Abstract
Alternative splicing (AS) plays a major role in the generation of proteomic diversity and in gene regulation. However, the role of the basal splicing machinery in regulating AS remains poorly understood. Here we show that the core snRNP (small nuclear ribonucleoprotein) protein SmB/B′ self-regulates its expression by promoting the inclusion of a highly conserved alternative exon in its own pre-mRNA that targets the spliced transcript for nonsense-mediated mRNA decay (NMD). Depletion of SmB/B′ in human cells results in reduced levels of snRNPs and a striking reduction in the inclusion levels of hundreds of additional alternative exons, with comparatively few effects on constitutive exon splicing levels. The affected alternative exons are enriched in genes encoding RNA processing and other RNA-binding factors, and a subset of these exons also regulate gene expression by activating NMD. Our results thus demonstrate a role for the core spliceosomal machinery in controlling an exon network that appears to modulate the levels of many RNA processing factors.
Keywords: alternative splicing, Sm proteins, snRNP, autoregulation, NMD, exon network
The production of multiple mRNA variants through alternative splicing (AS) is estimated to take place in transcripts from >95% of human multiexon genes (Pan et al. 2008; Wang et al. 2008). AS represents a mechanism for gene regulation and expansion of the proteome. The most widely studied trans-acting factors regulating AS are proteins of the SR (Ser/Arg-rich) and hnRNP (heterogeneous ribonucleoprotein) families, as well as numerous tissue-restricted AS factors (for review, see Chen and Manley 2009; Nilsen and Graveley 2010). These factors generally regulate AS by recognizing cis-acting sequences in exons or introns and by promoting or suppressing the assembly of the spliceosome at adjacent splice sites. In contrast to these AS regulatory factors, much less is known about how or the extent to which components of the basal or “core” splicing machinery modulate splice site decisions.
The spliceosome is a large RNP complex that carries out the removal of introns from pre-mRNAs. It comprises the U1, U2, U4/6, and U5 small nuclear RNPs (snRNPs) and several hundred protein factors (for review, see Wahl et al. 2009). Studies in yeast and metazoan systems have indicated that the levels of some of these core splicing components can affect splice site choice. Microarray profiling revealed transcript-specific effects on splicing in yeast strains harboring mutations in or deletions of core splicing components (Clark et al. 2002; Pleiss et al. 2007). Knockdown of several core splicing factors in Drosophila cells resulted in transcript-specific effects on AS reporters (Park et al. 2004). Deficiency of the snRNP assembly factor SMN (survival of motor neuron) in a mouse model of spinal muscular atrophy (SMA) resulted in tissue-specific perturbations in snRNP levels and splicing defects (Gabanella et al. 2007; Zhang et al. 2008). Tiling microarray profiling analysis of fission yeast RNA also revealed transcript-specific splicing defects of a temperature degron allele of SMN, and that some of the defects could be alleviated by strengthening the pyrimidine tract upstream of the branchpoint (Campion et al. 2010). However, the features that underlie the differential sensitivity of introns or alternative exons to particular defects in the core splicing machinery are not well understood.
Many splicing regulatory factors can regulate the AS of their own pre-mRNAs (autoregulation), and in some cases have been observed to affect the AS of pre-mRNAs encoding other AS factors (for review, see Lareau et al. 2007a; McGlincy and Smith 2008). Such autoregulation and cross-regulation are important for the establishment and maintenance of AS factor levels across different tissues and developmental stages. Autoregulation and cross-regulation of AS factors can produce protein isoforms with different functional properties (Dredge et al. 2005; Damianov and Black 2010), and have also been shown in many cases to regulate gene expression by producing alternative transcripts containing premature termination codons (PTCs) that are degraded by nonsense-mediated mRNA decay (AS-NMD). Consistent with their functional importance, regulated AS events involved in AS-NMD of splicing factors often lie within highly conserved or ultraconserved sequence regions (Lareau et al. 2007b; Ni et al. 2007; Yeo et al. 2007; Saltzman et al. 2008).
Using AS microarray profiling following knockdown of NMD factors, we previously identified highly conserved, PTC-introducing alternative exons in genes encoding multiple core splicing factors (Saltzman et al. 2008). These genes included SNRPB, which encodes the core snRNP component SmB/B′, a subunit of the heteroheptameric Sm protein complex that is assembled by the SMN complex onto the Sm-site of U1, U2, U4, and U5 snRNAs (for review, see Neuenkirchen et al. 2008). The SmB and SmB′ proteins are nearly identical and arise from alternative 3′ splice site (ss) usage in the terminal exon of SNRPB (Fig. 1A, top; Supplemental Fig. 1), leading to an additional repeat of a short proline-rich motif at the C terminus of SmB′ (van Dam et al. 1989). The PTC-introducing alternative exon in SNRPB lies within a region of the second intron that is highly conserved in mammalian genomes (Fig. 1A). Splice variants including this PTC-introducing exon accumulate when NMD is disrupted, as well as when expression of SmB is increased exogenously (Saltzman et al. 2008; this study). These results suggested that AS-NMD plays a role in the homeostatic regulation not only of AS regulatory factors, but of components of the basal splicing machinery as well. Moreover, the results further suggested that core spliceosomal components may regulate specific AS events in addition to their well-studied critical roles in constitutive splicing.
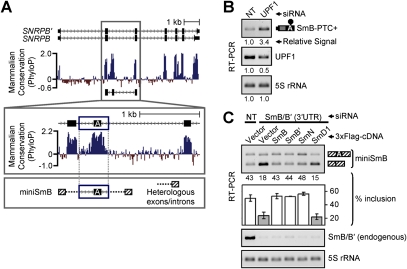
Figure 1.
The inclusion of a highly conserved PTC-introducing alternative exon in SNRPB is affected by SmB/B′ knockdown. (A, top panel) Diagram of the exon/intron structure of the SNRPB gene. The two encoded proteins, SmB and SmB′, arise from alternative 3′ ss usage in the final exon. A conservation plot (phyloP), is shown below (generated using the University of California at Santa Cruz genome browser (http://genome.ucsc.edu; Rhead et al. 2010). (Bottom panel) An expanded view of the region between the second and third SNRPB exons. The highly conserved area within this intron contains an alternative exon (“A”). This alternative exon and its conserved flanking intron regions (boxed in blue) were cloned into a minigene (miniSmB) in which they are flanked by heterologous intron and exon sequences. (B) RT–PCR assays confirm an increase in the level of the PTC-containing SNRPB variant when NMD is abrogated by knockdown of UPF1. See also Supplemental Figures 2 and 3. (C) Knockdown of SmB/B′ leads to more skipping of the SNRPB alternative exon in miniSmB. HeLa cells were transfected with a control nontargeting (NT) siRNA, or an siRNA targeting the 3′ untranslated region (UTR) of SmB/B′. Cells were then cotransfected with miniSmB and either an empty vector or a vector encoding a 3xFlag-tagged cDNA, as indicated above the gels. (Top panel) To assay alternative exon inclusion in miniSmB, RT–PCR assays were performed using primers specific for the flanking constitutive exons of the minigene (hatched boxes). Quantifications of the percent exon inclusion level are shown below the gel, and the average percent inclusion ± standard deviation calculated from at least three independent analyses is shown in the bar graph. The level of SmB/B′ mRNA (middle panel) or 5S rRNA (loading control, bottom panel) was assayed by RT–PCR.
In the present study, we investigated the role of the core spliceosomal machinery in AS, focusing on a detailed investigation of SmB/B′. Our results confirm that SmB/B′ functions in homeostatic autoregulation through the inclusion of a highly conserved PTC-introducing exon in its own pre-mRNA. This mode of regulation depends on a suboptimal 5′ss associated with the SNRPB PTC-introducing exon and appears to be controlled by changes in the level of U1 snRNP as a consequence of SmB/B′ depletion. We also show that knockdown of SmB/B′ leads to a striking reduction in the inclusion levels of many additional alternative exons, which are significantly enriched in functions related to RNA processing and RNA binding. Changes in the inclusion levels of a subset of these alternative exons also appear to control expression levels of the corresponding mRNAs by AS-NMD. Our results thus reveal an important role for the core spliceosomal machinery in establishing the inclusion levels of a specific subset of alternative exons, and further suggest that changes in the levels of these alternative exons control the expression of other RNA processing factors.
Results
Inclusion of a highly conserved PTC-introducing alternative exon in SNRPB pre-mRNA is affected by levels of core splicing factors
To initially explore the role of the core spliceosomal machinery in the regulation of AS, we determined the effect of SmB/B′ knockdown on the AS of the PTC-introducing SNRPB exon. This exon, together with its highly conserved flanking intronic sequences, was cloned into a minigene reporter plasmid containing upstream and downstream heterologous intron and constitutive exon sequences (Fig. 1A, miniSmB). Unlike endogenous SmB/B′ transcripts including the PTC-introducing exon (Fig. 1B), exon-included transcripts derived from the miniSmB reporter are not degraded by NMD, as neither the steady-state level nor the half-life of these transcripts is increased upon disruption of NMD (Supplemental Figs. 2, 3). Monitoring transcripts derived from miniSmB therefore allows an analysis of the splicing regulation of the PTC-introducing SNRPB exon in the absence of effects of NMD. HeLa cells were transfected with a control nontargeting (NT) siRNA or an siRNA to knock down SmB/B′, followed by transfection with the miniSmB reporter plasmid. Knockdown of SmB/B′ led to increased skipping of the SNRPB alternative exon in miniSmB (Fig. 1C). Loss of inclusion of the SNRPB alternative exon was rescued by expression of a Flag-epitope-tagged cDNA construct encoding SmB or SmB′ (Fig. 1C). It was also rescued by expression of SmN, a tissue-restricted paralog of SmB/B′ that is 93% identical to SmB′ (Supplemental Fig. 1). However, loss of inclusion of the SNRPB alternative exon upon SmB/B′ knockdown could not be rescued by expression of the related protein SmD1, another component of the heteroheptameric Sm ring (Fig. 1C). Thus, SmB/B′ knockdown leads to an increase in the exon-skipped miniSmB splice variant (Fig. 1C), which represents the protein-coding isoform. Reciprocally, our previous experiments have shown that increasing SmB/B′ protein levels leads to an increase in the exon-included PTC-containing splice variant (Saltzman et al. 2008). Together, these results support a role for the highly conserved SNRPB PTC-introducing alternative exon in the homeostatic autoregulation of SmB/B′ via AS-NMD. Our results further suggest that components of the core spliceosomal machinery can function in the regulation of AS, in addition to their well-established roles in constitutive splicing.
Knockdown of the core snRNP protein SmD1 affects the inclusion of the conserved SNRPB alternative exon
To investigate whether other proteins in the Sm heptameric complex might also affect the inclusion of the SNRPB alternative exon, SmD1 was knocked down and the effect on the AS of miniSmB was assayed by RT–PCR (Fig. 2A). As observed for SmB/B′ (Fig. 1C), knockdown of SmD1 resulted in more skipping of the miniSmB alternative exon. This effect was rescued by exogenous expression of SmD1, but not of SmB (Fig. 2A). Taken together with the results described above, these observations suggest that SmB and SmD1 affect AS in a similar but nonredundant manner. One possibility is that depletion of each component of the Sm heptameric complex similarly affects the overall levels and/or integrity of one or more snRNPs in a manner that affects the recognition of the SNRPB alternative exon.
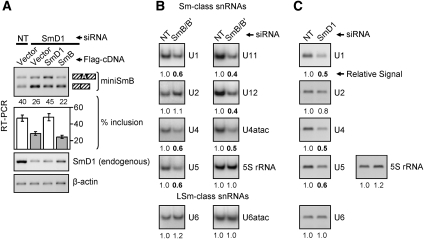
Figure 2.
Knockdown of SmD1 leads to more skipping of the SNRPB alternative exon in miniSmB (A), and knockdown of SmB/B′ (B) or SmD1 (C) affects snRNA levels. (A) HeLa cells were transfected with NT or SmD1-specific siRNAs. Cells were then cotransfected with miniSmB and with either an empty vector or a vector encoding the 3xFlag-tagged cDNA indicated above the gel. RT–PCR assays were performed using primers specific for the flanking constitutive exons of the minigene (top panel) or primers specific for SmD1 or β-actin transcripts (bottom panels). The quantifications and bar graph of percent inclusion levels are as in Figure 1. (B) Knockdown of SmB/B′ leads to a decrease in steady-state levels of three of four Sm-class snRNAs that form the major spliceosome (U1, U4, and U5, but not U2) and of the Sm-class snRNAs that form the minor spliceosome (U11, U12, and U4atac). (C) Knockdown of SmD1 shows similar effects, but also a slight decrease in U2 snRNA levels. The levels of LSm (Sm-like)-class snRNAs U6 and U6atac were not decreased in either the SmD1 or SmB/B′ knockdown.
Knockdown of Sm proteins affects the levels of Sm-class snRNAs
To investigate how the knockdown of SmB/B′ or SmD1 might affect SmB/B′ AS, we next determined whether reduced levels of either Sm protein affects the steady-state levels of spliceosomal snRNAs using RT–PCR assays. The relative levels of snRNAs measured in total cellular RNA are comparable with those in snRNPs immunoprecipitated with anti-Sm antibodies (Zhang et al. 2008). This is in agreement with previous studies indicating that the pool of “Sm-free” snRNAs is relatively small (Sauterer et al. 1988; Zieve et al. 1988). Consistent with an important role for Sm core assembly in snRNP stability (Jones and Guthrie 1990), knockdown of SmB/B′ led to a decrease in Sm-class snRNAs (U1, U4, U5, U11, U12, and U4atac), with the exception of U2 (Fig. 2B). Knockdown of SmD1 also led to a decrease in Sm-class snRNAs, including a slight decrease in U2 (Fig. 2C). The similar effects observed for both knockdowns are consistent with a shared role for Sm proteins in the inclusion of alternative exons through modulating the overall levels of one or more snRNPs.
Cis-acting elements regulating inclusion of the SNRPB alternative exon
To investigate the mechanism by which knockdown of Sm proteins and the associated reductions in snRNP levels affect AS of SmB/B′ pre-mRNA, we performed a detailed mutagenesis analysis of the miniSmB reporter. Recapitulation in this reporter (Fig. 1) of the Sm protein-dependent AS effects seen for endogenous transcripts (Saltzman et al. 2008) suggested that all of the cis-acting elements required for mediating regulation are contained within the highly conserved SNRPB PTC-introducing exon and/or its flanking intronic sequences. Linker-scanning mutagenesis was performed, in which successive 12-base segments of the alternative exon and its upstream and downstream flanking introns were deleted or substituted with a linker sequence. This strategy identified sequences in the exon and introns acting as splicing enhancers or silencers. These elements are concentrated near splice sites (Supplemental Fig. 4A). In most cases, knockdown of SmB/B′ led to a comparable increase in exon skipping of the mutated or deleted minigenes, as observed for the wild-type miniSmB (Supplemental Fig. 4B). These results suggest that these sequence motifs either act through different trans-acting factors, or are involved in regulation in a manner that is redundant with other sequence motifs. However, in several cases, the mutation and/or deletion of a particular sequence reduced or eliminated the impact of SmB/B′ knockdown on inclusion of the alternative exon. In particular, a deletion from the seventh to the 18th nucleotide downstream from the 5′ss led to an increase in the inclusion of the alternative exon. However, in contrast to wild-type miniSmB, knockdown of SmB/B′ had no detectable effect on the exon inclusion level of this mutant reporter (Supplemental Figs. 4, 5). Substitution of the same sequence with the 12-base linker also led to an increase in exon inclusion, but, in contrast to the deletion mutant, did not prevent increased exon skipping caused by SmB/B′ knockdown (Supplemental Figs. 4, 5). Examination of sequences adjacent to the 5′ss created by this pair of mutants revealed that the deletion but not the substitution created a sequence with increased potential for base-pairing to U1 snRNA. These results therefore suggest that regulation of the SNRPB alternative exon by Sm protein levels depends on sequences at or proximal to the exon 5′ss. Therefore, the role of 5′ss sequences in regulating the inclusion of the SNRPB alternative exon was investigated in greater detail.
Mutations that strengthen the 5′ss reduce the effects of Sm protein knockdown
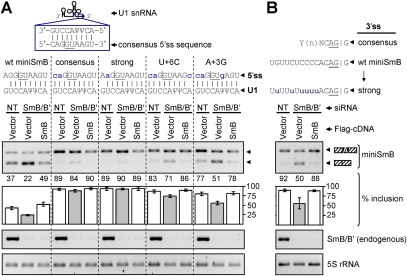
To determine the role of 5′ss sequences in Sm protein knockdown-dependent effects on miniSmB AS, 5′ss mutations were introduced into miniSmB, and the level of exon inclusion was assayed in control and SmB/B′ knockdown conditions (Fig. 3A). Two mutations that strengthen the 5′ss increase its inclusion level, yet, unlike wild-type miniSmB but similar to the deletion mutant described above that strengthens the 5′ss, show very little skipping when SmB/B′ is knocked down (Fig. 3A, consensus and strong). However, SmB/B′ knockdown sensitivity is restored by the introduction of additional mutations in the 5′ss consensus minigene that weaken the 5′ss (U+6C, A+3G) (Fig. 3A). In contrast, a mutation that strengthens the 3′ss (U10ACAG|G) also increases the inclusion level of the minigene, yet does not abrogate the effect of SmB/B′ knockdown (Fig. 3B). Similar results were obtained for two other mutations that increase the strength of the 3′ss (Supplemental Fig. 6). Thus, minigenes with a strong 5′ss or 3′ss have similarly high basal levels of exon inclusion (∼90%), yet the effect of SmB/B′ knockdown on exon skipping is only abrogated for minigenes that have mutations that strengthen the 5′ss. Taken together with the observation that SmB/B′ or SmD1 knockdown results in reduced levels of U1 snRNP (Fig. 2), our data provide evidence that the suboptimal 5′ss of the SNRPB PTC-introducing alternative exon is necessary for its sensitivity to Sm protein depletion.
Figure 3.
Mutations that strengthen the 5′ss, but not mutations that strengthen the 3′ss, reduce the effects of SmB/B′ knockdown on miniSmB AS. HeLa cells were transfected with NT or SmB/B′-specific siRNAs. Cells were then cotransfected with wild-type (wt) miniSmB or with minigenes harboring mutations in the 5′ss (A) or 3′ss (B) as indicated above the gel images, along with either empty vector or a vector encoding the 3xFlag-tagged SmB cDNA construct. (A) Increasing the 5′ss strength (consensus and strong) results in increased percent inclusion relative to wild type, as well as a marked reduction in the effect of SmB/B′ knockdown on percent inclusion. Secondary mutations introduced into the “consensus” construct to decrease the 5′ss strength (U+6C and A+3G) restore sensitivity to SmB/B′ knockdown. (B) Increasing the 3′ss strength results in increased percent inclusion relative to wild type, but no reduction in the effect of SmB/B′ knockdown. RT–PCR assays and quantifications of the percent exon inclusion are as in Figure 1C. (ψ) Pseudouridine.
A widespread role for core splicing factors in promoting the inclusion of alternative exons
To determine whether reducing the levels of SmB/B′, and thus Sm-class snRNPs, via SmB/B′ knockdown affects the inclusion of alternative exons from other genes, high-throughput RNA sequencing (RNA-seq) was performed on RNA from HeLa cells following knockdown of SmB/B′ using an siRNA pool, and on RNA from cells transfected with an NT siRNA pool as a control (Fig. 4A). To compare and assess the specificity and extent of the effects of SmB/B′ knockdown on AS with those of a relatively well-defined splicing regulator, we used another siRNA pool to knock down the SR family protein SRSF1 (also known as ASF, SF2, SFRS1) (Fig. 4A). A comparable knockdown efficiency of >85% was achieved in both factor knockdowns.
Figure 4.
Quantitative analysis of AS by RNA-seq reveals that knockdown of SmB/B′ leads to increased skipping of alternative exons. (A) Western blots indicate that SmB/B′ (top panels) or SRSF1 (bottom panels; also known as SF2, ASF, SFRS1) were efficiently depleted by siRNA transfections. (NT) NT siRNA. Serial dilutions of protein extract indicate that the blots are semiquantitative. (B) The percentage of alternative exons (left) or constitutive exons (right and inset) showing changes in inclusion levels upon knockdown of SmB/B′ or SRSF1 when compared with inclusion levels in cells transfected with a control NT siRNA are shown in a bar graph. (C) Distinct effects of SmB/B′ and SRSF1 knockdowns. The percent exon inclusion values in the control NT, SmB/B′, and SRSF1 knockdowns are shown for 268 alternative exons found to have a ≥30% inclusion change (increased skipping) in the SmB/B′ knockdown when compared with the control (NT). The AS events are ordered from top to bottom by the absolute difference in percent inclusion between the SRSF1 knockdown and the control (NT).
The RNA-seq reads (50 nucleotides [nt]) were mapped to exon–exon junctions in a database of EST/cDNA-supported cassette-type AS events (see the Materials and Methods). Counts of reads mapping to included versus skipped exon junctions were used to calculate the percent inclusion level of these alternative exons. Alternative exons meeting filtering criteria based on junction read coverage (n = 5752) (Supplemental Table 1) were analyzed, and the proportion of cassette alternative exons changing in inclusion level between each knockdown and the control was plotted (Fig. 4B, left).
Knockdown of SmB/B′ specifically reduced the inclusion levels of a large number of alternative exons, and, overall, affected the inclusion levels of more than twice the number of alternative exons than were affected by knockdown of SRSF1. Relative to the control knockdown, 18% (n = 1035) of alternative exons were ≥10% more skipped in the SmB/B′ knockdown, whereas only 0.8% (n = 48) were ≥10% more included. Moreover, all alternative exons showing a change in percent inclusion of ≥30% (n = 268) were more skipped in the SmB/B′ knockdown compared with the control. In contrast, knockdown of SRSF1 resulted in 7.4% (n = 423) of alternative exons changing by ≥10% inclusion, and 61% of these were more skipped while 39% were more included (Fig. 4B, left). Most alternative exons strongly affected by SmB/B′ knockdown were not similarly affected by knockdown of SRSF1, as shown in the heat map of the percent inclusion of alternative exons showing ≥30% change in the SmB/B′ knockdown (Fig. 4C). Thus, the SmB/B′ and SRSF1 knockdowns affected distinct sets of alternative exons, and nearly all changes in alternative exon inclusion following knockdown of SmB/B′ represent increased exon skipping.
To determine the effect of knockdown of SmB/B′ on the inclusion levels of constitutive exons, the RNA-seq reads were aligned to exon–exon junctions in a database of high-confidence internal constitutive exons using the same filtering criteria as applied for alternative exons (refer to the Materials and Methods; Supplemental Table 2). Only 1.9% of the constitutive exons (160 of 8626) showed ≥10% skipping when SmB/B′ was knocked down, compared with 18% (1035 of 5752) of alternative exons (P < 1 × 10−4; χ2) (Fig. 4B). These results indicate that a specific subset of alternative exons is particularly sensitive to reduced snRNP levels as a consequence of SmB/B′ knockdown, whereas constitutive exon inclusion is largely unaffected.
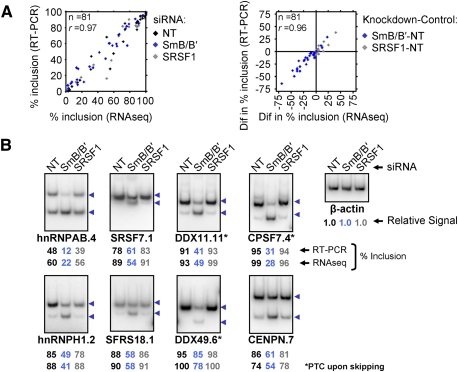
To assess the accuracy of alternative exon inclusion levels and knockdown-dependent changes detected by analysis of the RNA-seq data, AS events analyzed above (n = 5752) were divided into three equally sized groups based on their junction read coverage. Events showing more skipping, no change, or more inclusion when comparing the SmB/B′ knockdown with the control were selected from these three groups, and the alternative exon inclusion levels were measured by RT–PCR using primer pairs targeting the flanking constitutive exons (n = 28 events). The RNA-seq measurements for percent inclusion levels agreed very well with those from the RT–PCR data (r = 0.97) (Fig. 5A). Representative RT–PCR results are shown in Figure 5B, and all results are shown in Supplemental Figure 7. Twenty-one of these AS events were also assayed in two additional independent knockdowns of SmB/B′, and the knockdown-dependent AS changes were confirmed in all cases (Supplemental Fig. 8).
Figure 5.
Changes in alternative exon inclusion levels measured by RNA-seq are confirmed by RT–PCR assays. (A) Scatter plot showing agreement between percent inclusion of 27 alternative exons in the three knockdowns as measured by RT–PCR versus RNA-seq (left), and between differences in inclusion levels (knockdown relative to control NT) for the same 27 alternative exons (right). (B) Representative RT–PCR assays using primers annealing to flanking constitutive exons. For all RT–PCR assays, see Supplemental Figure 7. Gene names: (hnRNPAB) hnRNPA/B; (hnRNPH1) hnRNPH1; (SRSF7; also known as SFRS7, 9G8) serine/arginine-rich splicing factor 7; (SFRS18; also known as SRrp130) splicing factor, arginine/serine-rich 18; (DDX11) DEAD/H (Asp–Glu–Ala–Asp/His)-box polypeptide 11; (DDX49) DEAD-box polypeptide 49; (CPSF7) cleavage and polyadenylation-specific factor 7, 59 kDa; (CENPN) centromere protein N. The number following the period designates the AS event ID. (Names with an asterisk) PTC upon skipping.
Characteristics of SmB/B′ knockdown-dependent alternative exons
The results from analyzing the miniSmB reporter mutants indicated that the presence of a suboptimal 5′ss is an important determinant of the effect of SmB/B′ knockdown on alternative exon inclusion levels. To address whether this and/or other sequence features account more generally for the effects of SmB/B′ knockdown on exon inclusion levels, we next investigated the relationship between splice site strength and sensitivity to SmB/B′ knockdown by comparing the average splice site strength scores (Yeo and Burge 2004) of the affected alternative exons with those of other alternative and constitutive exons analyzed above by RNA-seq.
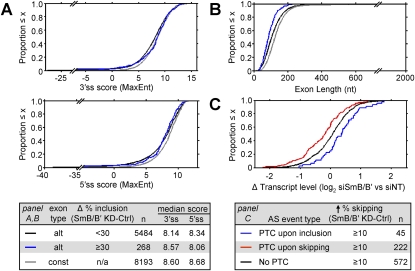
Consistent with previous results (Stamm et al. 2000; Clark and Thanaraj 2002; Itoh et al. 2004), the 3′ss (Fig. 6A, top panel) and 5′ss (Fig. 6A, middle panel) of the profiled alternative exons are, on average, weaker than those of the constitutive exons (3′ss: P = 4.5 × 10−27; 5′ss: P = 5.7 × 10−41, Wilcoxon rank sum test) (Fig. 6A, see legend, bottom panel). However, the average strength of the 3′ss of alternative exons whose inclusion is affected by the SmB/B′ knockdown is higher than that of the other profiled alternative exons (8.57 vs. 8.14; P = 0.02, Wilcoxon rank sum test), and is not significantly different from the average strength of the 3′ss of the constitutive exons (8.57 vs. 8.60) (Fig. 6A). In contrast, the average strength of the 5′ss of alternative exons affected by SmB/B′ knockdown was lower than that of the other profiled alternative exons, although this difference was not statistically significant (8.06 vs. 8.34) (Fig. 6A). In addition, alternative exons affected by SmB/B′ knockdown were, on average, shorter (median = 86 nt) than the other alternative exons (median = 104 nt; P = 3.9 × 10−11, Wilcoxon rank sum test) (Fig. 6B). Thus, alternative exons showing more skipping when SmB/B′ is knocked down are, on average, shorter and have a stronger 3′ss than other profiled alternative exons. These results are consistent with our SNRPB minigene mutagenesis results, in which the effect of SmB/B′ knockdown on exon inclusion was not reduced by mutations increasing the 3′ss strength, but was essentially eliminated by mutations increasing the 5′ss strength (Fig. 3).
Figure 6.
Characteristics of alternative exons affected by knockdown of SmB/B′. (A,B) Cumulative distribution function (CDF) plots of 3′ss scores (A, top), 5′ss scores (A, middle), and exon lengths (B) for alternative and constitutive exons profiled by RNA-seq (Fig. 4). Alternative exons that show a pronounced increase in skipping (≥30%) upon knockdown of SmB/B′ are plotted separately from other profiled alternative exons, as shown in the bottom panel of A. (C) CDF of the fold change in overall mRNA transcript level (log2 scale; SmB/B′ knockdown vs. control) of transcripts containing AS events that are more skipped upon knockdown of SmB/B′ compared with the control knockdown. Transcripts containing AS events that introduce a PTC upon exon inclusion or skipping or that do not introduce a PTC (No PTC) are plotted separately, as shown in the legend below the plot. (alt) Alternative; (const) constitutive.
Changes in transcript levels associated with SmB/B′ knockdown-dependent PTC-introducing alternative exons
To investigate the functional consequences of SmB/B′ knockdown-dependent AS changes, the capacity of these AS events to produce NMD-targeted isoforms that affect overall mRNA expression levels of the corresponding genes was next determined. The mRNA expression levels for genes containing SmB/B′ knockdown-sensitive alternative exons (≥10% more skipping) were measured by aligning RNA-seq reads to RefSeq transcripts (see the Materials and Methods). The fold change in expression level in the SmB/B′ knockdown compared with the control was plotted for AS events that do not introduce a PTC, and for events that introduce a PTC in the exon-included or exon-skipped isoform (Fig. 6C). For exons more skipped in the SmB/B′ knockdown that introduce a PTC upon skipping, the overall mRNA levels from the genes were, on average, reduced in the SmB/B′ knockdown, and the median fold change was significantly different from that of non-PTC-introducing events (P = 2.5 × 10−8, Wilcoxon rank sum test) (Fig. 6C). Examples of three such AS events are shown in Figure 5B (DDX49.6, DDX11.11, and CPSF7.4). Conversely, for exons more skipped in the SmB/B′ knockdown that introduce a PTC upon inclusion, the overall mRNA levels from the corresponding genes were, on average, higher in the SmB/B′ knockdown (P = 4 × 10−3) (Fig. 6C). These results are consistent with AS-NMD acting to both positively and negatively modulate transcript levels of these genes in response to reduced snRNP levels as a consequence of SmB/B′ depletion.
SmB/B′ knockdown affects AS events in RNA processing factor genes
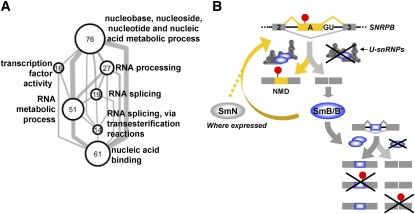
The functional categories represented in genes containing SmB/B′ knockdown-sensitive alternative exons were examined using Gene Ontology (GO) term enrichment (Fig. 7A; Supplemental Table 3). Genes containing alternative exons showing more skipping upon knockdown of SmB/B′ (≥30%) were significantly enriched for terms related to nucleic acid binding and RNA processing (Fig. 7A). These genes include spliceosome components; splicing regulatory factors such as SR, SR-related, and hnRNP family proteins; mRNA 3′-end processing factors; RNA helicases; and other RNA-binding proteins (e.g., Fig. 5B; Supplemental Figs. 7,8). Similar results were also obtained using Pathway Commons annotations (Cerami et al. 2011), which are compiled mostly from protein–protein interaction data (see the Materials and Methods; Supplemental Table 3). These results therefore support the conclusion that an important role for alternative exons affected by changes in the level of the core spliceosomal snRNP machinery is to coordinately control the expression of many RNA processing factors and other regulators of RNA.
Figure 7.
Model for the role of core snRNP proteins in AS regulation. (A) Enriched GO terms (P < 0.005 and FDR < 0.1) annotating genes containing exons affected by SmB/B′ knockdown (≥30% more skipping) are represented as a network of gene sets. Each node represents the set of genes annotated with the indicated GO term. Node size is proportional to the number of genes annotated by the term (indicated by the node label), and edge thickness is proportional to the number of genes in common between the sets. (B) Model summarizing our data. Levels of SmB/B′ are regulated through homeostatic feedback acting via AS-NMD. The related tissue-restricted Sm protein SmN also cross-regulates SmB/B′ (see the Discussion). SmB/B′ levels, through effects on functional snRNP concentrations, also promote inclusion of alternative exons in many other genes, a subset of which also regulate transcript levels via NMD.
Discussion
In this study, we identify a network of alternative exons in RNA processing factor genes that is controlled by the levels of the core spliceosomal machinery. We provide insight into how these exons are regulated, and their roles in both feedback and coordinated control of gene expression (summarized in Fig. 7B).
AS regulation by general splicing factors
Previous studies have shown that mutation, deletion, or knockdown of core spliceosomal and spliceosome assembly factors can result in altered splicing patterns in yeast (Clark et al. 2002; Pleiss et al. 2007; Kawashima et al. 2009; Campion et al. 2010), fly (Park et al. 2004), and mammalian cells (Massiello et al. 2006; Pacheco et al. 2006; Hastings et al. 2007; Zhang et al. 2008; Baumer et al. 2009). In addition, supporting our finding that knockdown of SmB/B′ and associated snRNP components affects AS levels, a recent genome-wide siRNA screen implicated SmB/B′ in the AS of two Bcl2 family apoptosis factors (Moore et al. 2010). Thus, as is well established for splicing regulatory factors, it is emerging that the relative concentration or activity of general splicing factors can affect splice site selection. Consistent with an important physiological role for the core spliceosomal machinery in the regulation of AS, components of snRNPs are differentially expressed in mammalian cells and tissues (Grosso et al. 2008; Castle et al. 2010), and several core splicing factors are differentially expressed during development and in tissues of the fly (Park et al. 2004). Evidence for critical physiological regulatory roles for core spliceosomal components and assembly factors have also emerged from the study of certain human diseases. For example, mutations in components of the U4/U6.U5 tri-snRNP particle are associated with retinitis pigmentosa (for review, see Mordes et al. 2006), mutations in the U4/U6 snRNP recycling factor SART3 (also known as p110) are associated with the skin disorder disseminated superficial actinic porokeratosis (ZH Zhang et al. 2005), and loss or mutation of the widely expressed snRNP assembly factor SMN1 causes SMA (see above; for review, see Burghes and Beattie 2009). Although the specific mechanisms and transcript targets that are responsible for these diseases are largely unknown, these studies point to the importance of maintaining appropriate expression of the core splicing machinery.
Through detailed mutagenesis of sequences surrounding a highly conserved PTC-introducing exon in the SNRPB gene, and the observation that knockdown of SmB/B′ results in reduced levels of U1 but not U2 snRNP, our results demonstrate that the strength of the 5′ss of an alternative exon is an important determinant of its sensitivity to depletion of this core spliceosomal component. Our global analysis of AS events displaying altered inclusion upon knockdown of SmB/B′ and the associated depletion of snRNPs also revealed an association of these alternative exons with relatively weak 5′ss and with 3′ss that were, on average, as strong as those of constitutive exons. Thus, the reduced inclusion levels of a large number of alternative exons as a consequence of Sm protein depletion may be mediated more generally by reduced rates of interaction between the 5′ss and U1 snRNP. These findings may relate to previous observations revealing that the Sm complex contributes to the stability of the U1 snRNA:pre-mRNA interaction in yeast (Zhang et al. 2001), and that proper Sm core assembly is essential for snRNP stability (Jones and Guthrie 1990; Zhang et al. 2008). Our results also relate to in vitro studies demonstrating that differential binding of U1 snRNP to stronger or weaker 5′ss can affect the inclusion levels of a reporter alternative exon (Kuo et al. 1991). Furthermore, our data support evidence that altering the kinetics of spliceosomal rearrangements can affect splice site selection (Query and Konarska 2004; Yu et al. 2008). Such kinetic competition between splice sites may provide a basis for the changes in the alternative exon inclusion levels that we observe in this study, where specific splice sites are no longer efficiently recognized when core splicing components, normally present at saturating levels, may become rate-limiting (Smith et al. 2008; Graveley 2009; Nilsen and Graveley 2010).
A network of coregulated AS events in RNA processing factor genes
Our finding that alternative exons affected by SmB/B′ knockdown are significantly enriched in genes encoding RNA processing/binding factors, and that some of these AS events regulate transcript levels via AS-NMD, suggest an important role for the core spliceosomal machinery in the regulation of AS events that likely function to maintain coordinated expression levels of many RNA processing factors. These results extend previous observations of feedback and cross-regulation among specific splicing factors via AS-NMD. For example, in addition to several reported examples of feedback regulation of splicing components and other RNA-binding proteins (for review, see Lareau et al. 2007a; McGlincy and Smith 2008), cross-regulation through AS-NMD has been found to occur between gene family members and paralogs of auxiliary splicing regulators, including HNRNPL and HNRPLL (Rossbach et al. 2009); PTBP1, PTBP2 (also known as nPTB/brPTB), and ROD1 (also known as PTBP3) (Wollerton et al. 2004; Boutz et al. 2007; Makeyev et al. 2007; Spellman et al. 2007); the fly homologs of the SR protein SRSF3 (also known as SRp20), Rbp1 and Rbp1-like (Kumar and Lopez 2005); the T-cell-restricted intracellular antigen-1 RNA-binding proteins TIA1 and TIAL1 (also known as TIAR) (Le Guiner et al. 2001; Izquierdo and Valcarcel 2007); CUGBP and ETR3-like family members CELF1 (also known as CUGBP1) and CELF2 (also known as CUGBP2) (Dembowski and Grabowski 2009); and the muscleblind-like factors MBNL1 and MBNL2 (Lin et al. 2006; Kalsotra et al. 2008). Interestingly, analogous to the observations in the present study, it was shown recently that the minor spliceosomal snRNP components U11-48K and U11/U12-65K (also known as SNRNP48 and RNPC3, respectively) are regulated post-transcriptionally through a feedback mechanism involving AS and AS-NMD (Verbeeren et al. 2010).
In addition to the examples summarized above, we propose that there is cross-regulation between SmB/B′ and its closely related paralog, SmN, encoded by the imprinted SNRPN locus that arose by duplication of the SNRPB gene in mammals (Rapkins et al. 2006). Unlike SmB/B′, which is widely expressed, SmN is expressed primarily in the brain and heart (Supplemental Fig. 1; McAllister et al. 1988, 1989). The expression of SmN is often disrupted in Prader-Willi syndrome (PWS), a disorder with a range of symptoms, including cognitive impairment (for review, see Cassidy and Driscoll 2009). However, in brain tissue from PWS individuals or mouse models lacking SmN expression, SmB expression is up-regulated through a previously unknown mechanism (Yang et al. 1998; Gray et al. 1999a,b). Our results strongly suggest that this apparent dosage compensation occurs by cross-regulation between SmN and SmB/B′ involving the highly conserved PTC-introducing exon we defined in SNRPB. In particular, we observed that SmN, SmB′, and SmB expressed from cDNAs display very similar activity in the restoration of inclusion levels of the SNRPB PTC exon when endogenous SmB/B′ is knocked down. It follows, therefore, that elevated SmN expression in the brain would lead to reduced levels of SmB/B′ by promoting inclusion of its PTC exon. This repression would be relieved when SmN expression is disrupted in PWS, allowing increased expression of SmB/B′. Such a mechanism could also relate to the concomitant reduction in SmB/B′ expression upon increased expression of SmN in the postnatal relative to the embryonic rodent brain (Grimaldi et al. 1993). Our results show that these highly similar proteins have overlapping functions, and therefore are capable of cross-regulation via AS-NMD in vivo. It is also interesting to consider that cross-regulation of these paralogs via AS-NMD may reduce the phenotypic severity of loss of SmN expression.
In summary, we uncovered a large set of alternative exons that are controlled by levels of the core spliceosomal machinery. Some of these exons affect mRNA levels by introducing PTCs that elicit NMD. A subset of the affected exons likely plays a critical role in maintaining balanced levels of splicing and other RNA-associated factors. These results thus provide new insight into regulated exon networks as well as the functions of core spliceosomal components.
Materials and methods
Cell culture, siRNA, and plasmid transfection
HeLa cells were grown in DMEM (Sigma) supplemented with 10% fetal bovine serum (Sigma) and penicillin/streptomycin (Gibco). For knockdowns, cells were transfected with siRNAs (On-TargetPlus-modified, Dharmacon) at a final concentration of 100 nM using Dharmafect1 (Dharmacon) according to the manufacturer's instructions. Plasmids were transfected using Lipofectamine2000 (Invitrogen) according to the manufacturer's instructions. For knockdown/minigene experiments, cells were transfected with siRNAs and then cotransfected with minigene plasmids and cDNA expression plasmids 2 d later. Cells were then harvested 2 d after the plasmid transfection. For RNA-seq experiments, cells were harvested 3 d post-siRNA transfection. Treatment with cycloheximide and mRNA half-life experiments are described in the legends for Supplemental Figures 2 and 3.
RNA and protein isolation, RT–PCR, and Western blotting
Total RNA was isolated using TRI reagent (Sigma) according to the manufacturer's instructions. RT–PCR assays were performed using 5–10 ng of input total RNA in a 10-μL reaction using the One-Step RT–PCR kit (Qiagen) with or without addition of α-32P-dCTP (Perkin-Elmer). Total protein lysates in radioimmunoprecipitation buffer supplemented with complete mini-EDTA-free protease inhibitor cocktail (Roche) were separated by SDS-PAGE, and Western blotting was performed with the monoclonal antibody Y12 to detect SmB/B′ (Lerner et al. 1981), with mAb96 to detect SRSF1 (Hanamura et al. 1998), or with anti-α-tubulin (Sigma, T6074). Primers for RT–PCR analysis of snRNAs were as described (Zhang et al. 2008). Primer sequences for AS events are available on request.
Plasmid construction
To construct miniSmB, the 124-nt SNRPB PTC-introducing alternative exon along with 124 nt of upstream intron and 122 nt of downstream intron was amplified by PCR from HeLa genomic DNA and cloned into the XhoI/NotI sites of the pET01/Exontrap vector (Mobitec). The amplified SNRPB fragment corresponds to human chr20:2395715-2396221 (Hg18; reverse strand). Mutations of miniSmB were introduced by site-directed mutagenesis or by overlap PCR using Phusion polymerase (NEB). For expression of 3xFlag-tagged proteins, the cDNAs encoding SmB, SmD1, or SmN from the human ORFeome collection (Open Biosystems) were cloned into pMT3989 (a gift from Marcia Roy, Tyers Laboratory) using Gateway LR Clonase II (Invitrogen). The SmB′ construct was generated by site-directed mutagenesis of SmB. All constructs were verified by sequencing.
Analysis of AS and transcript levels by RNA-seq
Total RNA was submitted to Illumina for the FastTrack mRNA-seq service, and 50-nt reads were generated (siNT, 4923MB; siSmB/B′ 2551 MB; siSRSF1: 2814 MB). Cassette AS events (n = 27,240) were mined by aligning EST/cDNA sequences to the genome essentially as described (Pan et al. 2004, 2005). The mRNA-seq reads were mapped to exon–exon junction sequences in this database of cassette AS events as described (Pan et al. 2008). Exon–exon junctions were filtered for coverage in all three samples by matching to one or both of the following two criteria, where exonA is the alternative exon, and exons C1 and C2 are the upstream and downstream flanking exons, respectively: (1) ≥20 reads matching the skipped junction (exonC1:exonC2), or (2) ≥20 reads matching the included junction with higher coverage and ≥15 reads matching the included junction with lower coverage (included junctions: exonC1:exonA and exonA:exonC2). Percent inclusion was calculated using the junction read counts as follows: avg(C1:A,A:C2)/[(C1:C2 + avg(C1:A,A:C2)]. In parallel, sequencing reads were also aligned to RefSeq transcripts, and transcript levels were estimated using the reads per kilobase of exon per million mapped reads (RPKM) calculation (Mortazavi et al. 2008). Data for filtered AS events (n = 5752) are provided in Supplemental Table 1. Sequencing read data were deposited in the NCBI Gene Expression Omnibus (accession number GSE26463).
A database of internal consecutive constitutive exon triplets (n = 33,319) was constructed using the Galaxy tool (http://main.g2.bx.psu.edu; Blankenberg et al. 2007; Blankenberg et al. 2010) as follows: Exons from University of California at Santa Cruz (UCSC) known genes that overlapped with genes in our AS database were selected following removal of exons that overlap sequences in the UCSC knownAlt track as well as removal of exons that overlap our cassette AS database. Reads were aligned to the exon–exon junctions and filtered as described above for the AS events. Data for filtered exon triplets (n = 8626) are provided in Supplemental Table 2.
Calculation of splice site strength
Strengths of splice sites for exons profiled by RNA-seq were calculated using maximum entropy models (Yeo and Burge 2004), available online at the Burge laboratory at Massachusetts Institute of Technology (http://genes.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq.html). The 5′ss scoring uses the last 3 nt of the exon and the first 6 nt of the downstream intron, while the 3′ss scoring uses the last 20 nt of the upstream intron and the first 3 nt of the exon.
GO analysis
Enrichment of GO (Ashburner et al. 2000) or Pathway Commons (Cerami et al. 2011) terms (Fig. 7A; Supplemental Table 3) was calculated for genes containing alternative exons changing by ≥30% inclusion in the SmB/B′ knockdown (n = 235) relative to all genes containing alternative exons passing our filtering criteria (n = 3173) using Web-based Gene Set Analysis Toolkit (WebGestalt, http://bioinfo.vanderbilt.edu/webgestalt) (B Zhang et al. 2005). A minimum of 10 genes per category were specified and enrichment P-values were calculated using the hypergeometric test, and were adjusted for multiple testing using the false discovery rate (FDR) (Benjamini and Hochberg 1995). A network of GO terms with P < 0.005 and FDR < 0.1 was constructed using the Enrichment Map plug-in (http://baderlab.org/Software/EnrichmentMap; Isserlin et al. 2010) for Cytoscape (Cline et al. 2007). Three nodes for GO terms with an identical set of 14 genes were collapsed into the single 14-gene node shown (Fig. 7A), and nodes were arranged using Cytoscape hierarchic layout.
Statistical analysis
To compare the frequency of SmB/B′ knockdown-dependent changes in percent inclusion of alternative versus constitutive exons profiled by RNA-seq, the χ2 test was used. Sample sizes are given in the text. To compare the median splice site strengths, the lengths of profiled exons, and the changes in transcript levels between subsets of AS events, the nonparametric Wilcoxon rank sum test was used. Sample sizes are shown in Figure 6.
Acknowledgments
We thank John Calarco for helpful comments on the manuscript. We thank Drs. Adrian Krainer, Timothy Hughes, and Mike Tyers for reagents. This work was supported by grants from the Canadian Cancer Society (formerly National Cancer Institute of Canada) and Canadian Institutes of Health Research (MOP-67011) to B.J.B., and in part by a grant to B.J.B. and others from Genome Canada funded through the Ontario Genomics Institute. A.L.S. performed the experiments and analyzed the data. Q.P. performed RNA-Seq read alignments and assisted with data analysis. A.L.S. and B.J.B. designed the experiments and wrote the manuscript.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.2004811.
Supplemental material is available for this article.
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene Ontology: Tool for the unification of biology. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumer D, Lee S, Nicholson G, Davies JL, Parkinson NJ, Murray LM, Gillingwater TH, Ansorge O, Davies KE, Talbot K 2009. Alternative splicing events are a late feature of pathology in a mouse model of spinal muscular atrophy. PLoS Genet 5: e1000773 doi: 10.1371/journal.pgen.1000773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y 1995. Controlling the false discovery rate—A practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 57: 289–300 [Google Scholar]
- Blankenberg D, Taylor J, Schenck I, He J, Zhang Y, Ghent M, Veeraraghavan N, Albert I, Miller W, Makova KD, et al. 2007. A framework for collaborative analysis of ENCODE data: Making large-scale analyses biologist-friendly. Genome Res 17: 960–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J 2010. Galaxy: A Web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol 19: 19.10.11–19.10.21 doi: 10.1002/0471142727.mb1910s89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutz PL, Stoilov P, Li Q, Lin CH, Chawla G, Ostrow K, Shiue L, Ares M Jr, Black DL 2007. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev 21: 1636–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burghes AH, Beattie CE 2009. Spinal muscular atrophy: Why do low levels of survival motor neuron protein make motor neurons sick? Nat Rev Neurosci 10: 597–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campion Y, Neel H, Gostan T, Soret J, Bordonne R 2010. Specific splicing defects in S. pombe carrying a degron allele of the survival of motor neuron gene. EMBO J 29: 1817–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy SB, Driscoll DJ 2009. Prader-Willi syndrome. Eur J Hum Genet 17: 3–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle JC, Armour CD, Lower M, Haynor D, Biery M, Bouzek H, Chen R, Jackson S, Johnson JM, Rohl CA, et al. 2010. Digital genome-wide ncRNA expression, including SnoRNAs, across 11 human tissues using polyA-neutral amplification. PLoS ONE 5: e11779 doi: 10.1371/journal.pone.0011779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami EG, Gross BE, Demir E, Rodchenkov I, Babur O, Anwar N, Schultz N, Bader GD, Sander C 2011. Pathway Commons, a Web resource for biological pathway data. Nucleic Acids Res 31: D685–D690 doi: 10.1093/nar/gkq1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Manley JL 2009. Mechanisms of alternative splicing regulation: Insights from molecular and genomics approaches. Nat Rev Mol Cell Biol 10: 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark F, Thanaraj TA 2002. Categorization and characterization of transcript-confirmed constitutively and alternatively spliced introns and exons from human. Hum Mol Genet 11: 451–464 [DOI] [PubMed] [Google Scholar]
- Clark TA, Sugnet CW, Ares M Jr 2002. Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science 296: 907–910 [DOI] [PubMed] [Google Scholar]
- Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, et al. 2007. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc 2: 2366–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damianov A, Black DL 2010. Autoregulation of Fox protein expression to produce dominant negative splicing factors. RNA 16: 405–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembowski JA, Grabowski PJ 2009. The CUGBP2 splicing factor regulates an ensemble of branchpoints from perimeter binding sites with implications for autoregulation. PLoS Genet 5: e1000595 doi: 10.1371/journal.pgen.1000595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dredge BK, Stefani G, Engelhard CC, Darnell RB 2005. Nova autoregulation reveals dual functions in neuronal splicing. EMBO J 24: 1608–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabanella F, Butchbach ME, Saieva L, Carissimi C, Burghes AH, Pellizzoni L 2007. Ribonucleoprotein assembly defects correlate with spinal muscular atrophy severity and preferentially affect a subset of spliceosomal snRNPs. PLoS ONE 2: e921 doi: 10.1371/journal.pone.0000921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR 2009. Alternative splicing: Regulation without regulators. Nat Struct Mol Biol 16: 13–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray TA, Saitoh S, Nicholls RD 1999a. An imprinted, mammalian bicistronic transcript encodes two independent proteins. Proc Natl Acad Sci 96: 5616–5621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray TA, Smithwick MJ, Schaldach MA, Martone DL, Graves JA, McCarrey JR, Nicholls RD 1999b. Concerted regulation and molecular evolution of the duplicated SNRPB′/B and SNRPN loci. Nucleic Acids Res 27: 4577–4584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi K, Horn DA, Hudson LD, Terenghi G, Barton P, Polak JM, Latchman DS 1993. Expression of the SmN splicing protein is developmentally regulated in the rodent brain but not in the rodent heart. Dev Biol 156: 319–323 [DOI] [PubMed] [Google Scholar]
- Grosso AR, Gomes AQ, Barbosa-Morais NL, Caldeira S, Thorne NP, Grech G, von Lindern M, Carmo-Fonseca M 2008. Tissue-specific splicing factor gene expression signatures. Nucleic Acids Res 36: 4823–4832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanamura A, Caceres JF, Mayeda A, Franza BR Jr, Krainer AR 1998. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA 4: 430–444 [PMC free article] [PubMed] [Google Scholar]
- Hastings ML, Allemand E, Duelli DM, Myers MP, Krainer AR 2007. Control of pre-mRNA splicing by the general splicing factors PUF60 and U2AF65. PLoS ONE 2: e538 doi: 10.1371/journal.pone.0000538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isserlin R, Merico D, Alikhani-Koupaei R, Gramolini A, Bader GD, Emili A 2010. Pathway analysis of dilated cardiomyopathy using global proteomic profiling and enrichment maps. Proteomics 10: 1316–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H, Washio T, Tomita M 2004. Computational comparative analyses of alternative splicing regulation using full-length cDNA of various eukaryotes. RNA 10: 1005–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo JM, Valcarcel J 2007. Two isoforms of the T-cell intracellular antigen 1 (TIA-1) splicing factor display distinct splicing regulation activities. Control of TIA-1 isoform ratio by TIA-1-related protein. J Biol Chem 282: 19410–19417 [DOI] [PubMed] [Google Scholar]
- Jones MH, Guthrie C 1990. Unexpected flexibility in an evolutionarily conserved protein–RNA interaction: Genetic analysis of the Sm binding site. EMBO J 9: 2555–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsotra A, Xiao X, Ward AJ, Castle JC, Johnson JM, Burge CB, Cooper TA 2008. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc Natl Acad Sci 105: 20333–20338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima T, Pellegrini M, Chanfreau GF 2009. Nonsense-mediated mRNA decay mutes the splicing defects of spliceosome component mutations. RNA 15: 2236–2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Lopez AJ 2005. Negative feedback regulation among SR splicing factors encoded by Rbp1 and Rbp1-like in Drosophila. EMBO J 24: 2646–2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo HC, Nasim FH, Grabowski PJ 1991. Control of alternative splicing by the differential binding of U1 small nuclear ribonucleoprotein particle. Science 251: 1045–1050 [DOI] [PubMed] [Google Scholar]
- Lareau LF, Brooks AN, Soergel DAW, Meng Q, Brenner SE 2007a. The coupling of alternative splicing and nonsense mediated mRNA decay. In Alternative splicing in the postgenomic era (eds. Blencowe BJ, Graveley BR), pp. 191–212 Landes Biosciences, Austin, TX [Google Scholar]
- Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE 2007b. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature 446: 926–929 [DOI] [PubMed] [Google Scholar]
- Le Guiner C, Lejeune F, Galiana D, Kister L, Breathnach R, Stevenin J, Del Gatto-Konczak F 2001. TIA-1 and TIAR activate splicing of alternative exons with weak 5′ splice sites followed by a U-rich stretch on their own pre-mRNAs. J Biol Chem 276: 40638–40646 [DOI] [PubMed] [Google Scholar]
- Lerner EA, Lerner MR, Janeway CA Jr, Steitz JA 1981. Monoclonal antibodies to nucleic acid-containing cellular constituents: Probes for molecular biology and autoimmune disease. Proc Natl Acad Sci 78: 2737–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Miller JW, Mankodi A, Kanadia RN, Yuan Y, Moxley RT, Swanson MS, Thornton CA 2006. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum Mol Genet 15: 2087–2097 [DOI] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T 2007. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell 27: 435–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massiello A, Roesser JR, Chalfant CE 2006. SAP155 Binds to ceramide-responsive RNA cis-element 1 and regulates the alternative 5′ splice site selection of Bcl-x pre-mRNA. FASEB J 20: 1680–1682 [DOI] [PubMed] [Google Scholar]
- McAllister G, Amara SG, Lerner MR 1988. Tissue-specific expression and cDNA cloning of small nuclear ribonucleoprotein-associated polypeptide N. Proc Natl Acad Sci 85: 5296–5300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister G, Roby-Shemkovitz A, Amara SG, Lerner MR 1989. cDNA sequence of the rat U snRNP-associated protein N: Description of a potential Sm epitope. EMBO J 8: 1177–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlincy NJ, Smith CW 2008. Alternative splicing resulting in nonsense-mediated mRNA decay: What is the meaning of nonsense? Trends Biochem Sci 33: 385–393 [DOI] [PubMed] [Google Scholar]
- Moore MJ, Wang Q, Kennedy CJ, Silver PA 2010. An alternative splicing network links cell-cycle control to apoptosis. Cell 142: 625–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordes D, Luo X, Kar A, Kuo D, Xu L, Fushimi K, Yu G, Sternberg P Jr, Wu JY 2006. Pre-mRNA splicing and retinitis pigmentosa. Mol Vis 12: 1259–1271 [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Neuenkirchen N, Chari A, Fischer U 2008. Deciphering the assembly pathway of Sm-class U snRNPs. FEBS Lett 582: 1997–2003 [DOI] [PubMed] [Google Scholar]
- Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O'Brien G, Shiue L, Clark TA, Blume JE, Ares M Jr 2007. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev 21: 708–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR 2010. Expansion of the eukaryotic proteome by alternative splicing. Nature 463: 457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco TR, Moita LF, Gomes AQ, Hacohen N, Carmo-Fonseca M 2006. RNA interference knockdown of hU2AF35 impairs cell cycle progression and modulates alternative splicing of Cdc25 transcripts. Mol Biol Cell 17: 4187–4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Misquitta C, Zhang W, Saltzman AL, Mohammad N, Babak T, Siu H, Hughes TR, Morris QD, et al. 2004. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol Cell 16: 929–941 [DOI] [PubMed] [Google Scholar]
- Pan Q, Bakowski MA, Morris Q, Zhang W, Frey BJ, Hughes TR, Blencowe BJ 2005. Alternative splicing of conserved exons is frequently species-specific in human and mouse. Trends Genet 21: 73–77 [DOI] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40: 1413–1415 [DOI] [PubMed] [Google Scholar]
- Park JW, Parisky K, Celotto AM, Reenan RA, Graveley BR 2004. Identification of alternative splicing regulators by RNA interference in Drosophila. Proc Natl Acad Sci 101: 15974–15979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C 2007. Transcript specificity in yeast pre-mRNA splicing revealed by mutations in core spliceosomal components. PLoS Biol 5: e90 doi: 10.1371/journal.pbio.0050090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Query CC, Konarska MM 2004. Suppression of multiple substrate mutations by spliceosomal prp8 alleles suggests functional correlations with ribosomal ambiguity mutants. Mol Cell 14: 343–354 [DOI] [PubMed] [Google Scholar]
- Rapkins RW, Hore T, Smithwick M, Ager E, Pask AJ, Renfree MB, Kohn M, Hameister H, Nicholls RD, Deakin JE, et al. 2006. Recent assembly of an imprinted domain from non-imprinted components. PLoS Genet 2: e182 doi: 10.1371/jounral.pgen.0020182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, et al. 2010. The UCSC Genome Browser database: Update 2010. Nucleic Acids Res 38: D613–D619 doi: 10.1093/nar/gkp939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossbach O, Hung LH, Schreiner S, Grishina I, Heiner M, Hui J, Bindereif A 2009. Auto- and cross-regulation of the hnRNP L proteins by alternative splicing. Mol Cell Biol 29: 1442–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltzman AL, Kim YK, Pan Q, Fagnani MM, Maquat LE, Blencowe BJ 2008. Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Mol Cell Biol 28: 4320–4330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauterer RA, Feeney RJ, Zieve GW 1988. Cytoplasmic assembly of snRNP particles from stored proteins and newly transcribed snRNA's in L929 mouse fibroblasts. Exp Cell Res 176: 344–359 [DOI] [PubMed] [Google Scholar]
- Smith DJ, Query CC, Konarska MM 2008. ‘Nought may endure but mutability’: Spliceosome dynamics and the regulation of splicing. Mol Cell 30: 657–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman R, Llorian M, Smith CW 2007. Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol Cell 27: 420–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm S, Zhu J, Nakai K, Stoilov P, Stoss O, Zhang MQ 2000. An alternative-exon database and its statistical analysis. DNA Cell Biol 19: 739–756 [DOI] [PubMed] [Google Scholar]
- van Dam A, Winkel I, Zijlstra-Baalbergen J, Smeenk R, Cuypers HT 1989. Cloned human snRNP proteins B and B′ differ only in their carboxy-terminal part. EMBO J 8: 3853–3860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbeeren J, Niemela EH, Turunen JJ, Will CL, Ravantti JJ, Luhrmann R, Frilander MJ 2010. An ancient mechanism for splicing control: U11 snRNP as an activator of alternative splicing. Mol Cell 37: 821–833 [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R 2009. The spliceosome: Design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollerton MC, Gooding C, Wagner EJ, Garcia-Blanco MA, Smith CW 2004. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol Cell 13: 91–100 [DOI] [PubMed] [Google Scholar]
- Yang T, Adamson TE, Resnick JL, Leff S, Wevrick R, Francke U, Jenkins NA, Copeland NG, Brannan CI 1998. A mouse model for Prader-Willi syndrome imprinting-centre mutations. Nat Genet 19: 25–31 [DOI] [PubMed] [Google Scholar]
- Yeo G, Burge CB 2004. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol 11: 377–394 [DOI] [PubMed] [Google Scholar]
- Yeo GW, Nostrand EL, Liang TY 2007. Discovery and analysis of evolutionarily conserved intronic splicing regulatory elements. PLoS Genet 3: e85 doi: 10.1371/journal.pgen.0030085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Maroney PA, Denker JA, Zhang XH, Dybkov O, Luhrmann R, Jankowsky E, Chasin LA, Nilsen TW 2008. Dynamic regulation of alternative splicing by silencers that modulate 5′ splice site competition. Cell 135: 1224–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Abovich N, Rosbash M 2001. A biochemical function for the Sm complex. Mol Cell 7: 319–329 [DOI] [PubMed] [Google Scholar]
- Zhang B, Kirov S, Snoddy J 2005. WebGestalt: An integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res 33: W741–W748 doi: 10.1093/nar/gki475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZH, Niu ZM, Yuan WT, Zhao JJ, Jiang FX, Zhang J, Chai B, Cui F, Chen W, Lian CH, et al. 2005. A mutation in SART3 gene in a Chinese pedigree with disseminated superficial actinic porokeratosis. Br J Dermatol 152: 658–663 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Lotti F, Dittmar K, Younis I, Wan L, Kasim M, Dreyfuss G 2008. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell 133: 585–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieve GW, Sauterer RA, Feeney RJ 1988. Newly synthesized small nuclear RNAs appear transiently in the cytoplasm. J Mol Biol 199: 259–267 [DOI] [PubMed] [Google Scholar]