Abstract
Bacteria cycle between periods when they perform individual behaviors and periods when they perform group behaviors. These transitions are controlled by a cell–cell communication process called quorum sensing, in which extracellular signal molecules, called autoinducers (AIs), are released, accumulate, and are synchronously detected by a group of bacteria. AI detection results in community-wide changes in gene expression, enabling bacteria to collectively execute behaviors such as bioluminescence, biofilm formation, and virulence factor production. In this study, we show that the transcription factor AphA is a master regulator of quorum sensing that operates at low cell density (LCD) in Vibrio harveyi and Vibrio cholerae. In contrast, LuxR (V. harveyi)/HapR (V. cholerae) is the master regulator that operates at high cell density (HCD). At LCD, redundant small noncoding RNAs (sRNAs) activate production of AphA, and AphA and the sRNAs repress production of LuxR/HapR. Conversely, at HCD, LuxR/HapR represses aphA. This network architecture ensures maximal AphA production at LCD and maximal LuxR/HapR production at HCD. Microarray analyses reveal that 300 genes are regulated by AphA at LCD in V. harveyi, a subset of which is also controlled by LuxR. We propose that reciprocal gradients of AphA and LuxR/HapR establish the quorum-sensing LCD and HCD gene expression patterns, respectively.
Keywords: quorum sensing, AphA, LuxR, Qrr sRNA, virulence
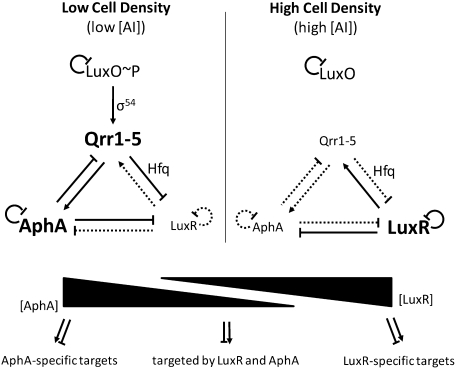
Quorum sensing is a mechanism of bacterial cell–cell communication. This process relies on the production, release, and detection of extracellular signaling molecules called autoinducers (AIs) that drive gene expression programs underlying collective behaviors such as virulence factor production, biofilm formation, and bioluminescence (Davies et al. 1998; Hammer and Bassler 2003; Novick 2003; Ng and Bassler 2009). The bioluminescent marine bacterium Vibrio harveyi produces three AIs that are detected by cognate two-component membrane-bound receptors (Ng and Bassler 2009). At low cell density (LCD), the extracellular concentration of AIs is low and the unliganded receptors function as kinases, shuttling phosphate to the response regulator called LuxO (Freeman and Bassler 1999). LuxO∼P activates transcription of five genes encoding homologous small noncoding RNAs (sRNAs) (Lenz et al. 2004; Tu and Bassler 2007). These sRNAs are called the Qrr sRNAs (Fig. 1). At LCD, with the assistance of Hfq, the Qrr sRNAs base-pair with the mRNA encoding the master quorum-sensing regulator LuxR and prevent its translation (Fig. 1; Tu and Bassler 2007). In the absence of LuxR, genes required for individual behaviors are expressed, while genes required for group behaviors are not. At high cell density (HCD), the extracellular concentration of AIs is high and the AIs are bound by their cognate receptors. AI binding switches the receptors from kinases to phosphatases (Neiditch et al. 2006; Swem et al. 2008), and this reverses the flow of phosphate through the circuit, leading to dephosphorylation of LuxO and cessation of qrr transcription (Fig. 1). In the absence of the Qrr sRNAs, luxR mRNA is translated, LuxR protein is produced, and LuxR controls the cascade of genes underlying collective behaviors (Swartzman et al. 1992; Henke and Bassler 2004; Waters and Bassler 2006; Ng and Bassler 2009).
Figure 1.
Model for reciprocal quorum-sensing control of AphA and LuxR. (Left) At LCD, low concentrations of AIs lead to phosphorylation of LuxO (LuxO∼P), which activates expression of the qrr1–5 genes encoding five redundant sRNAs (Qrr1–5). The Qrr sRNAs promote translation of the mRNA encoding AphA and inhibit translation of the mRNA encoding LuxR. AphA represses luxR, and AphA feeds back to repress qrr expression. (Right) At HCD, high concentrations of AIs reverse the phosphate flow in the circuit, leading to dephosphorylation of LuxO and cessation of Qrr sRNA production. AphA translation ceases and LuxR translation occurs. LuxR represses aphA, and LuxR feeds back to activate qrr expression. This network architecture results in reciprocal gradients of production of AphA and LuxR: Maximal AphA and minimal LuxR production occur at LCD, while minimal AphA and maximal LuxR production occur at HCD. AphA and LuxR are global transcriptional regulators, and they, individually and together, affect the expression of hundreds of target genes.
The Qrr sRNA pool dictates whether the LCD or HCD gene expression program is executed, due to the principal role the Qrr sRNAs play in regulating production of the master quorum-sensing regulator LuxR. Underscoring their importance, several regulatory feedback loops exist to precisely maintain the pool of Qrr sRNAs at appropriate levels. First, LuxR activates qrr expression, which increases Qrr sRNA production (Svenningsen et al. 2008; Tu et al. 2008). The Qrr sRNAs feed back to repress luxR mRNA translation, causing reduced LuxR production. This, in turn, reduces Qrr sRNA production. Second, the Qrr sRNAs inhibit translation of luxO, which decreases Qrr levels because qrr expression requires LuxO∼P (Svenningsen et al. 2009; Tu et al. 2010). Finally, the Qrr sRNAs also repress expression of genes encoding the AI synthases and cognate receptors (Mehta et al. 2009; S Teng, JN Schaffer, KC Tu, P Mehta, W Lu, NP Ong, BL Bassler, and NS Wingreen, in prep.). This loop fine-tunes AI:receptor ratios in response to changing AI concentrations, again altering LuxO∼P levels and changing the Qrr sRNA pool. Together, these feedback loops adjust the levels of Qrr sRNAs such that they precisely track with changes in AI levels.
We hypothesized previously that, in addition to LuxO∼P and LuxR, other regulators could control V. harveyi qrr gene expression (Tu and Bassler 2007; Tu et al. 2008). This idea came from our findings that the five qrr genes exhibit different expression profiles, and distinct DNA sequence motifs indicative of transcription factor-binding sites exist upstream of each qrr gene (Tu and Bassler 2007). Here, to identify putative regulatory factors, we performed a Tn5 mutagenesis screen for insertions altering qrr4 expression. This analysis identified AphA as a repressor of qrr4 (Fig. 1). AphA is a winged-helix transcription factor that controls virulence factor production in the closely related pathogen Vibrio cholerae (Skorupski and Taylor 1999; De Silva et al. 2005). We show that, in addition to repressing qrr4, AphA represses luxR. This finding, coupled with previous reports demonstrating that LuxR represses aphA (Lin et al. 2007; Pompeani et al. 2008), establishes that AphA and LuxR mutually repress each other. We also demonstrate that the Qrr sRNAs activate aphA. The regulatory architecture linking the Qrr sRNAs, LuxR, and AphA causes AphA to be maximally produced at LCD and LuxR to be maximally produced at HCD, with the Qrr sRNAs acting as the switch governing which of these transcription factors is expressed (Fig. 1). Microarray analyses show that AphA controls nearly 300 genes at LCD in V. harveyi, some of which overlap with known targets of LuxR. Targets of AphA include genes involved in type III secretion, flagella synthesis, and pilus production, and members of the quorum-sensing circuit (qrrs, luxR, and aphA). Finally, we show that AphA plays an analogous role in the quorum-sensing cascade of V. cholerae, further solidifying the link between quorum sensing and virulence (Kovacikova and Skorupski 2002; Kovacikova et al. 2003).
Results
AphA regulates qrr expression
Previous work suggested that factors in addition to LuxO∼P and LuxR could regulate expression of the qrr genes in V. harveyi (Tu and Bassler 2007; Tu et al. 2008). To identify such factors, we performed a random Tn5 mutagenesis screen in V. harveyi and assessed the mutants for altered fluorescence from a qrr4-gfp transcriptional fusion. Because the qrr genes are expressed at LCD (Lenz et al. 2004; Tu and Bassler 2007), we used a V. harveyi mutant locked in the LCD state as the parent strain for the mutagenesis (TL45). We reasoned that insertions inactivating repressors of qrr4 expression would result in mutants exhibiting increased GFP production, while mutants harboring insertions in activators of qrr4 expression would exhibit decreased GFP production. Importantly, the strain used for this experiment also lacked luxR, which enabled us to avoid identifying components that function indirectly through LuxR, a known activator of qrr4 (Tu et al. 2008).
We isolated 50 Tn5 insertion mutants displaying altered qrr4-gfp expression (∼200,000 mutants were screened). Fourteen insertions caused increased Qrr4-GFP production, and 36 insertions caused decreased fluorescence. Here, we focus on one transposon insertion mutant exhibiting increased Qrr4-GFP production (Fig. 2A). This Tn5 insertion mapped to V. harveyi gene VIBHAR_00046. This gene, called aphA, is conserved among vibrios and encodes a winged-helix transcription factor (Skorupski and Taylor 1999; De Silva et al. 2005). Introduction of a plasmid carrying the aphA gene together with its 380-base-pair (bp) upstream region decreased qrr4-gfp expression to the level observed in the parent strain (Fig. 2A). This result confirms that transposon disruption of aphA caused the observed increase in qrr4-gfp expression, suggesting that AphA is a repressor of qrr4. We examined whether AphA also affects expression of the other V. harveyi qrr genes by measuring qrr expression in strains containing and lacking aphA. Similar to the logic we used in the initial Tn5 screen, this experiment was performed in a locked LCD V. harveyi strain lacking luxR. Deletion of aphA increased expression of qrr2, qrr3, and qrr4 (Fig. 2B). Thus, AphA represses qrr2, qrr3, and qrr4 at LCD and repression is LuxR-independent. Expression of qrr1 remained unchanged in the presence and absence of aphA, indicating that AphA does not regulate qrr1. Although qrr5 expression is normally low (Tu and Bassler 2007), expression decreased modestly in the ΔaphA strain, indicating that AphA could activate qrr5 expression. However, this decrease did not occur in the ΔaphA strain containing LuxR (Supplemental Fig. S1). We are currently investigating the role of AphA (and potentially other factors) in regulation of qrr1 and qrr5.
Figure 2.
AphA regulates qrr gene expression. (A) Fluorescence from a chromosomal qrr4-gfp transcriptional fusion was measured in a locked LCD V. harveyi strain (TL45: ΔluxM, ΔluxS, ΔcqsS, ΔluxR, and Δqrr4∷gfp) and in the same strain carrying a Tn5 insertion in aphA (STRVh0095: TL45, aphA∷Tn5). The insertion mutant was complemented with a plasmid carrying aphA, denoted paphA (pSTR0502). Means and SEMs of measurements from quadruplicate overnight cultures are shown. (B) Qrr1, Qrr2, Qrr3, Qrr4, and Qrr5 sRNA levels were measured by quantitative RT–PCR (qRT–PCR) in locked LCD V. harveyi strains lacking luxR and containing aphA (KM810: luxO D47E, ΔluxR) (white bars) or lacking aphA (JV55: KM810, ΔaphA) (black bars). Means and SEMs for RNA collected from three independent cultures are shown. (C) Fluorescence from a qrr4-gfp transcriptional fusion on a plasmid (pKT1136) was measured in E. coli MC4100 carrying luxO D47E on the chromosome at the λatt site (STR0018). This strain contained an empty vector (pASK75) or a vector carrying aphA, denoted paphA (pSTR0538). Means and SEMs for triplicate overnight cultures are shown.
The above results show that, at LCD, AphA represses qrr2, qrr3, and qrr4. We used two strategies to determine whether AphA acts directly or indirectly on the qrr4 promoter. First, qrr4 expression was measured in the presence and absence of AphA in Escherichia coli carrying the V. harveyi luxO D47E gene on the chromosome. LuxO D47E is a LuxO∼P mimic that constitutively activates transcription of the qrr promoters (Freeman and Bassler 1999). We reasoned that, if AphA repressed qrr4-gfp expression in this E. coli strain, it would indicate that AphA acts directly on the qrr4 promoter, because no quorum-sensing components other than LuxO D47E are present in E. coli. Indeed, when AphA was introduced into E. coli, threefold repression of qrr4-gfp occurred, consistent with the results in V. harveyi (Fig. 2C). Second, we purified the AphA protein and examined binding to qrr4 promoter DNA. Five ∼50-bp fluorescently labeled probes covering the qrr4 promoter from −180 bp to +50 bp relative to the transcription start site were tested (Fig. 3A). Specific binding occurred only to a fragment carrying DNA overlapping the LuxO∼P-binding site (−126 to −76 relative to the transcription start site) (Fig. 3B). AphA bound this fragment with a Kd of ≈19 nM (Fig. 3C). Together, the above data show that AphA binds directly to the qrr4 promoter to repress transcription. We expect that AphA uses the same mechanism to repress qrr2 and qrr3.
Figure 3.
AphA binds to the qrr4, luxR, and aphA promoters. (A) Five fluorescently labeled DNA fragments were used to assess AphA binding. The schematic illustrates the qrr4 promoter, but the positions are similar for luxR and aphA. (B) Representative fluorescence anisotropy binding curves for the qrr4 promoter probe set from A are shown. Each point displays the mean and SEM of three separate binding reactions at each concentration of AphA. GraphPad software was used to fit one-site-specific nonlinear binding curves. (C) Summary of binding assays for AphA binding to probes in the qrr4, luxR, and aphA promoters. The promoter, the probe number, the position of the probe in base pairs relative to the predicted transcription start sites, and the calculated Kd in nanomolar for probes exhibiting binding are shown (see Supplemental Fig. S3 for individual binding curves). The Kds and SDs are interpolated from curves fit to the averages of three separate samples tested at each concentration.
AphA regulates luxR expression
The V. harveyi quorum-sensing circuit contains numerous regulatory feedback loops involving the Qrr sRNAs (Fig. 1; Ng and Bassler 2009). Above, we demonstrated that AphA represses the qrr genes (Fig. 2), and we know that the Qrr sRNAs repress luxR (Tu and Bassler 2007). Thus, we wondered what consequence altering AphA levels would have on luxR. Our prediction was that increased AphA would decrease qrr expression, promoting increased LuxR production. Quite to the contrary, when we overexpressed aphA, luxR expression decreased (Supplemental Fig. S2A). This result suggests that AphA represses luxR, and, furthermore, this repressive effect overrides any increased LuxR production stemming from AphA-directed repression of the qrr genes. luxR expression exhibited a similar decrease when aphA was overexpressed in a V. harveyi strain lacking the Qrr sRNAs (Δqrr1–5) (Fig. 4A), indicating that AphA repression of luxR does not depend on the Qrr sRNAs. AphA repression of luxR requires a functional AphA protein because, when a DNA-binding-defective AphA mutant protein (AphA K63E) (Kovacikova et al. 2004) was overexpressed, luxR expression was not altered (aphA*) (Fig. 4A).
Figure 4.
AphA represses expression of luxR and aphA. (A) luxR expression was monitored by qRT–PCR in a Δqrr1–5 (KT282: Δqrr1–5) strain carrying an empty vector (pJV17), a vector expressing V. harveyi aphA driven by an IPTG-inducible pTac promoter (paphA; pSTR0504), or the same vector expressing an aphA mutant defective for DNA binding (paphA*; pSTR0615) (Kovacikova et al. 2004). Averages and SEMs are shown. (B) Expression of V. harveyi chromosomal aphA was monitored by qRT–PCR in a locked LCD strain lacking luxR (KM812: luxO D47E, ΔluxR). The primers used in the qRT–PCR reactions were specific to the 5′ UTR, which is not encoded on the overexpression vector paphA (pSTR0504). Means and SEMs for three independent cultures are shown.
To examine whether AphA repression of luxR is direct, we measured AphA binding to DNA fragments spanning the luxR promoter region. Specific AphA binding (Kd ≈ 137 nM) occurred immediately downstream from the luxR transcription start site (+6 to +37 relative to the transcription start site) (Fig. 3C; Supplemental Fig. S3B). Together, these data indicate that AphA represses V. harveyi luxR expression by binding to the luxR promoter.
AphA represses its own expression
One mechanism bacteria commonly use to maintain appropriate levels of transcription factors is autorepression (Becskei and Serrano 2000; Rosenfeld et al. 2002; Nevozhay et al. 2009). Indeed, both of the known quorum-sensing transcription factors, LuxR and LuxO, display autorepression (Fig. 1; Chatterjee et al. 1996; Tu et al. 2010). Likewise, AphA autorepresses itself in V. cholerae (Lin et al. 2007). To address whether this is the case for AphA in V. harveyi, we measured aphA transcript levels using probes complementary to the aphA 5′ untranslated region (UTR). This strategy allowed us to exclusively follow chromosomally encoded aphA, since this upstream DNA was not present on the aphA overexpression construct. Overexpression of aphA in the locked LCD strain lacking luxR resulted in a twofold reduction in chromosomally encoded aphA expression (Fig. 4B), indicating that AphA modestly represses its own expression independently of LuxR. AphA autorepression also occurred in a strain lacking the Qrr sRNAs (Δqrr1–5) (Supplemental Fig. S2B), indicating that AphA repression of aphA does not occur through the Qrr sRNAs. Repression did not occur when the DNA-binding-defective aphA mutant (aphA*) was overexpressed (Fig. 4B; Supplemental Fig. S2B). We confirmed that AphA autorepression is direct using DNA-binding analyses. AphA specifically bound only to a fragment of DNA harboring the putative RNA polymerase binding site (−36 to +14 relative to the aphA transcription start site) with a Kd of ≈60 nM (Fig. 3C; Supplemental Fig. S3C).
LuxR and the Qrr sRNAs regulate aphA expression
LuxR has been shown previously to bind to the aphA promoter to repress aphA expression (Fig. 1; Lin et al. 2007; Pompeani et al. 2008). We confirmed this LuxR function by comparing aphA expression in V. harveyi strains producing low and high levels of LuxR. To test the effects of low levels of LuxR, we used a locked LCD strain (luxO D47E) in which the Qrr sRNAs are constitutively expressed and prevent production of LuxR. To test the effects of high levels of LuxR, we used two different locked HCD strains in which the Qrr sRNAs are not made and LuxR levels are maximal (ΔluxO and Δqrr1–5). Indeed, aphA expression was inversely correlated with LuxR levels: Low-level LuxR promoted high aphA expression (Fig. 5A, first bar), and high-level LuxR resulted in low aphA expression (Fig. 5A, second and third bars) consistent with the previous findings that LuxR represses aphA. However, levels of additional components (e.g., Qrr sRNAs) also differ between the LCD and HCD conditions. When this experiment was performed in these same LCD and HCD strains, but completely lacking luxR (ΔluxR), higher expression of aphA nonetheless occurred in the LCD strain compared with the HCD strains (Fig. 5A, cf. the fourth bar and the fifth and sixth bars). These results suggest that, in addition to negative regulation by LuxR, the Qrr sRNAs could activate aphA expression because, in the LCD strain, the Qrr sRNAs are constitutively produced, while in the HCD strain (through deletion of either luxO or the five qrr genes), the Qrr sRNAs are not present.
Figure 5.
The Qrr sRNAs activate aphA expression. (A) aphA expression was monitored by qRT–PCR in locked LCD strains containing luxR (KM83: luxO D47E) or lacking luxR (KM812: KM83, ΔluxR), strains locked at HCD due to deletion of luxO containing luxR (JAF78: luxO) or lacking luxR (KM806: JAF78, ΔluxR), and strains locked at HCD due to deletion of the five qrr genes containing luxR (KT282: Δqrr1–5) or lacking luxR (JS202: KT282, ΔluxR). Means and SEMs for RNA isolated from three independent cultures are shown. (B) Fluorescence from plasmid-encoded aphA-gfp (pYS069) or mutant aphA-gfp (pYS100) (deletion shown in C) transcriptional fusions were measured in E. coli MC4100 carrying an empty vector (pRHA109) or a vector expressing a rhamnose-inducible qrr4 gene (pSTR0227). GFP from at least two independent overnight cultures was assayed for each strain, and the means and SEMs are shown. (C) RNA alignment of the complement of the aphA 5′ UTR with V. harveyi Qrr 2–5. Sequence differences in Qrr1 (Tu and Bassler 2007) prevented it from aligning with the aphA mRNA and the other Qrr sRNAs. A stretch of the 5′ UTR (90 nucleotides) not involved in the RNA–RNA interaction was omitted for clarity. Boxed sequences with a star below are identical among the six RNAs. The translation initiation codon and the region deleted in the mutant aphA-gfp reporter are indicated above the aphA sequence.
To test for Qrr-directed activation of aphA, a plasmid carrying an inducible qrr4 gene was introduced into an E. coli strain containing a vector harboring aphA-gfp. Induction of qrr4 expression increased AphA-GFP production twofold (Fig. 5B). No other components of the V. harveyi quorum-sensing system are present in this E. coli strain, implying that Qrr sRNAs directly activate aphA expression. An alignment of the complement of the aphA 5′ UTR with the sequences of Qrr2–5 reveals a region of 30 noncontiguous complementary bases harboring stretches with up to eight consecutive complementary bases (Fig. 5C). These exact regions of the Qrr sRNAs are critical for regulation of other V. harveyi target mRNAs, including those encoding luxR and luxO (Tu and Bassler 2007; Tu et al. 2010). The homology between the 5′ UTR of the aphA mRNA and the Qrr sRNAs is more extensive than that in previously identified target mRNAs. Deletion of a portion of the aphA 5′ UTR eliminated Qrr4 activation of AphA-GFP production (Fig. 5B,C). We note, and Figure 5B shows, that this deletion also caused a modest increase in the basal level of expression. We are currently defining the mechanism by which the Qrr sRNAs enhance AphA production. Based on activation mechanisms defined for sRNAs in other bacterial systems, we suspect that Qrr sRNA binding to the target mRNA induces a conformation in the aphA 5′ UTR conducive to ribosome binding. Either of these events (Qrr sRNA binding or ribosome binding) could stabilize the target mRNA (Frohlich and Vogel 2009).
AphA controls expression of an LCD regulon
Our genetic analyses revealed four targets in the V. harveyi quorum-sensing circuit (qrr2, qrr3, qrr4, and luxR) whose transcription is controlled by AphA (Fig. 1). Given that AphA is a transcription factor, we wondered whether it controls the expression of additional V. harveyi genes. We performed microarray analyses to test this possibility. Our above findings demonstrate that AphA is maximally produced at LCD. For this reason, we compared the gene expression profiles of locked LCD V. harveyi strains possessing and lacking aphA. Again, as in our above analyses, we used ΔluxR strains to eliminate complications from indirect effects of LuxR. We identified 296 genes that were expressed at levels significantly above background (P < 0.0001) and whose expression changed more than twofold (Supplemental Table S1). Our results suggest that AphA activates 99 of these genes and represses the other 197 genes. Among the repressed group were qrr2, qrr3, and qrr4, confirming our above observations (Fig. 2). About half of the AphA-controlled genes (156) are classified as hypothetical genes. The remaining 140 genes have predicted functions, including in type III secretion, reductases/dehydrogenases, pilus production, flagellar structure, and gene expression (Table 1; Supplemental Table S1).
Table 1.
Classes of genes regulated by AphA more than twofold as determined by microarray analysis
See Supplemental Table S1 for a complete list of the genes.
Several of the AphA-controlled genes, such as the qrr genes and genes involved in type III secretion, are especially interesting to us because they are known to be regulated by LuxR (Henke and Bassler 2004; Waters and Bassler 2006; Tu et al. 2008). However, luxR was deleted in the strains used for the microarrays. Thus, we validated the microarray results and tested the role of LuxR by measuring AphA and/or LuxR regulation of a number of candidate targets at LCD (Fig. 6). One group of genes is regulated exclusively by AphA (for example, VIBHAR_06904) (Fig. 6A). A second class of genes is repressed by both AphA and LuxR (for example, VIBHAR_01726) (Fig. 6B). Finally, we identified a set of genes that is regulated by both AphA and LuxR, but by opposing means: One transcription factor represses expression, while the other activates expression (for example, VIBHAR_05035) (Fig. 6C). These results show that, first, AphA controls a large repertoire of genes at LCD that are not controlled by LuxR, and second, AphA and LuxR jointly regulate another set of genes.
Figure 6.
AphA and LuxR individually and jointly control target genes. RNA encoding candidate target genes was measured by qRT–PCR in the LCD-locked V. harveyi strains containing aphA (JAF548: luxO D47E), lacking aphA (JV50: JAF548, ΔaphA), lacking luxR (KM810: JAF548, ΔluxR), and lacking aphA and luxR (JV55: KM810, ΔaphA). Primers specific to VIBHAR_06904 (A), VIBHAR_01726 (B), and VIBHAR_05035 (C) were used. RNA was collected from three independent cultures, and means and SEMs are shown.
AphA has a role in the V. cholerae quorum-sensing circuit
In V. cholerae, AphA is involved in the regulatory cascade promoting virulence. Specifically, AphA activates tcpPH, and TcpPH is required for expression of toxT, which encodes a major virulence regulator in V. cholerae (Kovacikova and Skorupski 1999, 2001; Skorupski and Taylor 1999). V. cholerae AphA is 86% identical to V. harveyi AphA. Both V. harveyi and V. cholerae use the Qrr sRNAs to control expression of their corresponding master quorum-sensing regulators, LuxR and HapR, respectively (Lenz et al. 2004; Tu and Bassler 2007). In addition, LuxR/HapR represses aphA in both species (Kovacikova and Skorupski 2002; Lin et al. 2005; Pompeani et al. 2008). Given our above results demonstrating the central location of AphA in the V. harveyi quorum-sensing cascade, we wondered whether V. cholerae AphA plays an analogous role. That is, in V. cholerae, does AphA regulate qrr and hapR expression, and do the Qrr sRNAs regulate aphA expression? We test these possibilities below. First, as a control, we verified that, in our hands, V. cholerae AphA can activate a known target, vpsT (Yang et al. 2009), by measuring expression of a vpsT-lux reporter following overexpression of V. cholerae aphA. Overproduction of AphA resulted in dramatically increased vpsT-lux expression (∼12-fold) (Fig. 7A), consistent with previous findings that AphA activates vpsT expression (Yang et al. 2009).
Figure 7.
V. cholerae AphA represses expression of hapR and qrr4, and the V. cholerae Qrr sRNAs activate expression of aphA. (A) Bioluminescence from a vpsT-lux transcriptional fusion on a plasmid (pDL1711) was assayed in a V. cholerae locked HCD strain (WN865: ΔluxO) in the presence of an empty vector (pJV17) or a vector carrying V. cholerae aphA (pSTR0612). (B) Qrr2, Qrr3, and Qrr4 sRNA levels were measured by qRT–PCR in locked LCD V. cholerae strains lacking hapR and containing aphA (JC1796: luxO D47E, ΔhapR) (white bars) or lacking hapR and aphA (STRVc0044: JC1796, ΔaphA) (black bars). Means and SEMs for RNA collected from three independent cultures are shown. (C) hapR expression was measured directly using the QuantiGene Plex Reagent System (Panomics) (Tu et al. 2010). RNA was collected from an HCD-locked V. cholerae strain containing hapR (WN865: ΔluxO). The strain carried an empty vector (pJV17) or a vector overexpressing V. cholerae aphA, denoted paphA(Vc) (pSTR0612). (D) aphA RNA was measured by the same procedure in a locked HCD strain lacking hapR (WN868: WN865, ΔhapR) and a locked LCD strain lacking hapR (JC1796: luxO D47E, ΔhapR). In all cases, RNA from at least three independent cultures was analyzed. Means and SEMs are shown.
To examine the effect of AphA on V. cholerae qrr gene expression, we used the same strategy that we used above in V. harveyi. Specifically, to maximize qrr gene expression and eliminate indirect effects caused by HapR, we used an LCD-locked V. cholerae strain lacking hapR. In this strain, deletion of aphA increased expression of qrr2, qrr3, and qrr4 (Fig. 7B), showing that, in V. cholerae, as in V. harveyi, AphA represses qrr2, qrr3, and qrr4 expression independently of HapR. To test whether AphA controls hapR expression, we overexpressed aphA and measured hapR expression at HCD (when HapR levels are at their maximum). Indeed, overexpression of aphA repressed hapR expression (Fig. 7C). Finally, we measured aphA expression in V. cholerae strains lacking hapR that were locked at HCD or LCD. As a reminder, in the HCD strain, Qrr levels are low, while in the LCD strain, Qrr levels are high. This strategy allowed us to determine whether V. cholerae aphA is regulated by the Qrr sRNAs in the absence of HapR. Indeed, expression of aphA was threefold higher in the LCD-locked mutant than in the HCD-locked mutant (Fig. 7D), demonstrating that the Qrr sRNAs likely activate expression of V. cholerae aphA. We therefore conclude that AphA controls the flow of information through both V. harveyi and V. cholerae quorum-sensing circuits.
Discussion
The quorum-sensing bacterium V. harveyi produces, detects, and responds to three AIs to control collective behaviors. At LCD, LuxO∼P activates transcription of the qrr1–5 genes, encoding five redundant sRNAs (Qrr1–5), which repress translation of the master quorum-sensing regulator LuxR. Here we show that, at LCD, the Qrr sRNAs simultaneously activate production of another transcription factor, AphA. At HCD, the detection of AIs results in dephosphorylation of LuxO. Qrr sRNA production stops, which eliminates activation of aphA expression, and, in contrast, allows LuxR production to commence. Further reinforcement of this pattern comes from mutual repression of aphA and luxR. Specifically, AphA represses luxR expression at LCD and LuxR represses aphA expression at HCD. Additionally, AphA and LuxR both feed back to control qrr expression. The consequence of this network architecture is reciprocal control of AphA and LuxR. At LCD, AphA predominates: The Qrr sRNAs are present and activate translation of aphA, and AphA represses luxR (Fig. 1). At HCD, LuxR predominates: The Qrr sRNAs are not present to repress luxR or activate aphA, and, furthermore, LuxR represses aphA (Fig. 1). This same network architecture reciprocally controls the levels of AphA and HapR (a homolog of LuxR) in the V. cholerae quorum-sensing circuit.
Whether AphA, LuxR, or both are produced is determined by the pool of five Qrr sRNAs. The primary mechanism controlling Qrr sRNA production is via the cascade that phosphorylates or dephosphorylates LuxO in response to the absence or presence of AIs, respectively (Freeman and Bassler 1999). We predict that, at each particular concentration of LuxO∼P, there is a corresponding pool size of the Qrr sRNAs. This pool, in turn, establishes that particular amounts of AphA and LuxR are present. We are interested in understanding the mechanisms underlying Qrr sRNA activation (aphA) and repression (luxR and luxO) of translation of mRNA targets. We performed some studies of Qrr-directed repression of luxR and luxO translation (Lenz et al. 2004; Tu and Bassler 2007; Svenningsen et al. 2009; Tu et al. 2010). Repression occurs by Hfq-dependent Qrr sRNA base-pairing with the target mRNA over the ribosome-binding site and translation initiation codon. Binding at this site occludes the ribosome and exposes the target mRNAs to degradation. Regarding Qrr sRNA-directed activation of aphA expression, secondary structure predictions of the aphA mRNA indicate that stem–loops exist in the aphA 5′ UTR (data not shown). One of these putative stem–loops contains the predicted ribosome-binding site and start codon. This scenario is analogous to the 5′ UTR of sRNA-activated target mRNAs in other bacterial systems (Frohlich and Vogel 2009). At these targets, sRNAs base-pair with an mRNA region that does not overlap with the ribosome-binding site. Base-pairing disrupts inhibitory stem–loop structures, allowing the ribosome access to the mRNA, stabilizing the mRNA, or both (Majdalani et al. 1998; Hammer and Bassler 2007; Prevost et al. 2007; Landt et al. 2010).
Qrr sRNA control, coupled with mutual AphA–LuxR repression, likely allows V. harveyi to generate a variety of AphA:LuxR concentration ratios. Each blend of these two transcription factors will, likewise, drive a precise pattern of gene expression. If AphA and LuxR did not jointly control genes, AphA could repress or activate genes at LCD and LuxR could repress or activate genes at HCD. However, the AphA and LuxR regulons overlap, as shown by our microarray analyses, providing the cell with access to more sophisticated regulatory programs. Genes controlled by both LuxR and AphA can theoretically fall into four categories: AphA and LuxR both repress, AphA and LuxR both activate, AphA activates and LuxR represses, and AphA represses and LuxR activates. Additionally, each factor will have a specific strength of regulation at each promoter. Having two regulators of varying strength impinging on the same promoters could establish a combinatorial pattern of gene regulation in response to changing AI concentrations and different cell densities.
Type III secretion genes stand as one example of coregulation by LuxR and AphA. In this case, both regulators repress expression (VIBHAR_01726) (for example, Fig. 6B; Supplemental Table S1; Henke and Bassler 2004; Waters and Bassler 2006). Thus, production of the type III secretion apparatus is repressed at LCD and HCD, but by AphA in the first case and by LuxR in the second case. When, then, are type III secretion genes expressed? We propose that, as cells transition from LCD to HCD, at a specific concentration of extracellular AIs, there will simultaneously exist low levels of both AphA and LuxR, and this is the window of cell densities during which the type III secretion system is maximally expressed. Another gene coregulated by LuxR and AphA is the hypothetical protein VIBHAR_05035, which is repressed by AphA and activated by LuxR (Fig. 6C). This gene is repressed by two mechanisms at LCD (repression by AphA and lack of activation by LuxR). The gene is activated at HCD when AphA levels are low and LuxR levels are high. This regulatory wiring ensures that VIBHAR_05035 is expressed only at HCD. We identified two other genes likely to exhibit the VIBHAR_05035 pattern of regulation (VIBHAR_06262 and VIBHAR_06741) (Waters and Bassler 2006). We are currently characterizing the AphA and LuxR regulons to define the mode and strength of regulation by AphA, LuxR, and the two transcription factors together at each promoter. We anticipate that future experiments will identify examples of all four regulatory combinations described above.
The newly appreciated role of AphA as an LCD master regulator begins to account for some puzzling aspects of the quorum-sensing network. One outstanding puzzle is why the circuit uses five highly similar sRNAs (Tu and Bassler 2007). Obviously, genetic amplification of sRNAs immediately implies an increase of total sRNA production. Among other effects, this increased sRNA production accelerates the transition of cells from HCD to LCD (Mehta et al. 2008). We focus on this transition because other features of the network also seem engineered to achieve rapid sRNA production upon a switch to LCD. Specifically, phosphorylation of LuxO following removal of AIs is rapid, while the decrease of the sRNA activators LuxO and LuxR is slow, implying that maximal sRNA production occurs immediately following a switch to LCD (Tu and Bassler 2007; Teng et al. 2010; Tu et al. 2010). Physiologically, the transition from HCD to LCD can be as fast as expulsion of cells from a host or shedding from a biofilm, and so a rapid change in gene expression in this case may be highly advantageous. In contrast, the LCD-to-HCD transition presumably depends on cell growth and is therefore likely to be slow, requiring at least several cell generations. Rapid production of sRNAs following a switch to LCD is beneficial only if these sRNAs can, in turn, rapidly affect protein levels. This evident fact has presented a puzzle: The known targets of the sRNAs—e.g., LuxO and LuxR—are negatively regulated, implying that rapid production of sRNAs can only halt protein production, leaving the existing pools of protein to decay slowly via dilution by growth, as no proteolysis of LuxO or LuxR has been observed. The answer may lie in our discovery that aphA is positively regulated by the sRNAs: sRNA activation of aphA translation, with silent aphA transcripts that presumably already exist in the cell at HCD, likely enables a rapid, many-fold increase in the level of AphA immediately following the switch to LCD. Thus, positive regulation of AphA may provide the missing piece of the puzzle of why the multiple redundant sRNAs exist in the V. harveyi quorum-sensing network; namely, multiple sRNA genes means a higher total sRNA peak production rate, and, consequently, a faster accumulation of AphA at the HCD-to-LCD transition. What about the additional regulatory links we found to AphA? First, the negative regulation of aphA expression by LuxR, while reducing AphA production at the switch to LCD, is unlikely to affect the fold increase of AphA, and may be required to keep AphA levels low at HCD. Second, we expect that the negative feedback of AphA on sRNA production is present to ensure that the rapid production of sRNAs at the switch to LCD is reduced to a tolerable steady-state level once AphA accumulates (and notice that the decrease of LuxR at LCD, by reducing sRNA activation, has a parallel effect) (Tu et al. 2008). Third, repression of luxR by AphA helps keep LuxR levels low at LCD. Finally, the negative feedback of AphA on itself is likely to play a similar homeostatic role for AphA levels that LuxR autorepression plays for LuxR levels (Tu et al. 2008). Interestingly, the presence of AphA at LCD may contribute to the asymmetrically slow response of cells following addition of AI, since not only must LuxR accumulate, but AphA must also be diluted by growth for cells to transition to the HCD gene expression program. Overall, the V. harveyi network architecture seems to be strongly influenced by the dynamics of quorum-sensing transitions. We will directly explore these dynamics, in both wild-type cells and mutants engineered to change network architecture, in future experiments.
AphA has been well-studied for its role in controlling virulence in V. cholerae. Specifically, AphA, together with AphB, activates tcpPH expression, and TcpPH subsequently activates toxT, encoding a major V. cholerae virulence regulator (Kovacikova and Skorupski 1999, 2001; Skorupski and Taylor 1999; Kovacikova et al. 2004). In V. cholerae, AphA controls at least 15 additional target genes, some of which are involved in biofilm formation or motility (Kovacikova and Skorupski 2001; Kovacikova et al. 2003, 2005; Yang et al. 2009). Our results indicate that AphA controls a much larger regulon of genes than this at LCD in V. harveyi (and thus possibly also in V. cholerae). These target genes were revealed using V. harveyi strains locked at LCD and lacking luxR. Previous studies of AphA in V. cholerae examined cells under quite different conditions than the ones we used here. Specifically, the earlier V. cholerae microarray analyses were performed at a relatively HCD in strains possessing and lacking aphA (Kovacikova et al. 2005). Furthermore, HapR was wild type in these strains. Based on our results with V. harveyi and our preliminary results with V. cholerae, we suspect that HapR was the transcription factor that primarily functioned under the previous experimental conditions, and this could account for why so few genes were identified to be controlled by AphA (Supplemental Fig. S4). We predict that, at LCD, AphA regulates many additional V. cholerae genes. Some of these genes are likely to be critical for pathogenicity, given that V. cholerae produces its suite of virulence factors only at LCD (Kovacikova and Skorupski 2002; Miller et al. 2002).
We made progress in determining how V. harveyi LuxR controls gene expression by defining its DNA-binding site and decoding its regulon (Waters and Bassler 2006; Lee et al. 2008; Pompeani et al. 2008), but we understand comparatively little about AphA in V. harveyi. However, the mechanisms of AphA repression and activation have received attention in V. cholerae, which should guide our continued analyses (Kovacikova and Skorupski 2001; Kovacikova et al. 2003, 2004, 2005, 2010; De Silva et al. 2005; Yang et al. 2009). The preliminary results described here suggest that, in V. harveyi, AphA uses at least two mechanisms for repression. First, we take the case of qrr4: AphA binds to the qrr4 promoter where LuxO∼P binds. LuxO∼P binding is required for activation (Lenz et al. 2004; Tu and Bassler 2007), and thus AphA binding likely excludes LuxO∼P binding. Interestingly, AphA does not repress qrr1 or qrr5 expression. Differences in positioning of the LuxO∼P site at each of these promoters (Tu and Bassler 2007) and the strength of AphA binding could account for our findings. Second, we take the case of the luxR and aphA promoters: AphA binds in close proximity to the transcription start sites near where RNA polymerase binds. AphA occupancy likely prevents RNA polymerase from binding. This second mechanism is analogous to how AphA represses the alsD promoter in V. cholerae (Kovacikova et al. 2005).
Understanding LuxR/HapR and its regulon will provide insight into the HCD lifestyle of vibrios, presumably relevant when they occupy hosts. Likewise, understanding AphA and its regulon should give clues about the LCD lifestyle of virbios, presumably relevant when they are free-living in the environment. Based on the present findings, we predict that AphA-activated genes are beneficial for individual behaviors, while AphA-repressed genes are useful for collective behaviors. Additionally, placing AphA, an LCD regulator, at the center of the quorum-sensing circuit likely allows vibrios to optimally adjust from high expression of genes associated with a stressful stationary-phase environment to high expression of genes that adapt the bacteria to the nutrient-abundant rapid-growth phase. Consistent with this notion, we identified numerous AphA-repressed oxidases and reductases that could be involved in alleviating oxidative stress during stationary phase (Supplemental Table S1; Nystrom and Gustavsson 1998; Chang et al. 2002). Likewise, we identified an AphA-repressed protease and an oligopeptidase that could be involved in preventing protein accumulation during stationary phase (Supplemental Table S1; Gottesman 2003; Weichart et al. 2003). We suggest that AphA is used at LCD to promote individual behaviors and prevent expression of genes required for growth under stressful environmental conditions, whereas LuxR is present at HCD to promote collective behaviors and control the genetic program useful for nutrient-limiting situations.
Materials and methods
Bacterial strains and media
V. harveyi strain BB120 (BAA-1116) (Bassler et al. 1997) and derivatives were grown in Luria-Murine (LM) medium with shaking at 30°C. E. coli strains S17-1λpir (de Lorenzo and Timmis 1994), BL21(DE3) (Invitrogen), MC4100 (Casadaban 1976), and derivatives, and V. cholerae C6706 biovar El Tor (Thelin and Taylor 1996) were grown in Luria-Bertani (LB) medium with shaking at 30°C unless otherwise noted. Strains used in this study are described in Supplemental Table S2. Antibiotics (Sigma) were used at the following concentrations: 100 μg/mL ampicillin, 10 μg/mL chloramphenicol, 100 μg/mL kanamycin, 100 μg/mL gentamicin, 50 μg/mL polymyxin B, and 10 μg/mL tetracycline (except 5 μg/mL in liquid medium for V. harveyi). The Ptac promoter constructs were induced with 0.2 mM IPTG (in V. harveyi) or 0.05 mM IPTG (in V. cholerae). Levels of induction for wild-type aphA and the DNA-binding-defective mutant aphA (AphA K63E, aphA*) (Kovacikova et al. 2004) were similar (Supplemental Fig. S5). hfq mRNA levels were not significantly affected by overexpression of wild-type aphA or the DNA-binding-defective aphA mutant (data not shown). Induction of aphA(Vc) in V. cholerae with higher IPTG concentrations resulted in a growth defect that we cannot explain; however, this growth defect was not observed when aphA was overexpressed in V. harveyi. qrr4 on plasmid pSTR0227 was induced with 1 mM rhamnose. Plasmid constructs were introduced into electrocompetent E. coli S17-1λpir using 0.2 μM cuvettes (USA Scientific) using a Bio-Rad MicroPulser. Plasmids were transferred into V. harveyi and V. cholerae by conjugation between E. coli S17-1λpir and V. harveyi and V. cholerae cultures on LB plates, followed by isolation of exconjugants on LM or LB plates containing polymyxin B and the appropriate antibiotic selection for plasmid maintenance.
DNA manipulations and mutant construction
E. coli S17-1λpir was used for all cloning procedures. DNA manipulations were performed as in Sambrook et al. (1989). Restriction enzymes, T4 DNA ligase, T4 polynucleotide kinase, and Antarctic phosphatase were purchased from New England Biolabs. PCR reactions used iProof DNA polymase (Bio-Rad). Plasmids were constructed as described in Supplemental Table S3 using primers listed in Supplemental Table S4 that were purchased from Integrated DNA Technologies (IDT). QuickChange mutagenesis (Stratagene) was used to introduce the DNA-binding-defective mutation onto pSTR0504 (resulting in pSTR0615) using primers listed in Supplemental Table S4. All plasmids were confirmed by sequencing at Genewiz. luxO D47E was moved onto the E. coli chromosome using the λInCh method (Boyd et al. 2000) and contained a promoter mutation rendering it unresponsive to Qrr sRNA regulation (Tu et al. 2010). Mutants in V. harveyi were constructed using λ red recombineering in E. coli S17-1λpir∷pKD46 (Datsenko and Wanner 2000) on the pLAFR2 cosmid, which contained regions of the V. harveyi genome. The mutant cosmids (see Supplemental Table S3) were conjugated into V. harveyi, and mutant alleles were incorporated into the V. harveyi genome by homologous recombination (Bassler et al. 1993). Antibiotic markers were removed using FLP-mediated recombination by expressing FLP from a plasmid, pTL18 (Long et al. 2009). V. cholerae in-frame deletions were constructed as described (Skorupski and Taylor 1996).
Screen for regulators of qrr4 expression
Locked LCD V. harveyi (TL45) (Long et al. 2009) was conjugated with E. coli S17 λpir containing pRL27 (Larsen et al. 2002) overnight on LB plates, and the mixture was subsequently plated onto LM agar plates containing kanamycin and polymyxin B to select for exconjugants with Tn5 transposon insertions. A total of 2000–4000 colonies from at least six separate matings were pooled from the agar surface and resuspended in ∼4 mL of LM medium supplemented with kanamycin and polymyxin B. Aliquots were added to glycerol and frozen at −80°C or immediately used for FACS analysis (Waters and Bassler 2006). A 0.5-mL aliquot of the well-suspended mutant pool was diluted into 4 mL of LM supplemented with kanamycin and polymyxin B and grown for 1–2 h at 30°C with aeration. At least three separate 100-μL aliquots of the culture were diluted into 1 mL of filtered 1× PBS, and the cells were sorted using fluorescence-assisted cell sorting (Becton Dickinson FACSAria cell sorter). Approximately 200,000 events were screened corresponding to 200,000 cells containing Tn5 insertions, although not all insertions were unique. To screen for mutants in repressors of qrr4-gfp, cells exhibiting the highest levels of fluorescence were isolated (the top 1%). To screen for activators, cells exhibiting lowest fluorescence were isolated (the bottom 4%). These initial sorted populations were enriched by resorting. Dilutions of the final sorted population were plated onto LM agar supplemented with kanamycin and polymyxin B and incubated overnight at 30°C. Individual colonies were arrayed into 150 μL of LM supplemented with kanamycin and polymyxin B in separate wells of 96-well plates and grown with shaking overnight at 30°C. GFP was measured from the overnight cultures, and cultures exhibiting increased GFP were retained. A total of 50 mutants were obtained. The locations of the Tn5 insertions were mapped as described in the Supplemental Material.
RNA isolation
RNA used for microarray analyses and quantitative RT–PCR (qRT–PCR) was isolated from V. harveyi cultures at an OD600 = 1.1–1.2 using Trizol (Invitrogen) or by following the QIAgen RNesay Minikit with RNAprotect protocol (Qiagen). See the Supplemental Material for details.
qRT–PCR
Following RNA isolation from at least three independent cultures, cDNA was generated as described (Tu and Bassler 2007) with SuperScript III reverse transcriptase (Invitrogen) using 1–3 μg of RNA. Real-time PCR analyses were also performed as described (Tu and Bassler 2007) on an ABI Prism 7900HT Sequence Detection System using Sybr Green mix (ABI). The primers used are listed in Supplemental Table S4. Triplicate biological samples were measured and analyzed by a comparative CT method (Applied Biosystems) in which the relative amount of each target RNA was normalized to the same internal control RNA (hfq) and then to each other to determine the relative changes in RNA levels.
Direct RNA measurements
RNA levels were directly measured using the QuantiGene Plex 2.0 Reagent system (Panomics) as described (Tu et al. 2010). RNA from 100 μL of culture was processed following the manufacturer's protocol and analyzed with beads carrying probes specific to the targets indicated in the figure legends. RNA levels were normalized to the levels of hfq or recA RNA (both remained unchanged during the course of the experiments). qRT–PCR and analysis with QunatiGene Plex Reagent System gave similar results.
GFP and bioluminescence assays
OD600, GFP, and bioluminescence were all measured as described previously (Bassler et al. 1993; Lenz et al. 2004; Tu and Bassler 2007). Values reported are the mean and SEM of at least three biological replicates at an OD600 of 1.0–1.2, unless otherwise noted.
AphA purification
AphA was purified after overexpression from plasmid pSTR0606 encoding a 6-His tag, a thrombin cleavage sequence, and V. harveyi aphA under the T7 promoter as described in the Supplemental Material.
Fluorescence anisotropy
DNA binding was assessed by fluorescence anisotropy. Oligonucleotides (Supplemental Table S4) containing a 5′ fluoroscein tag were purchased from IDT to be used as substrates, and fluorescence anisotropy analyses were performed as described (see the Supplemental Material for details; Pompeani et al. 2008). The concentrations of AphA protein added are indicated in each graph. One-site-specific nonlinear binding curves were fit to the data using GraphPad software, and the Kd were interpolated from the plots. Three samples were averaged to obtain each point, and the Kd and SD for the fit line are shown.
Microarray analysis
Microarrays contained three 60-mer probes per ORF in the V. harveyi genome (GenBank strain BAA-1116), each spotted in duplicate, for a total of six probes per gene (Agilent custom array 2521087). RNA preparation, cDNA synthesis, hybridization conditions, and data acquisition are described in the Supplemental Material. Data were extracted with Agilent Feature Extractor and analyzed on the Princeton University Microarray Database (PUMAdb, http://puma.princeton.edu) based on Gollub et al. (2003). These data are publically available on the PUMAdb. Data were retrieved for probes that were above background (P < 0.0001) and differed more than twofold. Probes were averaged for each gene. Four arrays were performed comparing three independent cultures of each strain as well as a dye-swap comparison for one set of strains.
Acknowledgments
We thank Ned Wingreen for insightful comments and ideas, and members of the Bassler laboratory for helpful suggestions. We thank Zach Donnell and Wai-Leung Ng for strains. We also thank Donna Storton, Jessica Buckles, and John Matese for assistance with microarray experiments. PUMAdb is funded in part by NIH grant P50 GM071508. This work was supported by the Howard Hughes Medical Institute, National Institutes of Health (NIH) grant 5R01GM065859, NIH grant 5R01AI054442, National Science Foundation (NSF) grant MCB-0343821 to B.L.B, NIH fellowship F32AI085922 to S.T.R., and NIH fellowship F32GM089019 to J.C.V.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.2015011.
Supplemental material is available for this article.
References
- Bassler BL, Wright M, Showalter RE, Silverman MR 1993. Intercellular signalling in Vibrio harveyi: Sequence and function of genes regulating expression of luminescence. Mol Microbiol 9: 773–786 [DOI] [PubMed] [Google Scholar]
- Bassler BL, Greenberg EP, Stevens AM 1997. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J Bacteriol 179: 4043–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becskei A, Serrano L 2000. Engineering stability in gene networks by autoregulation. Nature 405: 590–593 [DOI] [PubMed] [Google Scholar]
- Boyd D, Weiss DS, Chen JC, Beckwith J 2000. Towards single-copy gene expression systems making gene cloning physiologically relevant: λ InCh, a simple Escherichia coli plasmid-chromosome shuttle system. J Bacteriol 182: 842–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban MJ 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage λ and Mu. J Mol Biol 104: 541–555 [DOI] [PubMed] [Google Scholar]
- Chang DE, Smalley DJ, Conway T 2002. Gene expression profiling of Escherichia coli growth transitions: An expanded stringent response model. Mol Microbiol 45: 289–306 [DOI] [PubMed] [Google Scholar]
- Chatterjee J, Miyamoto CM, Meighen EA 1996. Autoregulation of luxR: The Vibrio harveyi lux-operon activator functions as a repressor. Mol Microbiol 20: 415–425 [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280: 295–298 [DOI] [PubMed] [Google Scholar]
- de Lorenzo V, Timmis KN 1994. Analysis and construction of stable phenotypes in Gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol 235: 386–405 [DOI] [PubMed] [Google Scholar]
- De Silva RS, Kovacikova G, Lin W, Taylor RK, Skorupski K, Kull FJ 2005. Crystal structure of the virulence gene activator AphA from Vibrio cholerae reveals it is a novel member of the winged helix transcription factor superfamily. J Biol Chem 280: 13779–13783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman JA, Bassler BL 1999. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol Microbiol 31: 665–677 [DOI] [PubMed] [Google Scholar]
- Frohlich KS, Vogel J 2009. Activation of gene expression by small RNA. Curr Opin Microbiol 12: 674–682 [DOI] [PubMed] [Google Scholar]
- Gollub J, Ball CA, Binkley G, Demeter J, Finkelstein DB, Hebert JM, Hernandez-Boussard T, Jin H, Kaloper M, Matese JC, et al. 2003. The Stanford microarray database: Data access and quality assessment tools. Nucleic Acids Res 31: 94–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S 2003. Proteolysis in bacterial regulatory circuits. Annu Rev Cell Dev Biol 19: 565–587 [DOI] [PubMed] [Google Scholar]
- Hammer BK, Bassler BL 2003. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol 50: 101–104 [DOI] [PubMed] [Google Scholar]
- Hammer BK, Bassler BL 2007. Regulatory small RNAs circumvent the conventional quorum sensing pathway in pandemic Vibrio cholerae. Proc Natl Acad Sci 104: 11145–11149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke JM, Bassler BL 2004. Quorum sensing regulates type III secretion in Vibrio harveyi and Vibrio parahaemolyticus. J Bacteriol 186: 3794–3805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacikova G, Skorupski K 1999. A Vibrio cholerae LysR homolog, AphB, cooperates with AphA at the tcpPH promoter to activate expression of the ToxR virulence cascade. J Bacteriol 181: 4250–4256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacikova G, Skorupski K 2001. Overlapping binding sites for the virulence gene regulators AphA, AphB and cAMP-CRP at the Vibrio cholerae tcpPH promoter. Mol Microbiol 41: 393–407 [DOI] [PubMed] [Google Scholar]
- Kovacikova G, Skorupski K 2002. Regulation of virulence gene expression in Vibrio cholerae by quorum sensing: HapR functions at the aphA promoter. Mol Microbiol 46: 1135–1147 [DOI] [PubMed] [Google Scholar]
- Kovacikova G, Lin W, Skorupski K 2003. The virulence activator AphA links quorum sensing to pathogenesis and physiology in Vibrio cholerae by repressing the expression of a penicillin amidase gene on the small chromosome. J Bacteriol 185: 4825–4836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacikova G, Lin W, Skorupski K 2004. Vibrio cholerae AphA uses a novel mechanism for virulence gene activation that involves interaction with the LysR-type regulator AphB at the tcpPH promoter. Mol Microbiol 53: 129–142 [DOI] [PubMed] [Google Scholar]
- Kovacikova G, Lin W, Skorupski K 2005. Dual regulation of genes involved in acetoin biosynthesis and motility/biofilm formation by the virulence activator AphA and the acetate-responsive LysR-type regulator AlsR in Vibrio cholerae. Mol Microbiol 57: 420–433 [DOI] [PubMed] [Google Scholar]
- Kovacikova G, Lin W, Skorupski K 2010. The LysR-type virulence activator AphB regulates the expression of genes in Vibrio cholerae in response to low pH and anaerobiosis. J Bacteriol 192: 4181–4191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landt SG, Lesley JA, Britos L, Shapiro L 2010. CrfA, a small noncoding RNA regulator of adaptation to carbon starvation in Caulobacter crescentus. J Bacteriol 192: 4763–4775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen RA, Wilson MM, Guss AM, Metcalf WW 2002. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch Microbiol 178: 193–201 [DOI] [PubMed] [Google Scholar]
- Lee DH, Jeong HS, Jeong HG, Kim KM, Kim H, Choi SH 2008. A consensus sequence for binding of SmcR, a Vibrio vulnificus LuxR homologue, and genome-wide identification of the SmcR regulon. J Biol Chem 283: 23610–23618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL 2004. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118: 69–82 [DOI] [PubMed] [Google Scholar]
- Lin W, Kovacikova G, Skorupski K 2005. Requirements for Vibrio cholerae HapR binding and transcriptional repression at the hapR promoter are distinct from those at the aphA promoter. J Bacteriol 187: 3013–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Kovacikova G, Skorupski K 2007. The quorum sensing regulator HapR downregulates the expression of the virulence gene transcription factor AphA in Vibrio cholerae by antagonizing Lrp- and VpsR-mediated activation. Mol Microbiol 64: 953–967 [DOI] [PubMed] [Google Scholar]
- Long T, Tu KC, Wang Y, Mehta P, Ong NP, Bassler BL, Wingreen NS 2009. Quantifying the integration of quorum-sensing signals with single-cell resolution. PLoS Biol 7: e68 doi: 10.1371/journal.pbio.1000068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S 1998. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc Natl Acad Sci 95: 12462–12467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P, Goyal S, Wingreen NS 2008. A quantitative comparison of sRNA-based and protein-based gene regulation. Mol Syst Biol 4: 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P, Goyal S, Long T, Bassler BL, Wingreen NS 2009. Information processing and signal integration in bacterial quorum sensing. Mol Syst Biol 5: 325 doi: 10.1038/msb.2009.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MB, Skorupski K, Lenz DH, Taylor RK, Bassler BL 2002. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 110: 303–314 [DOI] [PubMed] [Google Scholar]
- Neiditch MB, Federle MJ, Pompeani AJ, Kelly RC, Swem DL, Jeffrey PD, Bassler BL, Hughson FM 2006. Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell 126: 1095–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevozhay D, Adams RM, Murphy KF, Josic K, Balazsi G 2009. Negative autoregulation linearizes the dose-response and suppresses the heterogeneity of gene expression. Proc Natl Acad Sci 106: 5123–5128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng WL, Bassler BL 2009. Bacterial quorum-sensing network architectures. Annu Rev Genet 43: 197–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick RP 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol 48: 1429–1449 [DOI] [PubMed] [Google Scholar]
- Nystrom T, Gustavsson N 1998. Maintenance energy requirement: What is required for stasis survival of Escherichia coli? Biochim Biophys Acta 1365: 225–231 [DOI] [PubMed] [Google Scholar]
- Pompeani AJ, Irgon JJ, Berger MF, Bulyk ML, Wingreen NS, Bassler BL 2008. The Vibrio harveyi master quorum-sensing regulator, LuxR, a TetR-type protein is both an activator and a repressor: DNA recognition and binding specificity at target promoters. Mol Microbiol 70: 76–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevost K, Salvail H, Desnoyers G, Jacques JF, Phaneuf E, Masse E 2007. The small RNA RyhB activates the translation of shiA mRNA encoding a permease of shikimate, a compound involved in siderophore synthesis. Mol Microbiol 64: 1260–1273 [DOI] [PubMed] [Google Scholar]
- Rosenfeld N, Elowitz MB, Alon U 2002. Negative autoregulation speeds the response times of transcription networks. J Mol Biol 323: 785–793 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T 1989. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Skorupski K, Taylor RK 1996. Positive selection vectors for allelic exchange. Gene 169: 47–52 [DOI] [PubMed] [Google Scholar]
- Skorupski K, Taylor RK 1999. A new level in the Vibrio cholerae ToxR virulence cascade: AphA is required for transcriptional activation of the tcpPH operon. Mol Microbiol 31: 763–771 [DOI] [PubMed] [Google Scholar]
- Svenningsen SL, Waters CM, Bassler BL 2008. A negative feedback loop involving small RNAs accelerates Vibrio cholerae's transition out of quorum-sensing mode. Genes Dev 22: 226–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenningsen SL, Tu KC, Bassler BL 2009. Gene dosage compensation calibrates four regulatory RNAs to control Vibrio cholerae quorum sensing. EMBO J 28: 429–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartzman E, Silverman M, Meighen EA 1992. The luxR gene product of Vibrio harveyi is a transcriptional activator of the lux promoter. J Bacteriol 174: 7490–7493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swem LR, Swem DL, Wingreen NS, Bassler BL 2008. Deducing receptor signaling parameters from in vivo analysis: LuxN/AI-1 quorum sensing in Vibrio harveyi. Cell 134: 461–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng SW, Wang Y, Tu KC, Long T, Mehta P, Wingreen NS, Bassler BL, Ong NP 2010. Measurement of the copy number of the master quorum-sensing regulator of a bacterial cell. Biophys J 98: 2024–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelin KH, Taylor RK 1996. Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infect Immun 64: 2853–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu KC, Bassler BL 2007. Multiple small RNAs act additively to integrate sensory information and control quorum sensing in Vibrio harveyi. Genes Dev 21: 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu KC, Waters CM, Svenningsen SL, Bassler BL 2008. A small-RNA-mediated negative feedback loop controls quorum-sensing dynamics in Vibrio harveyi. Mol Microbiol 70: 896–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu KC, Long T, Svenningsen SL, Wingreen NS, Bassler BL 2010. Negative feedback loops involving small regulatory RNAs precisely control the Vibrio harveyi quorum-sensing response. Mol Cell 37: 567–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters CM, Bassler BL 2006. The Vibrio harveyi quorum-sensing system uses shared regulatory components to discriminate between multiple autoinducers. Genes Dev 20: 2754–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichart D, Querfurth N, Dreger M, Hengge-Aronis R 2003. Global role for ClpP-containing proteases in stationary-phase adaptation of Escherichia coli. J Bacteriol 185: 115–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Frey EM, Liu Z, Bishar R, Zhu J 2009. The virulence transcriptional activator AphA enhances biofilm formation by Vibrio cholerae by activating expression of the biofilm regulator VpsT. Infect Immun 78: 697–703 [DOI] [PMC free article] [PubMed] [Google Scholar]