified and plotted. (B) Mutations of CDK target sites in ORC act in cis. Wild-type and mutant ORCs were partially purified on ARS1 DNA and phosphorylated with 20 nM CDK. Quantification of normalized phosphorylation levels is shown.
Mutation of Orc2 and Orc6 interfered with CDK phosphorylation. We incubated G1 extracts overexpressing wild-type or mutant ORC with ARS1-containing DNA coupled to magnetic beads to partially purify ORC. Analysis of CDK phosphorylation of DNA-bound mutant ORC showed that the presence of Orc6-4A or Orc2-6A eliminated phosphorylation of the corresponding subunits. The Orc6-rxl mutation resulted in an ∼40% reduction of Orc6 phosphorylation but had no effect on Orc2 phosphorylation (Fig. 2B; Supplemental Fig. S3). Interestingly, we observed that the Orc6-rxl protein had a reduced mobility that was similar to phosphorylated wild-type Orc6 that was unchanged by CDK phosphorylation of this protein (Supplemental Fig. S3, cf. lanes 1,2 and 5,6). All of the double and triple mutations were phosphorylated at levels consistent with the single mutations, with no evidence of mutations in one subunit altering phosphorylation of the other subunit.
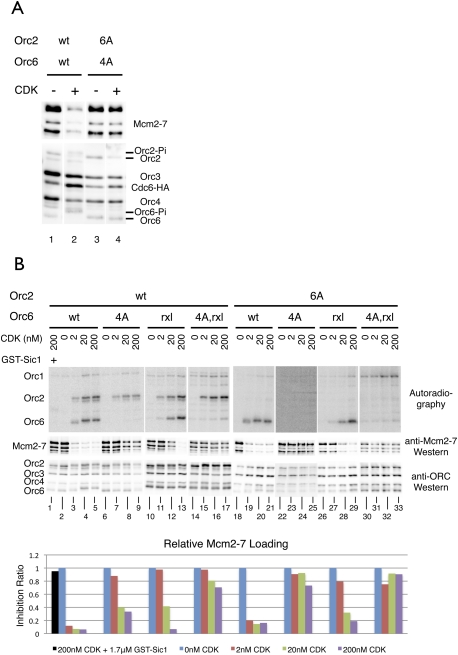
Although it is clear that CDK phosphorylation of ORC inhibits pre-RC formation (Tanny et al. 2006), the steps in the helicase loading reaction that are inhibited have been obscure. To address this question, we recapitulated CDK inhibition in an in vitro helicase loading reaction. We loaded wild-type ORC and ORCOrc2-6A Orc6-4A on ARS1 DNA, and treated it with purified CDK. After phosphorylation of ORC, CDK was washed away to prevent CDK phosphorylation of other helicase loading proteins. Subsequent addition of purified Cdc6 and G1-arrested cell extract lacking ORC drove helicase loading (Bowers et al. 2004). CDK phosphorylation of ORC dramatically reduced Mcm2–7 loading onto origin DNA (Fig. 3A, lanes 1,2). Mutation of the Orc2 and Orc6 phosphorylation sites eliminated CDK inhibition (Fig. 3A, lanes 3,4), indicating that CDK phosphorylation of ORC mediated the detected inhibition.
Figure 3.
CDK phosphorylation of ORC inhibits helicase loading in vitro. (A) Elimination of CDK phosphorylation sites on ORC prevents CDK inhibition of helicase loading. Pre-RC assembly was performed with wild-type ORC and ORCOrc2-6A Orc6-4A by preloading ORC on ARS1-containing DNA using ORC-overexpressing G1 whole-cell extracts (WCEs), treating with CDK, and washing, followed by incubation with ORC-depleted G1 WCE and purified Cdc6. DNA-associated proteins were analyzed by anti-Mcm (UM174, detects Mcm2, Mcm3, and Mcm6), anti-ORC (1108), and anti-HA (12CA5, for Cdc6-HA) immunoblotting. (B) Elimination of CDK target sites on ORC reduces CDK inhibition of helicase loading additively. Wild-type and mutant ORC was preloaded on ARS1 DNA and treated with CDK at the indicated concentrations in the presence of γ-P32-ATP. Pre-RC assembly reactions were carried out as in A. The extent of phosphorylation was revealed by autoradiography (top), and pre-RC assembly was revealed by immunoblotting (middle). (Bottom) The inhibition ratio of CDK was determined by quantification of Mcm2–7 levels and is expressed as the ratio of the amount of Mcm2–7 loaded in the presence of CDK relative to its absence.
Orc6 is the primary target for CDK inhibition
To assess the contribution of the different CDK targets on ORC to the inhibition of helicase loading, we treated wild-type and mutant ORC complexes with three different concentrations of CDK (Fig. 3B). As in the previous assays, we washed away CDK prior to initiating helicase loading to ensure that we were only measuring the consequences of CDK phosphorylation of ORC. We found that the DNA-bound Mcm2–7 observed in these experiments is a mixture of tightly DNA-bound and salt extraction-resistant “loaded” Mcm2–7 and more loosely DNA-bound and salt extraction-sensitive “associated” Mcm2–7 (Bowers et al. 2004). Importantly, salt extraction of pre-RC complexes formed with wild-type and mutant ORC showed the same relative levels of DNA-bound Mcm2–7 as those that were not salt-extracted (Supplemental Fig. S4). Thus, although we used non-salt-extracted reactions for the remaining analyses of CDK inhibition of helicase loading, we refer to the results as a measure of loaded Mcm2–7. We measured the level of CDK inhibition by determining the ratio of DNA-bound Mcm2–7 with and without CDK treatment (inhibition ratio) (Fig. 3B, bottom panel). Thus, the inhibition ratio will be 0 when CDK completely inhibits Mcm2–7 loading; if there is no CDK inhibition, this ratio will be 1. Treatment of wild-type ORC with the lowest amount of CDK (2 nM) resulted in an inhibition ratio of 0.12. A 10-fold increase in CDK concentration drove phosphorylation of Orc2 and Orc6 to completion and further decreased Mcm2–7 loading. Additional CDK led to no additional changes in either phosphorylation of ORC or loading of Mcm2–7. These effects were specific to CDK, as addition of GST-Sic1 fully alleviated CDK inhibition (Fig. 3B, cf. lanes 1 and 5).
Orc6 was the primary CDK target inhibiting helicase loading. CDK treatment of ORCOrc6-4A showed little inhibition at low CDK levels, and at the highest levels the inhibition ratio was only ∼0.3. Full CDK inhibition of Mcm2–7 loading by ORCOrc6-rxl was observed at only the highest CDK levels (Fig. 3B, cf. lanes 5 and 13), indicating that the Orc6-rxl defect could be overcome by increased CDK. In contrast, Mcm2–7 loading by ORCOrc6-4A,rxl was largely resistant to CDK treatment.
Elimination of Orc2 phosphorylation primarily reduced CDK inhibition of helicase loading in combination with Orc6 mutations. Mcm2–7 loading by ORCOrc2-6A remained sensitive to CDK treatment, although the extent of inhibition was less than wild-type ORC (Fig. 3B, lanes 18–21). However, when ORC included Orc2-6A and an Orc6 mutant, pre-RC formation became less sensitive to CDK compared with the complexes containing the Orc6 mutant alone (Fig. 3B, cf. lanes 22–33 and 6–17). When all the three motifs were mutated, Mcm2–7 loading was highly resistant to CDK phosphorylation, even at 200 nM CDK.
Orc6 phosphorylation inhibits Cdt1 recruitment in vitro
To determine the step in helicase loading that is inhibited by CDK phosphorylation of ORC, we analyzed several intermediate steps in the helicase loading reaction. We first measured the ATP hydrolysis and DNA-binding activities of ORC, because previous studies of Drosophila ORC found that CDK modification inhibited these activities (Remus et al. 2005). Using purified mock-treated yeast ORC or CDK-treated ORC (ORC-Pi) (Supplemental Fig. S2B), we found that there was no significant difference in ATP hydrolysis (ORC and ORC-Pi hydrolyzed 0.304 ± 0.032 and 0.341 ± 0.045 pmol of ATP/pmol of ORC per minute, respectively). Similarly, CDK phosphorylation did not affect ORC DNA binding measured by either ORC binding to bead-bound ARS1 DNA (Supplemental Fig. S3) or mobility shift assays (data not shown). Cdc6 recruitment to the origin also was not reduced by CDK treatment of ORC (Fig. 3A). Instead, we observed increased Cdc6 DNA association when ORC was phosphorylated. This increase most likely is due to Cdc6 stabilization in the absence of productive Mcm2–7 loading (Gillespie et al. 2001; Tsakraklides and Bell 2010).
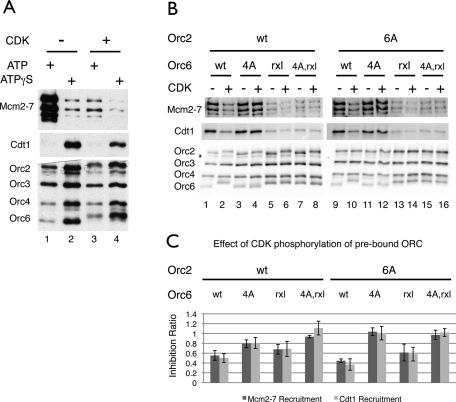
Previous studies indicate that Orc6 is required to recruit Cdt1 to the origin and contains two Cdt1-binding sites (Chen et al. 2007). Given the relative importance of Orc6 phosphorylation in CDK inhibition of Mcm2–7 loading, we asked whether CDK treatment altered Cdt1 origin recruitment. We measured the extent of Cdt1 recruitment by adding ATPγS to the helicase loading assay. This inhibits Cdc6 ATP hydrolysis and arrests helicase loading at an intermediate step after the initial recruitment of the Cdt1/Mcm2–7 complex but before the release of Cdt1 (Randell et al. 2006). To prevent ATPγS from interfering with CDK action, we first treated origin-bound ORC with CDK and ATP. After CDK phosphorylation, we washed away the CDK and ATP, and then initiated helicase loading in the presence of ATP or ATPγS (Fig. 4A). Interestingly, CDK phosphorylation of ORC led to a 50% decrease in the initial recruitment of Cdt1 and Mcm2–7 (Fig. 4A, lanes 2,4), in contrast to the >10-fold decrease in Mcm2–7 loading observed in the presence of ATP (Fig. 4A, lanes 1,3).
Figure 4.
CDK phosphorylation of ORC inhibits Cdt1 and Mcm2–7 recruitment in vitro. (A) CDK phosphorylation of wild-type ORC has different effects on helicase loading and Cdt1 recruitment. Helicase assembly reactions were performed as described in Figure 3 except ATPγS was substituted for ATP during the last incubation, as indicated. This prevents helicase loading but allows the detection of the initial recruitment of Cdt1/Mcm2–7 complexes. Cdt1 was detected using anti-Cdt1 antiserum HM5352. (B) Effect of ORC mutations on CDK inhibition of Cdt1 and Mcm2–7 recruitment. The initial recruitment of Cdt1/Mcm2–7 complexes was measured in the presence of ATPγS as in A using wild-type ORC and each ORC mutant after treatment with 20 nM CDK. (C) Inhibition ratios for Mcm2–7 and Cdt1 recruitment. Cdt1/Mcm2–7 recruitment assays were performed in triplicate for wild-type ORC and each of the ORC mutants using 20 nM CDK. Inhibition ratios were determined and plotted.
We tested each of the ORC mutants for their effect on the initial recruitment of Cdt1/Mcm2–7. Interestingly, all of the ORC complexes containing a mutation in the Orc6 RXL motif showed a threefold to fivefold reduction in both Cdt1 and Mcm2–7 recruitment with or without CDK treatment (Fig. 4B). Similarly, Mcm2–7 loading was consistently lower when the Orc6 subunit harbored the rxl mutation without CDK phosphorylation (Supplemental Fig. S5, cf. odd lanes; note that Fig. 3B should not be used for this comparison because the ORC mutants were not analyzed on the same gel). These observations suggest that, apart from its role in CDK targeting, the Orc6 RXL motif promoted efficient Cdt1/Mcm2–7 recruitment to ORC.
Mutation of ORC CDK target sites alleviated CDK inhibition of Cdt1/Mcm2–7 recruitment (e.g., Fig. 4B, cf. lanes 1,2 and 3,4). We determined the inhibition ratios for Mcm2–7 and Cdt1 recruitment in ATPγS after treating wild-type and mutant ORC with CDK (Fig. 4C). All of the complexes showed an inhibition ratio between 1 and 0.45. Consistent with both proteins being recruited as a complex, the extent of CDK inhibition of Cdt1 and Mcm2–7 recruitment paralleled each other for all ORC mutants. Importantly, although the extent of full CDK inhibition for Cdt1 and Mcm2–7 recruitment (inhibition ratio of ∼0.5) was distinct from Mcm2–7 loading (inhibition ratio <0.1), the relative effects of the ORC mutations on CDK inhibition of these events showed a similar pattern (cf. Figs. 3B [20 nM CDK] and 4B).
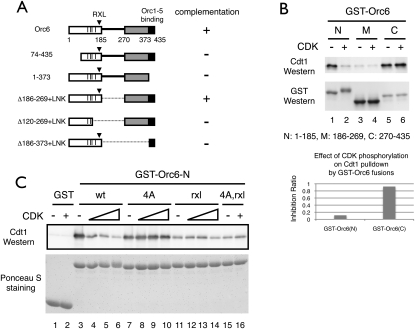
Phosphorylation of Orc6 N-terminal domain inhibits Cdt1 binding
The twofold reduction in Cdt1/Mcm2–7 recruitment after CDK treatment of ORC could be due to reduced affinity of a single Cdt1/Mcm2–7-binding site or complete inhibition of one of two binding sites for Cdt1/Mcm2–7 at the origin. Consistent with the latter possibility, previous studies identified two Cdt1-binding domains on Orc6 (Chen et al. 2007): one at the N terminus (1–185) and a second at the C terminus (270–435), separated by a D/E-rich region that did not bind Cdt1. Interestingly, the N-terminal-binding domain encompasses all four Orc6 CDK target sites (Nguyen et al. 2001) and the RXL motif (Wilmes et al. 2004).
Both the N- and C-terminal domains of Orc6 are required for cell growth. We tested a series of Orc6 mutations for their ability to complement an Orc6 deletion in vivo (Fig. 5A). Deletion of the last 62 amino acids, which mediate interaction of Orc6 with Orc1–5 (Chen et al. 2007), failed to support growth. In contrast, a mutant that replaced the 85-amino-acid D/E-rich region (186–269) with a 24-amino-acid flexible linker (Δ186–269 + LNK) complemented Orc6 function. Importantly, mutations that truncated either the N-terminal (Δ120–269 + LNK) or C-terminal (Δ186–373 + LNK) domains of Orc6 each failed to complement. Together with previous studies, these findings suggest that Orc6 is composed of two essential Cdt1-binding domains connected by a flexible linker.
Figure 5.
CDK phosphorylation of the essential N-terminal Cdt1-binding domain of Orc6 inhibits Cdt1 association. (A) The two Cdt1 interaction domains of Orc6 are both required to complement an ORC6 deletion in vivo. A schematic representation shows Orc6 domain structure as two Cdt1 interaction domains (open and gray boxes, with start and end amino acid positions shown below) joined by a D/E-rich region. Vertical lines denote CDK phosphorylation sites, while a triangle indicates the position of the RXL motif. A dashed line represents a 24-amino-acid flexible linker (LNK) made up of primarily glycine and serine [R(SGGGG)4SGS]. (B) CDK inhibits Cdt1 binding to the Orc6 N-terminal Cdt1-binding domain. GST fusion proteins of Orc6 fragments were bound to glutathione beads, washed, and treated with CDK and ATP. (Top panel) Purified Cdt1 protein was incubated with GST-Orc6. Bound Cdt1 was detected by immunoblotting. Immunoblotting with anti-GST antibody of glutathione bead-bound proteins is shown in the middle panel. (Bottom panel) A CDK inhibition ratio was determined for Cdt1 binding to the N- and C-terminal domains of Orc6. (C) CDK target site mutations prevent CDK inhibition of Cdt1 binding to the Orc6 N-terminal domain. Cdt1 binding to wild-type and the indicated mutant GST-Orc6 N-terminal domains was measured after phosphorylation without or with increasing amounts of CDK.
To determine if CDK preferentially inhibited binding to one of the two Cdt1-binding sites on Orc6, we measured Cdt1 binding to the different domains of Orc6 with and without CDK phosphorylation (Fig. 5B). Cdt1 bound the N- and C-terminal domains of Orc6 equally well but showed no affinity for the middle region. CDK phosphorylation dramatically reduced Cdt1 binding to the Orc6 N-terminal domain but had no effect on Cdt1 binding to the Orc6 C-terminal domain. We also tested Cdt1 binding to an N-terminal fragment of Orc2 that included all of the CDK target sites and did not observe binding (data not shown).
Elimination of the Orc6 CDK target sites prevented CDK inhibition of Cdt1 binding to the Orc6 N-terminal domain. Cdt1 binding to GST-Orc6N-4A was not inhibited by CDK (Fig. 5C). Binding of Cdt1 to GST-Orc6N-rxl and GST-Orc6N-4A,rxl was lower compared with wild type, consistent with the observation that the rxl mutation reduced the initial recruitment of Cdt1/Mcm2–7 (Fig. 4B). We did not observe significant inhibition of Cdt1 binding after CDK treatment of either GST-Orc6N-rxl or GST-Orc6N-4A,rxl under these conditions.
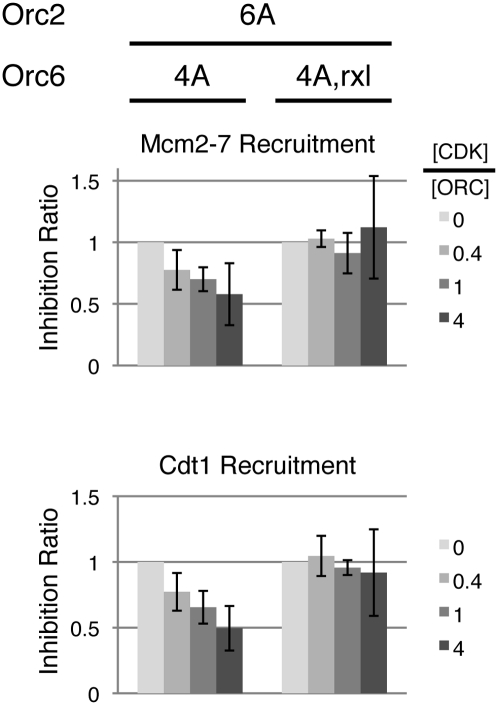
CDK binding to the RXL motif causes steric hindrance to helicase loading
Our observation that the Orc6 RXL motif facilitates helicase loading and prior findings that Clb5-Cdk1 (the form of CDK studied here) binds the same motif in Orc6 (Wilmes et al. 2004) suggested that this form of CDK could sterically inhibit the events of helicase loading. This mechanism of CDK inhibition would not have been detected in our previous experiments because CDK was washed away before the helicase loading or Cdt1/Mcm2–7 recruitment assays were performed. To assess the effect of Clb5-Cdk1 binding to ORC, we carried out the Cdt1/Mcm2–7 recruitment assay in the presence of this CDK. To ensure that any effect of Clb5-Cdk1 addition was independent of its phosphorylation of ORC, we studied two mutants of ORC that could not be phosphorylated by CDK. These mutants were distinguished by the presence or absence of the Orc6 RXL motif (ORCOrc2-6A Orc6-4A and ORCOrc2-6A Orc6-4A,rxl). As we observed with ATP, direct phosphorylation experiments using radioactive ATPγ35S showed that neither of the mutant complexes was phosphorylated at detectable levels (data not shown).
We tested three concentrations of Clb5-Cdk1, resulting in molar ratios of the kinase to ORC of 0.4, 1, and 4. The ratios of 0.4 and 4 corresponded to the 20 nM and 200 nM CDK concentrations in Figures 3 and 4. Despite their similar response to CDK treatment in previous experiments (Figs. 3, 4), these two mutants reacted differently when CDK was retained during Cdt1/Mcm2–7 recruitment (Fig. 6). The ORCOrc2-6A Orc6-4A mutant showed decreased Cdt1 and Mcm2–7 recruitment with increasing amounts of CDK. In contrast, Cdt1/Mcm2–7 recruitment was unaffected by even the highest level of CDK when the ORCOrc2-6A Orc6-4A,rxl mutant was tested under the same conditions. The dependence of this CDK inhibition on the presence of the Orc6 RXL motif and the proximity of this motif to the Cdt1-inhibiting CDK phosphorylation sites support a model in which CDK inhibits Cdt1/Mcm2–7 recruitment through steric inhibition, although we cannot eliminate a role for RXL-facilitated phosphorylation of other helicase loading proteins by CDK.
Figure 6.
CDK binding to the RXL motif sterically inhibits the initial recruitment of Cdt1/Mcm2–7. ORCOrc2-6A Orc6-4A and ORCOrc2-6A Orc6-4A,rxl were preloaded on ARS1 DNA and incubated with CDK in the presence of ATPγS. Cdt1 stabilization was assayed after addition of ORC-depleted G1 WCE and Cdc6 without removing CDK. Inhibition ratios were determined and averaged for three experiments.
Discussion
Two Cdt1-binding sites mediate Mcm2–7 helicase loading
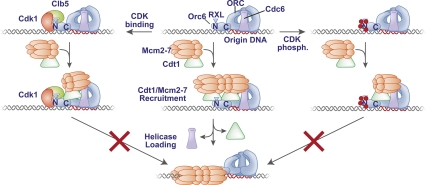
Our findings show that CDK can inhibit ORC function both by steric hindrance via direct binding to Orc6 and through phosphorylation of Orc2 and Orc6. In each case, CDK inhibits the same step in the helicase loading process: the initial recruitment of Cdt1/Mcm2–7 to the origin DNA. In vivo studies suggest that these two mechanisms work together to determine the extent of CDK inhibition of ORC. Phosphorylation of ORC by CDK leads to a partial loss of Cdt1/Mcm2–7 recruitment but a near complete inhibition of Mcm2–7 loading. Although it is possible that only the N-terminal Cdt1-binding site of Orc6 is important for helicase loading, our finding that the C-terminal binding domain is essential for Orc6 function suggests that this is not the case (Fig. 5A). Instead, we propose that CDK phosphorylation blocks binding of one of two Cdt1/Mcm2–7 complexes, and both are required for loading of a Mcm2–7 double hexamer on the origin DNA (Fig. 7). In support of this model, recent studies from our laboratory have identified a helicase loading intermediate containing two Cdt1 molecules, and have shown that a Cdt1 mutant that only allows one Cdt1-binding event prevents helicase loading (TJ Takara and SP Bell, in prep.).
Figure 7.
Model for the mechanisms of CDK-dependent inhibition of ORC. The middle panel illustrates the Cdt1/Mcm2–7 recruitment and helicase loading events. We propose that the two nonidentical Cdt1-binding sites on Orc6 facilitate the association of Cdt1/Mcm2–7 complexes in opposite orientations. The RXL motif of Orc6 stimulates the initial recruitment of Cdt1/Mcm2–7. This intermediate shows a Mcm2–7 double hexamer, although it is also possible that the two Mcm2–7 complexes do not interact at this stage. Helicase loading results in the release of Cdt1 and the assembly of a Mcm2–7 double hexamer around the DNA. Loading of the two hexamers could be coordinated or sequential, but binding to both sites is required for stable Mcm2–7 loading. (Right panel) We propose that CDK phosphorylation of Orc6 inhibits Cdt1/Mcm2–7 binding to the N-terminal Cdt1-binding domain of Orc6, but binding to the C-terminal domain is retained. The resulting complex is unable to load Mcm2–7 onto origin DNA. This could be due to the failure to form a Mcm2–7 double-hexamer intermediate or the inability to load Mcm2–7 in both orientations. A mechanism for the effect of Orc2 phosphorylation is unclear (and not illustrated), but is likely to inhibit the same step as Orc6 phosphorylation. (Left panel) We propose that Clb5-Cdk1 binding to the Orc6 RXL motif also inhibits the initial recruitment of Cdt1–Mcm2–7 to ORC. The proximity of the RXL motif to the Orc6 N-terminal, Cdt1-binding domain suggest that Clb5 binding to the Orc6 RXL motif inhibits Cdt1 binding to the Orc6 N-terminal domain. It is possible that Orc6-bound Clb5-Cdk1 can also inhibit Cdt1 binding to the C-terminal Cdt1-binding domain. We predict the resulting decrease in Cdt1/Mcm2–7 recruitment inhibits subsequent Mcm2–7 loading. Although not shown, the Clb5–Orc6–RXL interaction also facilitates phosphorylation of Orc6.
The presence of a pair of Cdt1-binding sites on ORC is consistent with the recent finding that only a head-to-head double-hexamer form of the Mcm2–7 complex is stably associated with origin DNA (Remus et al. 2009). The lack of loaded Mcm2–7 single hexamers on the DNA in these studies suggests that either both hexamers must be loaded coordinately or double-hexamer formation after loading is required for stable binding of Mcm2–7 to the origin DNA. Because the two regions of Orc6 that bind to Cdt1 show no similarity, it is likely that they interact with distinct regions of Cdt1. A nonidentical pair of Cdt1/Mcm2–7-binding sites on ORC could allow Cdt1/Mcm2–7 to be positioned in different orientations and provide an ideal scaffold to initiate head-to-head double-hexamer formation and cooperative loading (Fig. 7). Alternatively, it is possible that loading is not coordinated, but that each Cdt1/Mcm2–7-binding site is dedicated to loading the asymmetric Mcm2–7 in opposite orientations. In this model, eliminating one binding site would allow loading of Mcm2–7 in only one orientation, which would be expected to be unstable. We found previously that tethering Cdt1 to a 62-amino-acid C-terminal domain of Orc6 that bound the remaining ORC subunits recruited Mcm2–7 robustly but was inefficient for Mcm2–7 loading (Chen et al. 2007). Based on our present findings, we suspect that this defect is at least partly due to the recruitment of only one Mcm2–7 complex and, thus, the prevention of coordinated Mcm2–7 loading.
CDK inhibition of ORC function is mediated largely through Orc6
Our findings support a more direct and important role for Orc6 relative to Orc2 in the CDK inhibition of helicase loading. In both the rereplication and in vitro Cdt1/Mcm2–7 recruitment and helicase loading assays, only orc6 mutations show a defect on their own (Figs. 1, 3). In contrast, the orc2 mutation shows a defect only when combined with one or both orc6 mutations. Of the three mutations in ORC studied, the orc6-rxl mutation consistently showed the largest increases in rereplication (Fig. 1), either on its own or when it was combined with any other mutation(s).
A particularly important role for the Orc6 RXL motif is supported by our findings that it participates in both mechanisms of CDK inhibition of Cdt1/Mcm2–7 recruitment. First, the RXL motif facilitates CDK phosphorylation of Orc6 (Figs. 2B, 3B, cf. lanes 3,4 and 11,12). This is consistent with our finding that increased CDK levels could overcome the reduced CDK inhibition of helicase loading when ORC containing an Orc6-rxl mutant was tested (Fig. 3B, cf. lanes 2–5 and 10–13, and lanes 18–21 and 26–29). Importantly, in these experiments, CDK was removed prior to helicase loading and therefore could only affect the reaction by phosphorylating ORC. When we retained CDK during helicase recruitment, we observed a second function for the RXL motif. CDK addition resulted in a phosphorylation-independent but RXL-dependent inhibition of Cdt1/Mcm2–7 recruitment. A phosphorylation-independent role of the RXL motif is supported by the large increase in rereplication when this site is mutated in a strain lacking Orc2 and Orc6 phosphorylation sites (Fig. 1, cf. rows 1 and 3). Because previous studies suggest that Clb5 but not Clb2 can bind the Orc6 RXL motif (Wilmes et al. 2004), it is likely that the latter steric effect is specific to the Clb5-Cdk1 form of CDK.
In addition to its role in CDK regulation of helicase loading, the Orc6 RXL motif also plays a positive role in Cdt1 binding to ORC. Mutation of the Orc6 RXL motif interfered with Cdt1/Mcm2–7 recruitment (Fig. 4B), Mcm2–7 loading (Supplemental Fig. S5), and Orc6 binding to Cdt1 (Fig. 5C). Since Clb5 also binds the Orc6 RXL motif, it is likely that Clb5-Cdk1 and Cdt1 compete for binding to this motif (Fig. 7). Because previous studies found that Clb5 associates with the origin DNA only after initiation has occurred (Wilmes et al. 2004), the RXL motif is likely blocked until the helicase is activated, potentially through a second interaction with the Mcm2–7 complex (Cdt1 is not present at the origin after helicase loading) (Randell et al. 2006). The role of the Orc6 RXL motif in Mcm2–7 recruitment and loading also may explain why there is a difference in the CDK sensitivity of Mcm2–7 loading when ORCorc6-4A is compared with ORCorc6-4A-rxl (Fig. 3B, cf. lanes 6–9 and 14–17). If the only function of the Orc6 RXL motif was to facilitate CDK phosphorylation of Orc6 (Fig. 2), then mutation of the RXL motif in Orc6 lacking its phosphorylation sites should not cause further reduction in the CDK inhibition of Mcm2–7 loading. Instead, we see that the ORCorc6-4A-rxl is less sensitive to CDK inhibition than ORCorc6-4A. CDK can only phosphorylate Orc2 in these ORC mutants. To explain these findings, we propose that the Orc6-rxl mutation and Orc2 phosphorylation interfere with the same interaction. If this is the case, then including the Orc6-rxl mutation would reduce the effect of CDK phosphorylation of ORCorc6-4A-rxl relative to ORCorc6-4A by partially disrupting the interaction that Orc2 phosphorylation inhibits.
Although our studies did not define a clear mechanism by which Orc2 CDK phosphorylation inhibits helicase loading, several lines of evidence suggest that phosphorylation of Orc2 inhibits the same step in helicase loading as Orc6. First, mutations that eliminate Orc2 and Orc6 CDK phosphorylation sites show additive effects on rereplication in vivo and resistance to CDK in vitro. On the contrary, if Orc2 and Orc6 phosphorylation acted at different steps in the process, we would expect that there would be an epistatic relationship between the mutations. Second, the Orc2 phosphorylation mutant has little effect on its own, but enhances the ability of Orc6 mutations to bypass CDK regulation. Third, both Orc2 and Orc6 mutants altered the CDK inhibition of the initial recruitment of Cdt1/Mcm2–7 to ORC. The simplest explanation for our findings is that both Orc6 and Orc2 interact with the Cdt1/Mcm2–7 complex, but the Orc2 interaction is weaker. CDK phosphorylation-dependent elimination of the Orc2 interaction has little effect on its own, but in combination with the loss of the Orc6 interaction with Cdt1, the loss of the Orc2 interaction further reduces Cdt1/Mcm2–7 binding. Given the large size of both ORC (>400 kDa) and the Cdt1/Mcm2–7 complex (>600 kDa), it is not surprising that additional contacts would be involved in their interaction.
Correlation between in vitro assembly and in vivo rereplication
We observed good correspondence between our assays monitoring the effects of ORC mutations on pre-RC formation in vitro and rereplication in vivo. We found that, when Cdc6 is stabilized and Mcm2–7 is constitutively nuclear, CDK phosphorylation of Orc2, Orc6, and the Orc6 RXL motif contributes additively to rereplication levels. Our in vitro studies addressing the inhibition of Cdt1/Mcm2–7 recruitment and helicase loading by CDK phosphorylation of ORC showed a similar additive pattern.
Although the additive nature of the different ORC mutations on rereplication and our in vitro helicase loading assays was similar, the quantitative differences were more difficult to compare. Two considerations complicate a quantitative comparison. First, the in vitro helicase loading assays were done after washing CDK away from ORC, which did not allow the steric inhibition effect of Clb5 binding to the Orc6 RXL motif. Although it was possible that Clb5-Cdk1 could have been retained on the DNA-bound ORC after washing, direct assays did not detect any associated Clb5. We suspect that the association of Clb5-Cdk1 is relatively weak and requires the continuous presence of Clb5-Cdk1 in solution to be maintained. As a result, our helicase loading assays would be expected to underestimate the contribution of the Orc6 RXL motif to CDK regulation of helicase loading. Second, the rereplication assays measure reinitiation from all origins, whereas the in vitro assays monitor a single origin: ARS1. Previous studies showed that mutations in CDK targets involved in preventing rereplication have origin-specific effects (Green et al. 2006; Tanny et al. 2006). Thus, the effects on ARS1 are unlikely to precisely reflect the effects at all origins.
It is noteworthy that, unlike the indirect effects of CDK phosphorylation on Cdc6 stability and Mcm2–7 nuclear localization, our studies indicate that CDK phosphorylation of ORC intrinsically inhibits its function during helicase loading. A direct mechanism of inhibition has the potential to inactivate helicase loading more rapidly than the multistep events required for ubiquitin-dependent degradation or nuclear export. Rapid inactivation of ORC could be especially important during passage between G1 and S phase, when cells are transitioning between helicase loading and helicase activation. The use of two CDK-dependent mechanisms to inhibit ORC function may also make ORC a particularly effective target for CDK inhibition of helicase loading. Studies of rereplication induced by deregulation of a subset of CDK targets support a more important role for ORC in this process (Green et al. 2006). Elimination of both Cdc6 and Mcm2–7 regulation results in rereplication initiating from only a single origin of replication, suggesting that CDK inhibition of ORC alone is sufficient to inhibit detectable initiation from the remaining origins. In contrast, deregulation of ORC and Cdc6 allows significant rereplication (Green et al. 2006).
Materials and methods
Plasmids and yeast strains
Details of plasmid and yeast strain constructions are included in the Supplemental Material. See Supplemental Table S1 for strain genotypes.
Rereplication assay
Cells were grown in SC-Ura medium overnight, washed, and transferred to YPRaffinose medium at OD600 = 0.4 for 4 h. The GAL-ΔntCDC6-HA allele was induced by the addition of galactose to 3% final concentration. Samples were taken every hour for 4 h.
Protein expression and purifications
Detailed expression and purification protocols are included in the Supplemental Material.
CDK was purified from yeast strain ySC295, which overexpresses Clb5-Flag and Cdk1, by anti-Flag affinity purification. Flag-tagged ORC was purified from strain F1ORC1 as described previously (Tsakraklides and Bell 2010) and further purified on a 1-mL mono-Q column. GST-Orc6 fragments were purified as described previously (Chen et al. 2007).
CDK phosphorylation of ORC
We used concentrations of ORC (100 nM) and Clb5-Cdk1 (2–200 nM) based on estimations of their concentrations in growing yeast cells (Ghaemmaghami et al. 2003). Purified wild-type ORC (100 nM) was incubated with purified CDK at the indicated concentrations with 1 mM ATP and 30 μCi of γ-P32-ATP for 10 min at room temperature. Reactions were stopped with addition of TCA, and precipitated proteins were separated on an 8% SDS–polyacrylamide gel and silver-stained. The incorporation of 32P-phosphate was detected by autoradiography, quantitated on a PhosphorImager screen, and plotted for subunits Orc1, Orc2, and Orc6.
Preparation of whole-cell extracts (WCEs) and ORC-depleted extracts
Yeast WCE and ORC-depleted extract from strain ySC7 were prepared as described (Randell et al. 2006). To prepare WCEs from yeast cells overexpressing wild-type and mutant ORC, cells were grown in YP glycerol overnight to an OD600 of 0.6 at 30°C, induced by addition of galactose to 2% for 2 h before arresting with α-factor for 4 h. Cells were ground in a freezer mill as described (Tsakraklides and Bell 2010). Extracts were dialyzed against H300KGlut.
Preparation of photo-cleavable ARS1 DNA-coupled beads
Photo-cleavable ARS1-containing 1-kb DNA substrates were generated by PCR using Advantage 2 Polymerase (BD Biosciences) and the primers Ret101 (TGTGGAATTGTGAGCGGATA) and photo-cleavable 5′-biotin-labeled ARS1-3PCBio (5PCBio-CTGTTTTGTCCTTGGAAAAAAAGCACTACC) with pARS1/wild-type plasmid as described (Chen et al. 2007), and were purified with a Vivaspin 4-mL concentrator (MWCO 100,000).
Helicase loading and Cdt1/Mcm2–7 recruitment assays and CDK phosphorylation
Helicase loading assays were performed in three steps: Ten microliters of ORC overexpression extracts was incubated with 0.5 pmol of 1-kb photo-cleavable ARS1 DNA coupled to streptavidin beads in a 20-μL reaction as described previously (Randell et al. 2006), washed with H300KGlut, and treated with CDK in 10 μL of H300KGlut with 1 mM ATP (and γ-P32-ATP when needed) for 15 min at room temperature. DNA-bound proteins were washed and incubated with 10 μL of ORC-depleted ySC7 extract and 0.4 μg of purified Cdc6 in a 20-μL reaction including ATP for 15 min. Beads were washed three times with H300KGlut, resuspended in 30 μL of H300KGlut, and cleaved at short wavelength on a UV light box for 15 min. Released protein and DNA samples were precipitated with TCA and analyzed by SDS-PAGE and immunoblots. Proteins were quantified by comparing with known quantities of the same protein analyzed on the same immunoblot. Cdt1/Mcm2–7 recruitment assays were performed in the same manner except ATPγS was substituted for ATP in the last incubation.
GST pull-down assays
Lysates of GST-Orc6 fragments were incubated with glutathione Sepharose beads as described (Chen et al. 2007). The Orc-bound glutathione Sepharose beads were washed and resuspended in 50 μL of 2.5 mM ATP in H150 KCl with fivefold dilutions of CDK. After 30 min of incubation at room temperature, 0.75 μg of purified Cdt1 was added and incubated for 4 h at 4°C. Beads were washed with 0.25 mL of H150 three times and eluted in 0.2 mL of H150 containing 10 mM glutathione. One-fourth of the total proteins was assayed on a 10% SDS-PAGE.
Acknowledgments
We thank John Randell, Robyn Tanny, and Ryan Heller for plasmids and yeast strains, and Terry Orr-Weaver, Vasiliki Tsakraklides, Jue D. Wang, and Hannah G. Blitzblau for critical reading and helpful comments on this manuscript. The work described here was supported by the Howard Hughes Medical Institute. S.P.B. and S.C. are employees of the Howard Hughes Medical Institute.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.2011511.
Supplemental material is available for this article.
References
- Archambault V, Ikui AE, Drapkin BJ, Cross FR 2005. Disruption of mechanisms that prevent rereplication triggers a DNA damage response. Mol Cell Biol 25: 6707–6721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias EE, Walter JC 2007. Strength in numbers: Preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev 21: 497–518 [DOI] [PubMed] [Google Scholar]
- Bowers JL, Randell JC, Chen S, Bell SP 2004. ATP hydrolysis by ORC catalyzes reiterative Mcm2–7 assembly at a defined origin of replication. Mol Cell 16: 967–978 [DOI] [PubMed] [Google Scholar]
- Chen S, de Vries MA, Bell SP 2007. Orc6 is required for dynamic recruitment of Cdt1 during repeated Mcm2–7 loading. Genes Dev 21: 2897–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drury LS, Perkins G, Diffley JF 1997. The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J 16: 5966–5976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drury LS, Perkins G, Diffley JF 2000. The cyclin-dependent kinase Cdc28p regulates distinct modes of Cdc6p proteolysis during the budding yeast cell cycle. Curr Biol 10: 231–240 [DOI] [PubMed] [Google Scholar]
- Elsasser S, Chi Y, Yang P, Campbell JL 1999. Phosphorylation controls timing of Cdc6p destruction: A biochemical analysis. Mol Biol Cell 10: 3263–3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrin C, Clarke P, Zech J, Lurz R, Sun J, Uhle S, Li H, Stillman B, Speck C 2009. A double-hexameric MCM2–7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci 106: 20240–20245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS 2003. Global analysis of protein expression in yeast. Nature 425: 737–741 [DOI] [PubMed] [Google Scholar]
- Gillespie PJ, Li A, Blow JJ 2001. Reconstitution of licensed replication origins on Xenopus sperm nuclei using purified proteins. BMC Biochem 2: 15 doi: 10.1186/1471-2091-2-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green BM, Morreale RJ, Ozaydin B, Derisi JL, Li JJ 2006. Genome-wide mapping of DNA synthesis in Saccharomyces cerevisiae reveals that mechanisms preventing reinitiation of DNA replication are not redundant. Mol Biol Cell 17: 2401–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K, Diffley JF, Kearsey SE 1999. G1-phase and B-type cyclins exclude the DNA-replication factor Mcm4 from the nucleus. Nat Cell Biol 1: 415–422 [DOI] [PubMed] [Google Scholar]
- Liku ME, Nguyen VQ, Rosales AW, Irie K, Li JJ 2005. CDK phosphorylation of a novel NLS-NES module distributed between two subunits of the Mcm2–7 complex prevents chromosomal rereplication. Mol Biol Cell 16: 5026–5039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen VQ, Co C, Irie K, Li JJ 2000. Clb/Cdc28 kinases promote nuclear export of the replication initiator proteins Mcm2–7. Curr Biol 10: 195–205 [DOI] [PubMed] [Google Scholar]
- Nguyen VQ, Co C, Li JJ 2001. Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature 411: 1068–1073 [DOI] [PubMed] [Google Scholar]
- Randell JC, Bowers JL, Rodriguez HK, Bell SP 2006. Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2–7 helicase. Mol Cell 21: 29–39 [DOI] [PubMed] [Google Scholar]
- Remus D, Diffley JF 2009. Eukaryotic DNA replication control: Lock and load, then fire. Curr Opin Cell Biol 21: 771–777 [DOI] [PubMed] [Google Scholar]
- Remus D, Blanchette M, Rio DC, Botchan MR 2005. CDK phosphorylation inhibits the DNA-binding and ATP-hydrolysis activities of the Drosophila origin recognition complex. J Biol Chem 280: 39740–39751 [DOI] [PubMed] [Google Scholar]
- Remus D, Beuron F, Tolun G, Griffith JD, Morris EP, Diffley JF 2009. Concerted loading of Mcm2–7 double hexamers around DNA during DNA replication origin licensing. Cell 139: 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclafani RA, Holzen TM 2007. Cell cycle regulation of DNA replication. Annu Rev Genet 41: 237–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny RE, MacAlpine DM, Blitzblau HG, Bell SP 2006. Genome-wide analysis of re-replication reveals inhibitory controls that target multiple stages of replication initiation. Mol Biol Cell 17: 2415–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsakraklides V, Bell SP 2010. Dynamics of pre-replicative complex assembly. J Biol Chem 285: 9437–9443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmes GM, Archambault V, Austin RJ, Jacobson MD, Bell SP, Cross FR 2004. Interaction of the S-phase cyclin Clb5 with an ‘RXL’ docking sequence in the initiator protein Orc6 provides an origin-localized replication control switch. Genes Dev 18: 981–991 [DOI] [PMC free article] [PubMed] [Google Scholar]