Abstract
Small RNA (sRNA)-induced mRNA degradation occurs through binding of an sRNA to a target mRNA with the concomitant action of the RNA degradosome, which induces an endoribonuclease E (RNase E)-dependent cleavage and degradation of the targeted mRNA. Because many sRNAs bind at the ribosome-binding site (RBS), it is possible that the resulting translation block is sufficient to promote the rapid degradation of the targeted mRNA. Contrary to this mechanism, we report here that the pairing of the sRNA RyhB to the target mRNA sodB initiates mRNA degradation even in the absence of translation on the mRNA target. Remarkably, even though it pairs at the RBS, the sRNA RyhB induces mRNA cleavage in vivo at a distal site located >350 nucleotides (nt) downstream from the RBS, ruling out local cleavage near the pairing site. Both the RNA chaperone Hfq and the RNA degradosome are required for efficient cleavage at the distal site. Thus, beyond translation initiation block, sRNA-induced mRNA cleavage requires several unexpected steps, many of which are determined by structural features of the target mRNA.
Keywords: RyhB, small RNA, RNAse E, RNA degradosome, mRNA decay, translation initiation, Hfq
The degradation of mRNAs in bacteria has been exhaustively studied for more than four decades. It is now well established that mRNA stability depends on several factors, such as, but not limited to, translation initiation and elongation, secondary structures in the 5′-untranslated region (UTR) and 3′-UTR, and polyadenylation (for review, see Coburn and Mackie 1999; Kushner 2004; Carpousis et al. 2009; Belasco 2010). In the bacterium Escherichia coli, the main pathway of mRNA degradation depends on the RNA degradosome, which is a protein complex composed of endoribonuclease E (RNase E), a 3′–5′ polynucleotide phosphorylase (PNPase), an RNA helicase (RhlB), and enolase. Although many enzymes can initiate mRNA decay, the most important is RNase E, which cleaves within accessible RNA structures such as the 5′-UTR or the intercistronic region of polycistronic messages (Carpousis 2007). Following initial mRNA cleavage by RNase E, the resulting RNA fragments are degraded by exoribonucleases such as PNPase. When the RNA fragments are highly structured, exoribonucleases depend on the ATP-dependent RNA helicase RhlB to efficiently remove these structures (Py et al. 1996). Interestingly, RNase E is an enzyme that lacks sequence specificity and, while it cleaves generally in AU-rich single-strand regions, it is dependent on the region adjacent to the cleavage site (Mackie and Genereaux 1993; McDowall et al. 1995). Although several RNase E cleavage sites have been identified so far (Kaberdin 2003), it has been impossible to predict RNase E cleavage sites based on RNA sequences or structures.
Whereas each step of mRNA decay is important for normal turnover, the initial cleavage of mRNAs by RNase E is considered to be the limiting factor (Kushner 2002). This is explained by the fact that RNase E activity increases in the presence of monophosphorylated 5′ ends, which are generated by the initial cleavage (Mackie 1998, 2000). Until recently, RNase E was thought to initiate the degradation of the targeted RNA. However, a newly discovered pyrophosphatase, named RppH, was demonstrated in E. coli to dephosphorylate the 5′-end triphosphate of the first nucleotide in the mRNA, similar to the decapping of mRNAs in eukaryotes (Celesnik et al. 2007; Deana et al. 2008). Such types of enzymes are likely to cause dephosphorylation of some mRNAs first, thereby creating a monophosphorylated 5′ end that stimulates RNase E cleavage within the mRNA.
Although RNase E is the central component of the RNA degradosome, the composition of this protein complex can vary considerably. Recently, the RNA degradosome was shown to interact with the protein Hfq, an RNA chaperone involved in mRNA and small RNA (sRNA) interaction (Morita et al. 2005). The same study also demonstrated that two sRNAs—SgrS and RyhB, both involved in rapid degradation of specific target mRNAs—were bound to Hfq in a complex formed by Hfq and the RNA degradosome (Morita et al. 2005). These observations suggested that the RNA degradosome, the RNA chaperone Hfq, and the sRNA together form a ribonucleoprotein specialized in sRNA-induced mRNA degradation.
sRNA-induced mRNA degradation was first demonstrated by the trans-expressed sRNA RyhB acting on target mRNAs such as sodB, acnB, fumA, and sdhCDAB (Massé and Gottesman 2002; Massé et al. 2003a). A follow-up of these experiments indicated that RyhB triggered the degradation of at least 18 transcripts, which made RyhB the sRNA with the most known target mRNAs (Massé et al. 2005). Furthermore, it was demonstrated that the RNA degradosome and the RNA chaperone Hfq play essential roles in this mechanism (Massé and Gottesman 2002; Massé et al. 2003b; Kawamoto et al. 2005; Morita et al. 2005). We also demonstrated that removal of the C-terminal region of RNase E resulted in inhibition of sRNA-induced mRNA degradation (Massé et al. 2003a). It was later shown that the C-terminal region of RNase E was responsible for the binding to Hfq RNA chaperone (Morita et al. 2005). A recent study has shown that RyhB alone was sufficient to stop mRNA translation in the absence of mRNA degradation (Morita et al. 2006). Because these experiments were performed in the absence of mRNA degradation, when the RNA degradosome is inactivated, these results suggested that the target mRNA degradation was only secondary to the sRNA action.
A previous attempt at describing the RyhB-induced cleavage in vitro suggested the presence of two RNase E sites in the upstream part of sodB target mRNA (Afonyushkin et al. 2005). Remarkably, the addition of RyhB did not induce significant cleavage of sodB during in vitro cleavage assays (Afonyushkin et al. 2005). The interpretation of these puzzling results suggested that RyhB did not act as an inducer of cleavage but merely blocked initiation of translation. Recently, another sRNA, SgrS, has been shown to act, to some extent, similarly to RyhB. The sRNA SgrS regulates the levels of the glucose transporter ptsG mRNA according to glucose availability (Morita et al. 2005). Notably, the target mRNA ptsG must be localized at the membrane for the SgrS sRNA to act (Kawamoto et al. 2005). While it is still unclear why SgrS action depends on membrane localization, one likely explanation is the observation that the RNA degradosome is associated with the cytoplasmic membrane (Liou et al. 2002). However, none these studies indicate how or where the initial sRNA-induced cleavage occurred on target mRNAs. Beside RyhB and SgrS, other sRNAs such as RybB (Papenfort et al. 2006), SraD or MicA (Rasmussen et al. 2005, Udekwu et al. 2005), OmrA, and OmrB (Guillier and Gottesman 2008) use RNase E to promote the decay of their respective targets. Similar sRNAs have been characterized in Vibrio sp. (Lenz et al. 2004). Notably, all of these sRNAs regulate their targets by binding at the ribosome-binding site (RBS). The bulk of these observations suggests that, at least for these sRNAs, blocking the initiation of translation is a prerequisite for efficient degradation of mRNA.
Here, we report that RyhB induces mRNA degradation by promoting a distal downstream RNase E-dependent cleavage site within the ORF of the target mRNA sodB. Using in vivo and in vitro techniques, we mapped a prominent cleavage site located 350 nucleotides (nt) downstream from the sodB pairing site with RyhB. Mutagenesis of this cleavage site protects the target mRNA sodB from RyhB-induced cleavage. Using a modified sodB mRNA with prematurely stopped translation, we still observed a RyhB-induced mRNA cleavage within sodB, at the same cleavage site. Thus, blocking translation alone is not sufficient for sRNA-induced degradation of a target mRNA. These results shed light on some long-standing questions regarding the mechanism of sRNA-induced mRNA degradation.
Results
Minimal sequence requirement for sRNA-induced mRNA degradation in vivo
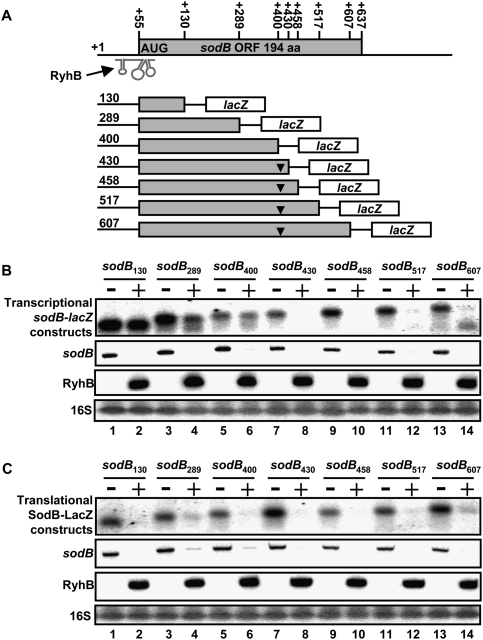
The sRNA RyhB pairs with its target mRNAs at the RBS, thereby blocking the initiation of translation (Morita et al. 2006). Whereas the effect of RyhB pairing on translation is well established, the subsequent steps leading to the initiation of mRNA degradation still remain obscure. Thus, we sought to determine the minimal target mRNA sequence necessary for RyhB-induced mRNA degradation. We designed several transcriptional sodB-lacZ fusions (Fig. 1A), with various lengths that spanned the sodB transcript from nucleotide +130 to +607 (relative to the transcriptional +1). Each fusion was tested for RyhB sensitivity, as shown in Figure 1B. While the fusions of 430 nt and longer were sensitive to RyhB expression, fusions of 400 nt and shorter demonstrated resistance to RyhB expression. As a control, the endogenous sodB mRNA was monitored under the same conditions (Fig. 1B, second panel). These data indicate that the region between +400 and +430 of sodB mRNA encompasses a region that is required for RyhB-dependent mRNA degradation (Fig. 1A).
Figure 1.
Minimal sequence requirement for sRNA-induced mRNA degradation in vivo. (A) Description of the different constructs used to determine the minimal sodB to generate RyhB-induced mRNA degradation. (B) Northern blots with a lacZ probe showing the effect of RyhB expression (for 10 min) on various transcriptional sodB-lacZ constructs and the endogenous sodB transcript. The expression of the endogenous sodB transcript and RyhB is also shown. 16S ribosomal RNA was used as a loading control. (C) Northern blots using a lacZ probe showing the effect of RyhB expression (for 10 min) on various translational SodB-LacZ constructs. The expression of the endogenous sodB transcript and RyhB is also shown. 16S ribosomal RNA was used as a loading control.
We then performed a similar experiment on translational SodB-LacZ fusions. Contrary to transcriptional fusions, in which both sodB and lacZ ORFs have independent translation initiation starts, the corresponding translation fusions contained a single translation initiation site (from sodB RBS) and produced single-hybrid SodB-LacZ peptides. These SodB-LacZ fusions were assayed under the same conditions as described above for sensitivity to RyhB. Results showed that each SodB-LacZ fusion assayed, from +130 to +607, was sensitive to RyhB expression (Fig. 1C). Even though constructs shorter than sodB430 (sodB130, sodB289, and sodB400) did not harbor a putative cleavage site within sodB, they remained sensitive to RyhB-induced cleavage. These results suggest the presence of additional cleavage sites downstream from nucleotide 430 of sodB, most likely in the lacZ section of the fusion (see the Discussion for details).
RyhB-dependent degradation of other target mRNAs
The results shown in Figure 1 suggested that RyhB induced a cleavage at a distal site in the target mRNA sodB. We then addressed the question of whether these observations were strictly specific to sodB or could be applied to other RyhB target mRNAs in general. To verify this, we monitored the effect of RyhB on other target mRNAs such as fumA and iscRSUA (Massé and Gottesman 2002; Massé et al. 2005; Desnoyers et al. 2009). We used constructs similar to sodB, displaying various mRNA lengths fused to lacZ. Figure 2 demonstrates the effect of RyhB on fumA target mRNA (see Supplemental Fig. S1 for the determination of RyhB–fumA pairing). A sequence between +301 and +399 was required to induce cleavage (Fig. 2A). This result indicated that the cleavage site was located at least 230 nt downstream from the RyhB pairing site. In addition, we tested the iscRSUA transcript, which was demonstrated to interact with RyhB at the RBS of iscS (see Supplemental Fig. S2 for schematic description of RyhB–iscS pairing) (Desnoyers et al. 2009). In this study, whereas the iscRS1268 fused to lacZ was sensitive, the shorter iscRS687 was resistant to RyhB. Thus, we concluded that the iscRSUA polycistronic mRNA required the sequence located >19 nt downstream from the RyhB pairing site to promote cleavage (Fig. 2B). The combined results of Figures 1 and 2 suggest that RyhB-induced mRNA cleavage is a general mechanism requiring a sequence further downstream from the RyhB pairing site.
Figure 2.
RyhB-induced mRNA degradation of various target mRNAs. Northern blots with a lacZ probe showing the effect of RyhB expression (for 10 min) on various transcriptional fumA-lacZ constructs (A) and transcriptional iscRS-lacZ constructs (B). RyhB pairs at nucleotides 664–670 of iscRS, which corresponds to the RBS of iscS. Whereas the iscRS687 fusion extends 19 nt into the iscS ORF, the iscRS1268 fusion extend 600 nt into iscS ORF. Expression of endogenous sodB transcript and RyhB is also shown. 16S ribosomal RNA was used as loading control.
Both the RNA degradosome and RNA chaperone Hfq are essential for the RyhB-induced degradation of the minimal target sodB430-lacZ
We demonstrated previously that the RNA degradosome is an essential factor in RyhB-mediated mRNA degradation (Massé et al. 2003a). To test whether this was the case in our minimal sodB430-lacZ construct, we tested the effect of RyhB expression in the RNA degradosome mutant rne131, which lacks residues 586–1061 that constitute the C terminus of RNase E (Kido et al. 1996; Vanzo et al. 1998). As shown in Figure 3A, the C-terminal domain of RNase E is essential for RyhB-mediated degradation of both transcriptional and translational sodB430-lacZ fusions (cf. lanes 2 and 4, and lanes 6 and 8).
Figure 3.
Both the RNA degradosome and RNA chaperone Hfq are essential for the RyhB-induced degradation of the minimal target sodB430-lacZ. (A) Northern blots using a lacZ probe showing the effect of RyhB expression (for 10 min) on transcriptional sodB430-lacZ fusions (left panel) in wild-type cells (lanes 1,2) and rne131 cells (lanes 3,4), and translational SodB430-LacZ fusions (right panel) in wild-type cells (lanes 5,6) and rne131 cells (lanes 7,8). (B) Effect of RyhB expression on β-galactosidase activity of transcriptional sodB430-lacZ fusions in wild-type and rne131 cells, and translational SodB430-LacZ fusions in wild-type and rne131 cells. (C) Northern blots using a lacZ probe showing the effect of RyhB expression (for 10 min) on transcriptional sodB430-lacZ fusions (left panel) in wild-type cells (lanes 1,2) and hfq cells (lanes 3,4), and translational SodB430-LacZ fusions (right panel) in wild-type cells (lanes 5,6) and rne131 cells (lanes 7,8). (D) Effect of RyhB expression on β-galactosidase activity of transcriptional sodB430-lacZ fusions (left panel) in wild-type and hfq cells, and translational SodB430-LacZ fusions (right panel) in wild-type cells and hfq cells.
In addition, we examined whether the inactivation of the RNA degradosome allowed RyhB to block translation by measuring the β-galactosidase activity. Whereas there was only a slight reduction of β-galactosidase activity from the transcriptional fusion (25% reduction), RyhB strongly reduced the translational fusion (75% reduction) (Fig. 3B). Although translation was strongly repressed, this condition was not sufficient to initiate the degradation of the transcriptional sodB430-lacZ construct. These observations are in agreement with previous results (Morita et al. 2006), and indicate that RyhB can efficiently block translation even in the absence of RNA degradation.
Because the RNA chaperone Hfq is involved in RyhB activity and stability, as well as sRNA transactions in general, we tested the effect of the hfq mutation on both transcriptional and translational sodB430-lacZ fusions. As shown in Figure 3C, the hfq mutation prevented RyhB-induced degradation of the minimal transcriptional sodB430-lacZ construct. In contrast to the transcriptional fusion, RyhB slightly repressed the mRNA level of the translational fusion even in the absence of Hfq (Fig. 3C, right panel). In addition, we tested the effect of RyhB on β-galactosidase activities of both transcriptional and translational sodB fusions. As demonstrated in Figure 3D, the effect of RyhB on the β-galactosidase translational SodB-LacZ fusion was similar to the decreased mRNA level (Fig. 3C, lanes 7,8). These results suggest that, in the absence of Hfq, RyhB can still pair with sodB and partly block the initiation of translation.
The putative cleavage site of sodB430 promotes the degradation of a RyhB-resistant sodB130 construct
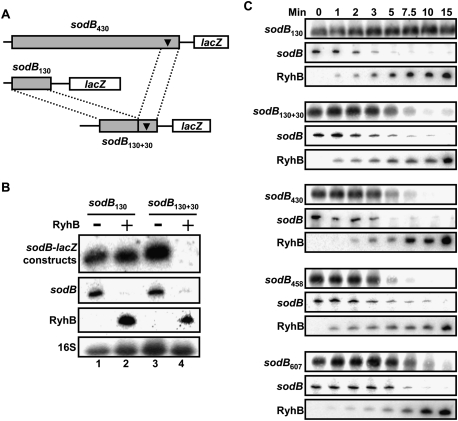
The results in Figures 1B and 3A suggest that RyhB induces an RNA degradosome-dependent mRNA repression that requires the region encompassing nucleotides 400 and 430 of sodB mRNA. We sought to determine if this RNA region was sufficient for a RyhB-resistant sodB130-lacZ construct to become sensitive to RyhB. We created a sodB130 + 30 construct by fusing the 30-nt-long region that was essential for RyhB-mediated mRNA repression of sodB430 into sodB130 (Fig. 4A). We then expressed RyhB in cells carrying the sodB130 or sodB130 + 30 constructs. Data showed that the sodB130 + 30 construct became sensitive to RyhB expression as compared with sodB130 (Fig. 4B, top panel). This observation suggests that, even when the repression site is out of its endogenous context, it still responds efficiently to RyhB.
Figure 4.
The putative cleavage site of sodB430 promotes the degradation of a RyhB-resistant sodB130 construct. (A) Schematic description of sodB430 and sodB130 + 30, with the putative cleavage site represented by the black triangle. (B) Northern blots using a lacZ probe showing the effect of RyhB expression (for 10 min) on sodB130-lacZ and sodB130 + 30-lacZ transcriptional fusions. (C) Northern blots using a lacZ probe showing the effect of RyhB during a time-course expression on sodB130-lacZ, sodB130 + 30-lacZ, sodB430-lacZ, sodB458-lacZ, and sodB607-lacZ.
The sRNA RyhB pairs at the RBS and the first AUG codon of sodB mRNA. Because the closest RyhB-induced cleavage site (between sodB400 and sodB430) is ∼350 nt downstream from the interaction site, we sought to determine if a shorter distance downstream from the RyhB–sodB pairing site would modify the kinetics of cleavage. To address this, we monitored the mRNA levels of various sodB-lacZ constructs. As shown in Figure 4C, we did not observe any significant difference in the time course of degradation whether the putative cleavage site was close to the RyhB pairing site (sodB130 + 30) or located further downstream (sodB430). The fact that larger fusions (sodB458 and sodB607) did not exhibit faster cleavage kinetics suggests that the minimal sodB430 construct was already optimal for sRNA-mediated mRNA degradation.
In vitro mapping and mutagenesis of the minimal sodB430 cleavage site
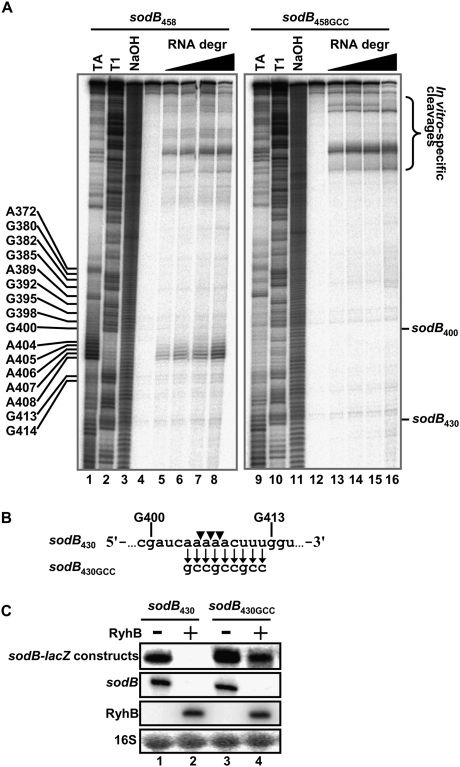
The data from Figures 1B and 4B suggest the presence of a putative cleavage site within the region between nucleotides 400 and 430 of sodB mRNA. We aimed to map the precise site by an in vitro cleavage reaction using purified RNA degradosomes. Notably, these experiments were performed in the absence of RyhB. As demonstrated in Figures 5A (left panel), the addition of purified RNA degradosomes to a 3′-end radiolabeled sodB458 transcript induced a cleavage in the region near position 407 of sodB mRNA (see Supplemental Fig. S3 for in vitro determination of the cleavage site in fumA). This result correlates with in vivo data suggesting the presence of a cleavage site between nucleotides 400 and 430 of sodB. The nucleotide sequence of the cleavage site at sodB407 correlates with previously characterized AU-rich RNase E and RNA degradosome cleavage sites on various RNA substrates (Carpousis 2007; Belasco 2010). The RNA degradosome also catalyzed additional cleavages toward the 5′ end of the sodB transcript (denoted by the bracket in the right margin of Fig. 5A).
Figure 5.
In vitro mapping and mutagenesis of the minimal sodB430 cleavage site. (A) Effect of increasing amounts of purified RNA degradosomes (0.5, 0.76, 1.0, and 1.5 ng/μL final) on 32P-pCp 3′-end radiolabeled sodB458 (left panel, lanes 4–8) and sodB458GCC (right panel, lanes 12–16). (Lanes 1,9) RNase TA ladder. (Lanes 2,10) RNase T1 ladder. (Lanes 3,11) NaOH ladder. (Lanes 4,12) Radiolabeled RNA alone. (B) Local nucleotide sequence (between G400 and G413) of the cleavage site from sodB430 (top line) and the mutated sodB430GCC (bottom). (C) Northern blots using a lacZ probe on sodB430-lacZ and sodB430GCC-lacZ transcriptional fusions in the absence or presence of RyhB expression (for 10 min).
To determine the importance of the cleavage site at position 407 within the context of RyhB-induced mRNA degradation, we introduced a 9-nt-long mutation at this site, keeping the sodB ORF intact (see Fig. 5B). This construct was designated as sodB430GCC. We then determined the effect of the RNA degradosome on the sodB430GCC construct. The data showed that cleavages toward the 5′ end of this transcript are virtually identical to those in the sodB430 transcript (Fig. 5A, right panel). However, no cleavage occurred in the 407 region of the sodB430GCC transcript at any concentration of RNA degradosomes tested. We next monitored the in vivo effect of RyhB on the sodB430GCC construct. Whereas RyhB clearly destroyed the sodB430-lacZ construct within 10 min of expression (Fig. 5C, lane 2), it had little effect on the sodB430GCC-lacZ construct (Fig. 5C, lane 4). This clearly indicates that the cleavage site at sodB407 is essential for RyhB-induced mRNA degradation in vivo.
Translational block alone is not sufficient to induce full degradation of sodB430 mRNA in the absence of RyhB
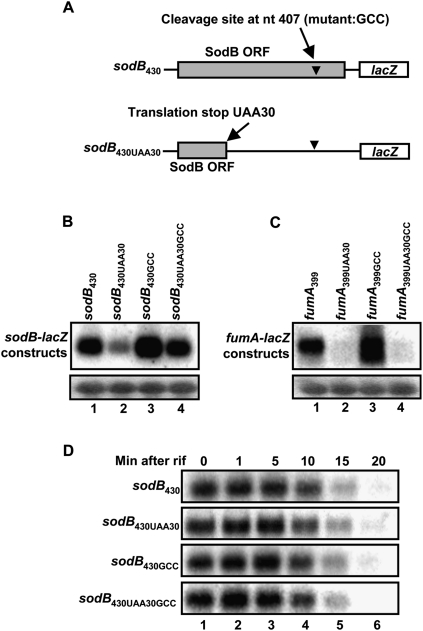
Because our previous results indicated that a RyhB-induced cleavage site was present within the ORF of sodB, we asked whether active translation could protect the cleavage site from ribonuclease attack. To address this issue, we introduced a UAA stop codon at the 30th amino acid in the ORF of sodB430, thus creating the sodB430UAA30 fusion (see Fig 6A). Northern blots assays were used to determine the steady-state mRNA levels, without expression of RyhB. The results suggested that blocking translation reduced the mRNA levels, but this was not sufficient to cause full degradation of sodB430 transcripts (Fig. 6B, cf. lanes 1 and 2). Furthermore, in the case of constructs with prematurely terminated translation, the RNA levels of sodB430UAA30GCC (without a cleavage site) contained slightly more mRNA than sodB430UAA30 (with a cleavage site). These observations suggested that the cleavage site at position 407 was partially responsible for turnover in the absence of translation (see the Discussion for details). This result is in marked contrast to that observed in the case of the fumA399-lacZ construct, which was totally degraded when translation was blocked ahead of the RyhB-induced cleavage site by adding a UAA stop at the 30th codon (Fig. 6C, lanes 1,2). These results suggest that, at least in the case of sodB, translation block by an sRNA such as RyhB is not sufficient to fully account for the observed mRNA degradation.
Figure 6.
Translation block alone in the absence of RyhB is not sufficient to cause full degradation of sodB mRNA. (A) Schematic description of the different sodB constructs used to monitor the effect of translation on mRNA steady-state level and stability. The gray bar represents sodB ORF. The arrowhead is the cleavage site at position 407. UAA30 is the stop codon introduced to prematurely terminate translation. GCC represents the mutation introduced to inactivate the cleavage site at position 407. (B) Northern blots using a lacZ probe showing the steady-state levels of sodB430, sodB430UAA30, sodB430GCC, and sodB430UAA30GCC constructs. (C) Northern blots using a lacZ probe showing the steady-state level of fumA399, fumA399UAA30, fumA399GCC, and fumA399UAA30GCC constructs. (D) Northern blots using a lacZ probe showing the stability of sodB430, sodB430UAA30, sodB430GCC, and sodB430UAA30GCC constructs. Rifampicin (250 μg/mL) was added at time 0 before total RNAs were extracted at the indicated time points. Control 16S are shown in Supplemental Figure S8.
We also assessed the half-life (in the absence of RyhB expression) of various constructs displaying active translation beyond the cleavage site (sodB430 and sodB430GCC) or with translation prematurely stopped (sodB430UAA30 and sodB430UAA30GCC). All constructs displayed roughly the same stability, with a half-life of ∼12 min (Fig. 6D). This is significantly longer than the RyhB-induced degradation kinetics of the sodB430 construct, which was fully degraded after 5–7 min of RyhB expression (Fig. 4C). These data strongly suggest that blocking translation cannot fully explain RyhB-induced mRNA degradation (see the Discussion for details).
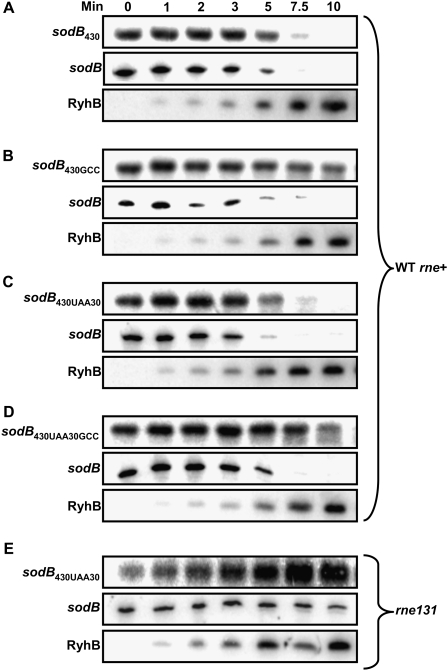
RyhB-induced mRNA cleavage of the target mRNA sodB430-lacZ in the absence of translation
While the previous data (Fig. 6B) indicated that prematurely stopped translation contributed to destabilization of the sodB430 construct, this was is not sufficient to fully degrade the construct. This observation suggests that RyhB could promote degradation of the sodB430 construct even in the absence of translation. To further address this, we expressed RyhB in strains containing the construct sodB430UAA30, which harbors a stop signal at the 30th codon. As shown in Figure 7C, the expression of RyhB promoted rapid degradation of sodB430UAA30, which was still dependent on the cleavage site at residue 407 (cf. sodB430UAA30 [Fig. 7C] and sodB430UAA30GCC [Fig. 7D]). Although the translation was blocked far upstream of the cleavage site, the construct sodB430UAA30 was nevertheless sensitive to RyhB, with degradation kinetics that are similar to those of the wild-type sodB430 construct (half-life ≤7.5 min) (Fig. 7A) and the endogenous sodB mRNA. These rapid degradation kinetics of the sodB430UAA30 construct by RyhB suggest an active mechanism of degradation as compared with passive degradation. Indeed, when we monitored the effect of RyhB on sodB430UAA30 in rne131 cells (Fig. 7E), the transcript was not degraded. These data demonstrate that both the sRNA RyhB and the RNA degradosome promote mRNA cleavage in the absence of translation elongation at the cleavage site.
Figure 7.
RyhB-induced mRNA cleavage in prior absence of translation on the target mRNA sodB430-lacZ. Northern blots using a lacZ probe showing the effect of RyhB expression in wild-type cells on sodB430 (A), sodB430GCC (B), sodB430UAA30 (C), and sodB430UAA30GCC (D). (E) Northern blots using a lacZ probe showing the effect of RyhB expression on sodB430UAA30 in rne131 cells.
Discussion
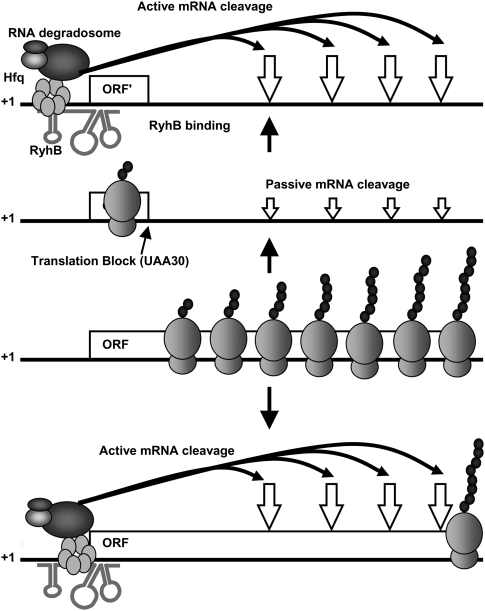
Our work unveils key determinants in the mechanism of sRNA-induced mRNA degradation. First, we demonstrate that pairing of the sRNA promotes the cleavage at a distal site, sometimes hundreds of nucleotides downstream within the target mRNA. Second, prematurely terminated translation of a target mRNA is not sufficient to induce the full degradation of a target mRNA. Third, the sRNA-induced cleavage can initiate even in the absence of translation on the target mRNA. We propose a model (Fig. 8) in which RyhB and Hfq act first on the target mRNA by competing with 30S ribosome subunits and blocking the initiation of translation. The following steps include the recruitment of the RNA degradosome and cleavage at the distal site deep within the ORF. Surprisingly, the cleavage site is located as far as 350 nt downstream from the RyhB pairing site. This suggests a mechanism that allows translating ribosomes to clear off the ORF before the RNA degradosome proceeds to cleave the mRNA (see the model in Fig. 8). In this way, the mRNA cleavage happens after the passage of elongating ribosomes, thereby preventing accumulation of cleaved transcripts harboring stalled ribosomes. Such a mechanism seems widely employed, since we describe three different target mRNAs (sodB, fumA, and iscRSUA) where RyhB induces distal mRNA degradation (Figs. 1, 2). We assume that a significant fraction of sRNAs that binds at the RBS of their target mRNAs will function similarly to RyhB.
Figure 8.
Model for sRNA-induced mRNA degradation (see the text for details).
In fact, a large number of Hfq-binding sRNAs such as MicA (Rasmussen et al. 2005; Udekwu et al. 2005), SgrS (Morita et al. 2005), RybB (Papenfort et al. 2006), OmrA, and OmrB (Guillier and Gottesman 2008) bind to the RBS of their specific target mRNAs to repress translation and decrease the mRNA level. We think that these sRNAs share, at least in part, the same mechanism as the one described here for RyhB. The effects of RyhB-induced target mRNA degradation have been widely investigated (Massé et al. 2003a, 2005; Geissmann and Touati 2004; Afonyushkin et al. 2005; Morita et al. 2005; Desnoyers et al. 2009). However, none of these studies have investigated whether RyhB sRNA acts solely by blocking translation initiation, thereby inducing mRNA degradation (passive degradation model), or whether RyhB promotes the cleavage and degradation of the target mRNA independently of translation (Fig. 8, active cleavage model).
The distant location of downstream cleavage sites may contribute to optimal sRNA-induced mRNA decay. Indeed, when RyhB pairs with a target mRNA and blocks translation initiation, the last translating ribosome can reach and pass the cleavage site before the degradosome initiates mRNA cleavage. Once the mRNA is cleaved, the resulting generation of a 5′-P (monophosphorylated) end accelerates by a 30-fold factor the RNase E activity toward the 3′ end of the transcript (Mackie 1998; Jiang and Belasco 2004; Garrey et al. 2009). Consequently, the cleaved mRNA will be fully degraded more rapidly. When the mRNA is cleaved once, the following rapid RNase E cleavages, concomitantly with exonuclease action, fully destroy the remaining RNA (Carpousis 2007; Belasco 2010). In this context, if RNase E cleavage sites are located upstream of the ORF, the activated RNase E could cleave within the ORF before ribosomes have completed translation, thus trapping ribosomes in a prematurely terminated ORF. If this were the case, the tmRNA (SsrA) would likely help such a stalled ribosome (Keiler et al. 1996). In contrast, a distal cleavage site may prevent RNase E cleavage before ribosomes have cleared the upstream part of the mRNA.
We show here that, even with a prematurely terminated translation of the target mRNA (Fig. 7C, construct sodB430UAA30), the sRNA RyhB promotes an RNA degradosome-dependent cleavage. A similar result was demonstrated previously for the sRNA SgrS and the target mRNA ptsG (Morita et al. 2006). However, in the previous study, Morita et al. (2006) used the antibiotic kasugamycin to block cell-wide translation and then express the sRNA SgrS. This is in contrast with our results, where a stop codon was inserted in the ORF of sodB mRNA, thus specifically blocking sodB translation. The UAA stop codon was used because it is the natural stop codon of the sodB ORF as well as being the most abundant and strongest termination signal in E. coli (Poole et al. 1995; Crawford et al. 1999). By terminating translation at the 30th codon, we prevented any bias in the sequence and structure of sodB mRNA at both the RyhB pairing site (at the RBS) and the cleavage site at position 407. A point of concern would be the possibility that the AUU codon at the 30th amino acid of sodB430UAA30 was leaky. If this were the case, then ribosomes reading through the stop codon would contribute to the stabilization of the mRNA. To verify this possibility, we introduced a stop codon followed by a frameshift, which would fully prevent any ribosome read-through. As shown in Supplemental Figure S4, we observed the same results as presented in Figure 6 with the single AUU stop at the 30th codon. Thus, both of the sodB constructs with blocked translation were most likely free of ribosomes at the position 407. Furthermore, even though translation was terminated upstream, the cleavage site at position 407 of sodB was still critical for efficient RyhB-induced degradation (Fig. 7D).
An argument suggesting the importance of the cleavage site at residue 407 was the significant effect that inactivating the sodB407 cleavage site had on the steady-state level of sodB, whether the transcript was translated (cf. sodB430 and sodB430GCC in Fig. 6B) or not (cf. sodB430UAA30 and sodB430UAA30GCC in Fig. 6B). Nonetheless, in the absence of translation, the sodB430UAA30 transcript was present at a significant level, which was in contrast to the fully degraded fumA399UAA construct (Fig. 6C). One reason explaining why the untranslated sodB430UAA30 construct was not fully degraded is the presence of secondary structures in the RNA in the vicinity of residue 407 (Supplemental Fig. S5). Because RNase E requires ssRNA, the presence of secondary structures may hinder the ribonuclease attack. To test this, we performed in vitro RNA degradosome cleavage using an oligonucleotide located 5 nt downstream from the cleavage site to remove the RNA structure. The presence of the oligonucleotide clearly improved the specific cleavage at position 407 (Supplemental Fig. S5), which suggests that local RNA structures hinder the action of the RNA degradosome. Thus, the presence of inhibitory RNA structures in the vicinity of position 407 in sodB may explain why this site was not fully attacked in the absence of translation as compared with fumA.
Surprisingly, while the steady-state levels of the constructs varied significantly (Fig. 6B), they all displayed the same stability (Fig. 6D). To explain this, we suggest that the construct with prematurely terminated translation (sodB430UAA30) comprised two distinct populations of transcripts: a first population of transcripts with a rapid turnover (<1 min), and a second population that is more stable. The rapid decay of the first population of transcripts likely depends on cleavage at nucleotide 407, since the construct sodB430UAA30GCC displayed a steady-state level similar to that of the wild-type sodB430. This suggests that, when translation is prematurely stopped (sodB430UAA30), the cleavage site at nucleotide 407 is exposed and cleavage occurs rapidly.
While the group of short transcriptional sodB-lacZ fusions (sodB130, sodB289, and sodB400) displayed resistance to RyhB (Fig. 1B), the equivalent SodB-LacZ translational fusions were all RyhB-sensitive. These results suggest that the RyhB-induced cleavage within SodB-LacZ translational fusions occurs in the lacZ gene regardless of the sodB length. Thus, these data indicate that the sRNA pairs to an RNA sequence and promotes cleavage within a foreign gene other than the original target mRNA. In addition, it is very likely that the sRNA-induced translation block alone is sufficient to destabilize the lacZ transcript, as observed previously (Iost and Dreyfus 1995).
Earlier studies with sodB-lacZ fusions suggested that RyhB triggered degradation of sodB within the region upstream of nucleotide 192 (Geissmann and Touati 2004). Our data are in disagreement with this observation. We explain these previous results by the use of a lacZ transcriptional fusion that contained a potential RNase E-dependent cleavage site in the linker region between the sodB and lacZ mRNAs. Here, the use of a shorter linker (pFRΔ construct) that did not contain an RNase E-dependent cleavage provided a tool to focus on the sequence in sodB mRNA.
In another study performed by a different group, Afonyushkin et al. (2005) attempted to reproduce RyhB-induced mRNA degradation in vitro. They used a sodB transcript identical to the transcript used by Geissmann and Touati (2004) (sodB192) to characterize a RyhB-induced cleavage site at nucleotide +67, which is just 6 nt downstream from the RyhB pairing site. These in vitro results do not corroborate our findings. Whereas we also detect cleavage within the 5′ end of the sodB458 substrate (Fig. 5A, in vitro-specific cleavages), we did not consider them significant, as these cleavages did not correlate with the RyhB-induced in vivo cleavage. Our results indicate that cleavages in the 5′ end of sodB are not significant in vivo, at least within the context of RyhB-induced cleavage. To test whether RyhB was pairing at position 407 to induce local cleavage, we tested in vitro pairing between sodB and RyhB. The results clearly showed (Supplemental Fig. S6) that, whereas RyhB bound to the previously known pairing site (Supplemental Fig. S6, left panel), there was no evidence suggesting RyhB pairing at position 407 (Supplemental Fig. S6, right panel).
Another possible reason explaining the RyhB-induced cleavage of sodB192 in vitro (Afonyushkin et al. 2005) is the use of a 5′-monophosphate-labeled target mRNA, which differs from the in vivo 5′-triphosphate substrate. As mentioned above, 5′-monophosphate substrate has been shown to promote significantly RNase E-dependent cleavage (Mackie 1998, 2000). In contrast to this, the in vitro mRNA substrates used here were labeled at their 3′ end (pCp) (see the Materials and Methods), which keeps the 5′-triphosphate intact and prevents degradation by the 3′–5′ PNPase of the RNA degradosome. Thus, we believe that this type of in vitro target mRNA probably reproduces more closely the in vivo substrate that is cleaved through RNase E (Massé et al. 2003a). To confirm the cleavage site at sodB407, we performed one additional in vitro approach. We incubated a T7 RNAp-generated sodB458 transcript with purified RNA degradosome followed by primer extension, which stops at any RNase E-generated cleavage. As shown in Supplemental Figure S7, whereas the absence of RNA degradosome prevented any cleavage, incubation of sodB458 in the presence of the RNA degradosome resulted in a major cleavage at the sodB407 site. Although this approach still did not prevent in vitro-specific cleavage in the upstream region of the mRNA, it confirmed our data on the specific cleavage at sodB407.
Whereas RyhB and all other previously mentioned sRNAs bind on their targets at or close to the RBS, a recent study has provided evidence that pairing at the RBS was not a universal step in the mechanism of sRNA-dependent mRNA silencing. Indeed, the sRNA MicC in Salmonella typhimurium induces an RNase E-dependent mRNA cleavage in the ompD mRNA even though the sRNA pairs within the ORF, far downstream from the RBS (Pfeiffer et al. 2009). Hfq binds in vitro to the target ompD close to MicC pairing within the ORF. This is similar to a previous study where the RNA chaperone Hfq was shown to bind close to the sRNA pairing site to melt the mRNA structure and promote sRNA pairing (Geissmann and Touati 2004). In contrast to previously described sRNA systems, MicC does not alter translation of the target (Pfeiffer et al. 2009). These results may be explained by the low translation rate of the target mRNA ompD, which would allow the sRNA MicC to pair at the ORF without competing with ribosomes.
Our study uncovered many critical determinants of sRNA-induced mRNA degradation. However, additional steps have to be elucidated to reproduce the complete reaction in vitro. Additional factors such as protein components or RNA structures still need to be characterized to provide a comprehensive view of the whole process. This is not surprising, given the complexity of the system. A major question that remains to be addressed is whether the sRNA must block translation in order to induce mRNA degradation. This question is of importance in view of the recent description of the sRNA MicC, which promotes degradation of ompC mRNA without blocking its translation (Pfeiffer et al. 2009). Further work is needed to unravel the relationship between the activity of sRNAs and the translation activity of their target mRNAs. Studies along these lines of research are currently ongoing in our laboratory.
Materials and methods
Bacterial strains and plasmids
Derivatives of E. coli MG1655 were used in all experiments. The DH5α bacterial strain was used for routine cloning procedures. Strains constructed by P1 transduction were selected for the appropriate antibiotic-resistant marker. Except as otherwise indicated, for cells carrying pFRΔ, pRS1551, pNM12, and pBAD-ryhB, ampicillin was used at a final concentration of 50 μg/mL. See Supplemental Tables S1 and S2 for a complete description of strains and oligonucleotides used in this study.
For the construction of sodB-lacZ transcriptional and translational fusions, a PCR fragment made with the oligonucleotides EM423 (forward for all sodB constructions), EM424 (sodB130), EM516 (sodB289), EM515 (sodB400), EM527 (sodB430), EM528 (sodB458), EM514 (sodB517), and EM511 (sodB607) was digested by EcoRI and BamHI and ligated into EcoRI/BamHI-digested pFRΔ (for transcriptional fusions) and EcoRI/BamHI-digested pRS1551 (for in-frame translational fusions). The vector pFRΔ is a derivative of the original transcriptional vector, pRS1553 (Simons et al. 1987), which harbors a transcriptional terminator in the linker between the cloned gene and the lacZ reporter gene (Repoila and Gottesman 2001). All of the transcriptional fusions were constructed from the pFRΔ vector (without transcriptional terminator) and were in-frame with a TAA translation stop (gatccGGCATTTTAA), which was located 26 nt upstream of the RBS of lacZ.
To generate sodB130 + 30, a PCR product of 30 nt was made with oligonucleotides EM625–EM626, then digested with BamHI and ligated into BamHI-digested pFRΔ-sodB130. To produce sodB430 modifications, two independent PCR reactions were performed using the sodB430 or sodB430GCC (to construct sodB430UAA30GCC) fusions as a template with the oligonucleotides sodB430UAA30 (EM1050–EM195 and EM194–EM1051) and sodB430GCC (EM630–EM195 and EM194–EM631). The two PCR products were then mixed to serve as the template for a third PCR (EM194–EM195, oligonucleotides in the pFRΔ). The resulting PCR product was then digested by BamHI and EcoRI and ligated into EcoRI/BamHI-digested pFRΔ (for transcriptional fusions).
For the construction of fumA-lacZ transcriptional fusions, a PCR fragment made with the following oligonucleotides: EM533 (forward for all fumA constructions), EM534 (fumA126), EM560 (fumA301), EM559 (fumA399), EM558 (fumA507), EM556 (fumA703), and EM555 (fumA801). A PCR fragment for the iscRS-lacZ fusion was made with EM359 (forward for iscRS constructions), EM629 (iscRS687), and EM587 (iscRS1268). These PCR products were digested by EcoRI and BamHI and ligated into EcoRI/BamHI-digested pFRΔ. To generate fumA399 modifications, two independent PCR reactions were performed using the fumA399 or fumA399GCC (to construct fumA399UAA30GCC) fusion as template with the following oligonucleotides: fumA399UAA30 (EM1028–EM195 and EM194–EM1029) and fumA399GCC (EM1114–EM195 and EM194–EM1115). The two PCR products were then mixed to serve as the template for a third PCR (EM194–EM195, oligonucleotides in the pFRΔ sequence). The resulting PCR product was then digested by BamHI and EcoRI and ligated into EcoRI/BamHI-digested pFRΔ (for transcriptional fusions).
The transcriptional and translational fusions were delivered in a single copy into the bacterial chromosome of different strains—EM1455 (Δara714 leu+ ΔryhB∷cat) or JF133 (rne131 zce-726∷Tn10 ΔryhB∷cat)—at the λ att site as described previously (Simons et al. 1987). Stable lysogens were screened for single insertion of recombinant λ by PCR (Powell et al. 1994).
E. coli strain KP604 [BL21 (DE3) pLysS/pET21b-rne-Flag] was used for purification of RNase E-Flag degradosomes. The plasmid pET21b-rne-Flag was constructed by PCR amplification of GM402 (harboring pGM102 with the complete rne gene; a gift from George Mackie) with primers EM476 and EM478, and then digested with NdeI–XhoI. The resulting fragment containing the rne gene was then inserted into pET21b digested with NdeI–XhoI.
Purification of RNase E-Flag degradosomes
RNase E-Flag degradosomes were prepared as described previously (Regonesi et al. 2006), with some modifications. E. coli. KP604 (BL21 [DE3] pLysS/pET21b-rne-Flag) culture was grown in 500 mL of Luria-Bertani (LB) broth with ampicillin (50 μg/mL) and chloramphenicol (30 mg/mL) at 30°C until it reached an OD600 value of 0.5–0.6. Expression of RNase E-Flag was induced with 1mM IPTG for 3 h at 30°C.
RNA extraction and Northern blot analysis
Cells were grown at 37°C on LB medium and total RNA was extracted using the hot-phenol procedure (Aiba et al. 1981). Arabinose (0.1%) was added when indicated. Half-life determination of RNA was performed by addition of 250 μg/mL rifampicin. After total RNA extraction, 5 μg of total RNA was loaded on a polyacrylamide gel (5%–10% acrylamide 29:1, 8 M urea) and 20–30 μg was loaded on an agarose gel (1%, 1× MOPS). After migration, the RNA was transferred to a Hybond-XL membrane (Amersham Biosciences) or a Biodyne B membrane (Pall) and cross-linked by UV (1200 J). The membrane was prehybridized with 50% formamide, 5× SSC, 5× Denhardt's reagent, 1% SDS, and 100 μg/mL sheared salmon sperm DNA for 4 h at 60°C. Then, the radiolabeled RNA probe was added directly in the prehybridization buffer with the membrane and incubated 16 h at 60°C. Before exposure to a phosphor screen, the membrane was washed three times with 1× SSC/0.1% SDS and once with 0.1× SSC/0.1% SDS. The phosphor screen was analyzed on a Storm 860 (Molecular Dynamics) or a Typhoon Trio (GE Healthcare), and quantification was performed using the ImageQuant software (Molecular Dynamics).
Internally radiolabeled RNAs generated by in vitro RNA synthesis
The radiolabeled probes used for Northern blot analysis were transcribed with a purified T7 RNA polymerase from a PCR product to generate the antisense transcript of the gene of interest. Transcription was performed in T7 transcription buffer (80 mM HEPES-KOH at pH 7.5, 24 mM MgCl2, 40 mM DTT, 2 mM spermidine), 400 μM NTPs (A, C, and G), 10 μM UTP, 3 μL of α-32P-UTP (3000 Ci/mmol), 20 U of RNase OUT (Invitrogen), 5 μg of T7 RNA polymerase, and 1 μg of DNA template. After 4 h of incubation at 37°C, the mixture was treated with 2 U of Turbo DNase (Ambion) and extracted once with phenol-chloroform. Nonincorporated nucleotides were removed with a G-50 Sephadex column. The primers used for generating DNA templates for in vitro RNA synthesis were EM470–EM471 (lacZ), EM188–EM189 (sodB), EM190–EM191 (ryhB), and EM293–EM294 (16S rRNA).
RNAs generated by in vitro RNA synthesis
Unlabeled RNAs were transcribed in vitro with purified T7 RNA polymerase from a PCR product. Transcription was performed in T7 transcription buffer (80 mM HEPES-KOH at pH 7.5, 24 mM MgCl2, 40 mM DTT, 2 mM spermidine), 500 μM NTPs (A, C, G and U), 40 U of RNase OUT (Invitrogen), 2 U of inorganic pyrophosphatase (Roche), 10 μg of T7 RNA polymerase, and 2 μg of DNA template. After 4 h of incubation at 37°C, the mixture was treated with 2 U of Turbo DNase (Ambion), and the RNA was extracted once with phenol-chloroform and precipitated with isopropanol. The RNA products were purified from a denaturing 6% polyacrylamide, 8 M urea gel and precipitated before use. The primers used for generating DNA templates for in vitro RNA synthesis were EM90–EM528 (sodB458), and the primers were digested with BamHI. To produce sodB458GCC, two independent PCR reactions were performed using the sodB458 DNA as template with the following oligonucleotides: EM630–EM528 and EM90–EM631. The two PCR products were then mixed to serve as the template for a third PCR (EM90–EM528). The resulting PCR product was then digested by BamHI.
3′-end labeling of RNA
For the 3′-end labeling of RNA with pCp, we transcribed RNA with T7 RNA polymerase from a PCR product as described above. Then, 60 pmol of in vitro transcribed RNA was mixed with 10% DMSO, 20 U of RNase OUT (Invitrogen), 1 mM ATP (Fermentas), 20 U of T4 RNA Ligase (Fermentas), 1× T4 RNA Ligase buffer (Fermentas), and 4 μL of cytidine 3′, 5′-bis(phosphate) (5′-32P) at 3000 Ci/mmol in a final volume of 40 μL. The mixture was incubated for 60 min at 37°C. The reaction was stopped by addition of 1 vol of loading buffer II (95% formamide, 18 mM EDTA, 0.025% SDS, xylene cyanol, bromophenol blue) (Ambion). The labeled RNA was then purified from a denaturing 6% polyacrylamide, 8 M urea gel and precipitated before use.
RNA degradosome degradation assay
To determine the RNase E cleavage sites on an RNA transcript, 5 nM 3′end-labeled sodB and 0.0066 μg/μL tRNAs were mixed and heated for 2 min at 90°C. The mixture was slowly cooled until it reached 37°C before adding the degradation buffer (13.3 mM Tris-Cl at pH 7.5, 0.33 mM DTT, 73.34 mM NH4Cl, 3.33 mM magnesium acetate, 0.1 mM EDTA, 0.7% glycerol), followed by incubation for 40 min at 37°C. Then, purified RNase E-Flag degradosomes (0.5, 0.76, 1.0, and 1.5 ng/μL final) or RNA degradosome buffer only (5 mM Tris-Cl at pH 7.5, 50% glycerol, 75 mM NaCl) was added, followed by incubation for 30 min at 37°C. The reaction was stopped with 1 vol of phenol, and samples were separated on an 8% polyacrylamide, 8 M urea gel.
Ribonuclease-generated RNA ladder
Ribonucleases T1 (0.1 U) (Ambion) and TA (0.5 U) (Jena Biosciences) were used for 5 min and 0.5 min, respectively, at 37°C in the sequence buffer (Ambion). The alkaline ladder was performed in the alkaline buffer (Ambion) for 5 min at 90°C.
β-Galactosidase assays
Kinetic assays for β-galactosidase activity were performed as described previously (Prévost et al. 2007) using a SpectraMax 250 microtitre plate reader (Molecular Devices). Briefly, overnight bacterial cultures were incubated at 37°C in LB medium with ampicillin at a final concentration of 50 μg/mL and diluted 1000-fold into 50 mL of fresh LB medium with ampicillin at 37°C. Cultures were grown with agitation to an OD600 of 0.1 before inducing RyhB expression by adding arabinose to a final concentration of 0.1% (strains carrying pBAD-ryhB or the control vector pNM12). Specific β-galactosidase activities (around OD600 of 0.8 and 1.0) were calculated using the formula Vmax/OD600. The reported results represent data from at least three independent experiments.
Acknowledgments
We thank George Mackie, François Bachand, Sherif Abou-Elela, Daniel Lafontaine, Frederieke Brouwers, and Gilles Dupuis for helpful suggestions throughout the course of this work. This work was funded by operating grant MOP69005 to E.M. from the Canadian Institutes for Health Research (CIHR). G.D. is a PhD scholar from the FQRNT (Fonds Québécois de la Recherche sur la Nature et les Technologies). E.M. is a CIHR New Investigator and FRSQ (Fonds de la Recherche en Santé du Québec) Junior II scholar.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.2001711.
Supplemental material is available for this article.
References
- Afonyushkin T, Vecerek B, Moll I, Blasi U, Kaberdin VR 2005. Both RNase E and RNase III control the stability of sodB mRNA upon translational inhibition by the small regulatory RNA RyhB. Nucleic Acids Res 33: 1678–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiba H, Adhya S, de Crombrugghe B 1981. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem 256: 11905–11910 [PubMed] [Google Scholar]
- Belasco JG 2010. All things must pass: Contrasts and commonalities in eukaryotic and bacterial mRNA decay. Nat Rev Mol Cell Biol 11: 467–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpousis AJ 2007. The RNA degradosome of Escherichia coli: An mRNA-degrading machine assembled on RNase E. Annu Rev Microbiol 61: 71–87 [DOI] [PubMed] [Google Scholar]
- Carpousis AJ, Luisi BF, McDowall KJ 2009. Endonucleolytic initiation of mRNA decay in Escherichia coli. Prog Mol Biol Transl Sci 85: 91–135 [DOI] [PubMed] [Google Scholar]
- Celesnik H, Deana A, Belasco JG 2007. Initiation of RNA decay in Escherichia coli by 5′ pyrophosphate removal. Mol Cell 27: 79–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn GA, Mackie GA 1999. Degradation of mRNA in Escherichia coli: An old problem with some new twists. Prog Nucleic Acid Res Mol Biol 62: 55–108 [DOI] [PubMed] [Google Scholar]
- Crawford DJ, Ito K, Nakamura Y, Tate WP 1999. Indirect regulation of translational termination efficiency at highly expressed genes and recoding sites by the factor recycling function of Escherichia coli release factor RF3. EMBO J 18: 727–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deana A, Celesnik H, Belasco JG 2008. The bacterial enzyme RppH triggers messenger RNA degradation by 5′ pyrophosphate removal. Nature 451: 355–358 [DOI] [PubMed] [Google Scholar]
- Desnoyers G, Morissette A, Prévost K, Massé E 2009. Small RNA-induced differential degradation of the polycistronic mRNA iscRSUA. EMBO J 28: 1551–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrey SM, Blech M, Riffell JL, Hankins JS, Stickney LM, Diver M, Hsu YH, Kunanithy V, Mackie GA 2009. Substrate binding and active site residues in RNases E and G: Role of the 5′-sensor. J Biol Chem 284: 31843–31850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissmann TA, Touati D 2004. Hfq, a new chaperoning role: Binding to messenger RNA determines access for small RNA regulator. EMBO J 23: 396–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillier M, Gottesman S 2008. The 5′ end of two redundant sRNAs is involved in the regulation of multiple targets, including their own regulator. Nucleic Acids Res 36: 6781–6794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iost I, Dreyfus M 1995. The stability of Escherichia coli lacZ mRNA depends upon the simultaneity of its synthesis and translation. EMBO J 14: 3252–3261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Belasco JG 2004. Catalytic activation of multimeric RNase E and RNase G by 5′-monophosphorylated RNA. Proc Natl Acad Sci 101: 9211–9216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaberdin VR 2003. Probing the substrate specificity of Escherichia coli RNase E using a novel oligonucleotide-based assay. Nucleic Acids Res 31: 4710–4716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto H, Morita T, Shimizu A, Inada T, Aiba H 2005. Implication of membrane localization of target mRNA in the action of a small RNA: Mechanism of post-transcriptional regulation of glucose transporter in Escherichia coli. Genes & Dev 19: 328–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler KC, Waller PR, Sauer RT 1996. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271: 990–993 [DOI] [PubMed] [Google Scholar]
- Kido M, Yamanaka K, Mitani T, Niki H, Ogura T, Hiraga S 1996. RNase E polypeptides lacking a carboxyl-terminal half suppress a mukB mutation in Escherichia coli. J Bacteriol 178: 3917–3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushner SR 2002. mRNA decay in Escherichia coli comes of age. J Bacteriol 184: 4658–4665, discussion 4657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushner SR 2004. mRNA decay in prokaryotes and eukaryotes: Different approaches to a similar problem. IUBMB Life 56: 585–594 [DOI] [PubMed] [Google Scholar]
- Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL 2004. The small RNA chaperone hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118: 69–82 [DOI] [PubMed] [Google Scholar]
- Liou GG, Chang HY, Lin CS, Lin-Chao S 2002. DEAD box RhlB RNA helicase physically associates with exoribonuclease PNPase to degrade double-stranded RNA independent of the degradosome-assembling region of RNase E. J Biol Chem 277: 41157–41162 [DOI] [PubMed] [Google Scholar]
- Mackie GA 1998. Ribonuclease E is a 5′-end-dependent endonuclease. Nature 395: 720–723 [DOI] [PubMed] [Google Scholar]
- Mackie GA 2000. Stabilization of circular rpsT mRNA demonstrates the 5′-end dependence of RNase E action in vivo. J Biol Chem 275: 25069–25072 [DOI] [PubMed] [Google Scholar]
- Mackie GA, Genereaux JL 1993. The role of RNA structure in determining RNase E-dependent cleavage sites in the mRNA for ribosomal protein S20 in vitro. J Mol Biol 234: 998–1012 [DOI] [PubMed] [Google Scholar]
- Massé E, Gottesman S 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci 99: 4620–4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E, Escorcia FE, Gottesman S 2003a. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev 17: 2374–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E, Majdalani N, Gottesman S 2003b. Regulatory roles for small RNAs in bacteria. Curr Opin Microbiol 6: 120–124 [DOI] [PubMed] [Google Scholar]
- Massé E, Vanderpool CK, Gottesman S 2005. Effect of RyhB small RNA on global iron use in Escherichia coli. J Bacteriol 187: 6962–6971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowall KJ, Kaberdin VR, Wu SW, Cohen SN, Lin-Chao S 1995. Site-specific RNase E cleavage of oligonucleotides and inhibition by stem–loops. Nature 374: 287–290 [DOI] [PubMed] [Google Scholar]
- Morita T, Maki K, Aiba H 2005. RNase E-based ribonucleoprotein complexes: Mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev 19: 2176–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T, Mochizuki Y, Aiba H 2006. Translational repression is sufficient for gene silencing by bacterial small noncoding RNAs in the absence of mRNA destruction. Proc Natl Acad Sci 103: 4858–4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K, Pfeiffer V, Mika F, Lucchini S, Hinton JC, Vogel J 2006. σE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol Microbiol 62: 1674–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer V, Papenfort K, Lucchini S, Hinton JC, Vogel J 2009. Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat Struct Mol Biol 16: 840–846 [DOI] [PubMed] [Google Scholar]
- Poole ES, Brown CM, Tate WP 1995. The identity of the base following the stop codon determines the efficiency of in vivo translational termination in Escherichia coli. EMBO J 14: 151–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell BS, Rivas MP, Court DL, Nakamura Y, Turnbough CL Jr 1994. Rapid confirmation of single copy λ prophage integration by PCR. Nucleic Acids Res 22: 5765–5766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prévost K, Salvail H, Desnoyers G, Jacques JF, Phaneuf E, Massé E 2007. The small RNA RyhB activates the translation of shiA mRNA encoding a permease of shikimate, a compound involved in siderophore synthesis. Mol Microbiol 64: 1260–1273 [DOI] [PubMed] [Google Scholar]
- Py B, Higgins CF, Krisch HM, Carpousis AJ 1996. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature 381: 169–172 [DOI] [PubMed] [Google Scholar]
- Rasmussen AA, Eriksen M, Gilany K, Udesen C, Franch T, Petersen C, Valentin-Hansen P 2005. Regulation of ompA mRNA stability: The role of a small regulatory RNA in growth phase-dependent control. Mol Microbiol 58: 1421–1429 [DOI] [PubMed] [Google Scholar]
- Regonesi ME, Del Favero M, Basilico F, Briani F, Benazzi L, Tortora P, Mauri P, Deho G 2006. Analysis of the Escherichia coli RNA degradosome composition by a proteomic approach. Biochimie 88: 151–161 [DOI] [PubMed] [Google Scholar]
- Repoila F, Gottesman S 2001. Signal transduction cascade for regulation of RpoS: Temperature regulation of DsrA. J Bacteriol 183: 4012–4023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons RW, Houman F, Kleckner N 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53: 85–96 [DOI] [PubMed] [Google Scholar]
- Udekwu KI, Darfeuille F, Vogel J, Reimegard J, Holmqvist E, Wagner EG 2005. Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev 19: 2355–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanzo NF, Li YS, Py B, Blum E, Higgins CF, Raynal LC, Krisch HM, Carpousis AJ 1998. Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes Dev 12: 2770–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]