Abstract
Motivation: With recent advances in sequencing, structural and functional studies of RNA lag behind the discovery of sequences. Computational analysis of RNA is increasingly important to reveal structure–function relationships with low cost and speed. The purpose of this study is to use multiple homologous sequences to infer a conserved RNA structure.
Results: A new algorithm, called Multilign, is presented to find the lowest free energy RNA secondary structure common to multiple sequences. Multilign is based on Dynalign, which is a program that simultaneously aligns and folds two sequences to find the lowest free energy conserved structure. For Multilign, Dynalign is used to progressively construct a conserved structure from multiple pairwise calculations, with one sequence used in all pairwise calculations. A base pair is predicted only if it is contained in the set of low free energy structures predicted by all Dynalign calculations. In this way, Multilign improves prediction accuracy by keeping the genuine base pairs and excluding competing false base pairs. Multilign has computational complexity that scales linearly in the number of sequences. Multilign was tested on extensive datasets of sequences with known structure and its prediction accuracy is among the best of available algorithms. Multilign can run on long sequences (> 1500 nt) and an arbitrarily large number of sequences.
Availability: The algorithm is implemented in ANSI C++ and can be downloaded as part of the RNAstructure package at: http://rna.urmc.rochester.edu
Contact: david_mathews@urmc.rochester.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
RNA sequences have been discovered to play remarkably diverse roles, such as catalysis (Fedor and Williamson, 2005; Nissen et al., 2000), gene expression regulation (Batey, 2006; Lee, 1993) and sequence recognition (Kiss-Laszlo, 1996; Vendeix et al., 2008). Recent studies show that there are a large number of RNA transcripts that do not encode proteins (Ravasi et al., 2006; Sharma et al., 2010; The Encode Consortium, 2007; The Fantom Consortium, 2002). The functional and structural roles of these RNAs are of great interest. With current techniques, however, it is slow and expensive to experimentally determine structures for the majority of those RNA. Thus, structural and functional analysis of RNA molecules has lagged behind the rate of sequencing, leaving a large gap needing to be filled.
Structure prediction is an attractive tool for studying all the currently available sequences. RNA secondary structure, defined as the sum of canonical base pairs (A-U, G-U and G-C) is commonly predicted and these predictions have been used to design structures (Aguirre-Hernandez et al., 2007; Diamond et al., 2001; Dirks et al., 2004), discover functional RNA sequences in genomes (Torarinsson et al., 2006; Uzilov et al., 2006; Washietl et al., 2005a,b), study folding (Li et al., 2007), design siRNA sequences (Long et al., 2007; Lu and Mathews 2008; Tafer et al., 2008) and facilitate comparative sequence analysis (Mathews et al., 1997). The prediction of lowest free energy structures with a dynamic programming algorithm is a popular approach for making such predictions (Mathews and Turner, 2006). In this approach, a nearest neighbor energy model with a set of thermodynamic parameters derived from optical melting experiments on small RNA models (Mathews et al., 2004; Turner and Mathews, 2010; Xia et al., 1998) is used to evaluate possible structures and the dynamic programming algorithm guarantees that the most stable structure will be found. Only 73% or fewer known base pairs are predicted by free energy minimization for sequences shorter than 700 nt (Dowell and Eddy, 2004; Mathews et al., 2004; Mathews and Turner, 2006). The prediction is worse for longer sequences, with 20–60% average accuracies for small and large subunit rRNA (Dowell and Eddy, 2004; Mathews et al., 2004).
When multiple homologous sequences are available, evolutionary conservation can be used to improve RNA secondary structure prediction accuracy. RNA sequences often change during evolution, but RNA structures are generally conserved in order to preserve function. The set of structures that multiple different sequences can adopt is smaller than the number of structures each can adopt independently, so determining the conserved structure is more accurate. A number of computational approaches have been applied to this problem, as previously reviewed (Bernhart and Hofacker, 2009; Mathews, 2006). Some of the algorithms require a fixed initial alignment as input (Bernhart et al., 2008). The drawback to this is that the initial alignment, based on nucleotide matching, may not reflect the correct alignment because of compensating changes in the sequence. This imperfect alignment can seriously restrict prediction accuracy. Sankoff (1985) introduced a dynamic programming algorithm to find optimal consensus structures and sequence alignment simultaneously without being constrained to a fixed alignment. The computational complexity scales O(N3S) in time and O(N2S) in memory, where N is the average length of the sequences and S is the number of sequences. Dynalign is a variant of Sankoff's algorithm that uses the complete nearest neighbor thermodynamic model to find the lowest free energy structure common to two RNA sequences (Mathews and Turner, 2002). An X-Dynalign was reported to simultaneously fold and align three sequences (Masoumi and Turcotte, 2005). It improves prediction accuracy, but is extremely computationally demanding even for sequences as short as 5S rRNA. A profile alignment algorithm using Dynalign was also explored to find strict common base pairs in all the input sequences (Bellamy-Royds and Turcotte, 2007). Its performance on 5S rRNA and tRNA was reported to be comparable with other benchmarked methods, but depends highly on the quality of the guide tree that is used to guide the progressive alignment (Bellamy-Royds and Turcotte, 2007).
In this contribution, a new algorithm, called Multilign, is introduced to solve the high-computational complexity of predicting structures common to three or more sequences. Multilign is based on multiple Dynalign calculations. It departs from previous greedy approaches that build upon fixed decisions made early in the calculation. Instead, Multilign uses a single index sequence and performs Dynalign calculations with the index sequence and each other sequence in turn. Base pairs for the index sequence that are in low free energy structures are allowed in subsequent Dynalign calculations. This progressively refines the set of allowed pairs utilizing comparison with each sequence. Multilign works well for an arbitrary number of long sequences, e.g. > 1500 nt, with a memory requirement the same as Dynalign and a time requirement that scales linearly with sequence number. In accuracy, Multilign performs as well as the best available methods.
2 METHODS
2.1 Progressive templating
The progressive calculations used by Multilign are illustrated in Figure 1. A sequence is chosen as the index sequence. This sequence is then utilized for pairwise structure prediction with each other sequence in the set using Dynalign, one after another. In each calculation, the energy dot plot is calculated (Mathews, 2005). This plot determines, for each possible pair in each sequence, the lowest total free energy for a conserved structure and alignment that contains that pair. During the progressive calculations, subsequent calculations only allow the set of pairs for the index sequence that were found in low free energy structures in prior calculations. The allowed pairs are determined using a threshold described below.
Fig. 1.
Multilign algorithm flowchart, illustrating progressive calculations of S sequences. The index sequence is used with each other sequence as input to Dynalign. The cycle is repeated for multiple iterations, using the same index sequence for all iterations. A structure is predicted for each sequence with the final iteration. The computational complexity for this algorithm is O(N4) in memory and O(ISN6) in time for I iterations of calculation for average sequence length of N.
As the Multilign calculation proceeds, the energy dot plot for the index sequence becomes less crowded with possible pairs because few pairs are in low free energy structures with all previous Dynalign calculations. This relies on the hypothesis that the true base pairs are in low free energy structures predicted by all Dynalign calculations, but the competing false base pairs are not. Supplementary Figure S1 shows that the set of relatively low free energy structures contains the vast majority of true base pairs for a diverse set of RNA families. An example of the removal of false pairs by Multilign is shown in Figure 2. After calculations with all other sequences, the folding space of the index sequence is well constrained, but the folding spaces for the other sequences is not as well defined. Thus, subsequent iterations in the same progressive manner and with the same index sequence can be used to constrain the folding of the other sequences until a common structure is determined.
Fig. 2.
Energy dot plots of 5S rRNA sequence from Methanobacterium thermoautotrophicum A1 (Szymanski et al., 1999) predicted by Dynalign or Multilign. In each plot, a dot indicates a base pair between nucleotides with indices as indicated along the x and y axes. The lower triangle shows the base pairs in the known structure and the upper triangle shows all base pairs possible for secondary structures with folding free energy change within the percentage intervals above the minimum free energy structure as annotated by color. In (A) and (B), the base pairs were predicted by Dynalign with M.thermoautotrophicum B 5S rRNA and Methanococcus voltae [lnk] 5S rRNA as the second sequences, respectively. In (C), the base pairs were predicted by Multilign together with other nine 5S rRNA sequences from M.thermoautotrophicum A2, M.thermoautotrophicum B, M.thermoformicicum, Methanobrevibacter ruminantium, Methanothermus fervidus, M.thermolithotrophicus, M.vannielii [Lnk], M.vannielii [Xtr], M.voltae [Lnk]. For these calculations, the default settings of Multilign were used.
2.2 Determination of parameters
A threshold is used to select pairs that will be allowed for the index sequence in subsequent Dynalign calculations. The optimal threshold is one that allows just all true base pairs and few false positive base pairs in subsequent calculations. There is, however, no universal cutoff for all types of RNA as shown in Supplementary Figure S1, either in absolute free energy or in percentage of free energy change. Empirically, it was found that a threshold that allows a specified number of pairs (MaxPairs) in conjunction with a percentage cutoff (maxdsvchange) provided the best average performance. The cutoff maxdsvchange allows all pairs found in structures with folding free energy within a maximum percentage interval above the lowest free energy structure. The criterion for allowing pairs at each step is the one that allows the most pairs in subsequent calculations.
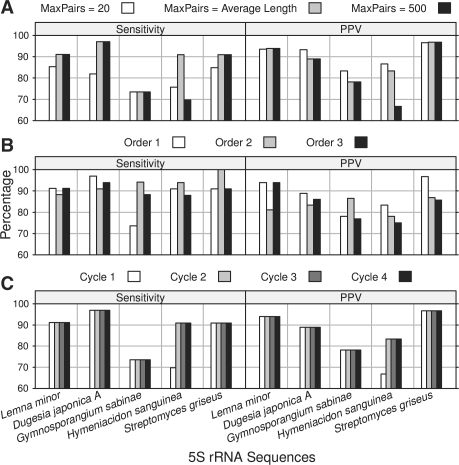
Other factors that may influence the Multilign prediction accuracy include the choice of the index sequence, the order of the pairwise Dynalign calculations, the number of iterations and the total number of sequences used. How these factors impact the prediction result were tested (Section 3 and Fig. 3).
Fig. 3.
The impact of parameters on the performance of 5S rRNA secondary structure prediction by Multilign. Results in (A) are predicted with MaxPairs setting the number of allowed base pairs to 20, the average length of the five sequences (= 120 in this test), or 500. (B) shows the results when structures are predicted for three different orders, chosen at random. This randomization also changes the sequence that is chosen to be the index sequence. (C) shows the prediction accuracy from 1 to 4 iterations.
The default settings are a maxdsvchange of 1%, a MaxPairs equal to the average length of all the input sequences, the index sequence as the first of the input sequences and the number of iterations set to two. The default settings were used for the benchmarks here. The index sequence was chosen at random.
2.3 Benchmark
Default options and parameters are used for all the programs except RNAshapes. For RNAshapes prediction on tRNA and 5s rRNA, the options, ‘-t 3 –c 50’ were used to have less abstract levels and wider energy ranges. For the other RNA families with longer sequences, default parameters (‘-t 5 –c 10’) were used to reduce memory requirement. All the calculations were done on cluster compute nodes each having two quad-core Intel Xeon 3.0 GHz processors with 16 GB of RAM. Multilign has both serial and parallel versions available, where the parallel version works on shared memory architectures. The parallel version was used in the benchmark. ClustalW2 (Larkin et al., 2007) was used to predict an alignment for RNAalifold (Bernhart et al., 2008), which requires a sequence alignment as input.
Secondary structures determined by comparative sequence analysis were used as reference structures for testing Multilign. These structures are accurate; it has been shown that > 97% of base pairs in ribosomal RNA secondary structures predicted by comparative sequence analysis exist in high-resolution crystal structures (Gutell et al., 2002). The dataset includes types of RNA used in previous benchmarks (Harmanci et al., 2008; Mathews et al., 1999, 2004), including tRNA (Sprinzl and Vassilenko, 2005), 5S rRNA (Szymanski et al., 1999), Signal Recognition Particle (SRP) RNA (Larsen et al., 1998), RNase P RNA (Brown, 1999) and small subunit rRNA (Gutell, 1993). For SRP RNA, sequences shorter than 200 nt were removed from the dataset since they do not accommodate a consensus structures with longer sequences.
From the sequence pool, 400 tRNA, 100 5S rRNA, 20 SRP RNA, 60 RNase P RNA and 40 small subunit rRNA were randomly selected without replacement. The sequences in each RNA type were divided into groups of 5, 10 or 20 sequences in a randomly ordered list. All methods except Dynalign and the single-sequence free energy minimization method (Fold in RNAstructure package; Reuter and Mathews, 2010) ran on these divided groups. Fold was run on each sequence. Dynalign also ran on the divided groups of sequences but in a different style because it only predicts structures of two sequences at a time. For each group, the first sequence (used as the index by Multilign) in the group is predicted along with each of the other sequences by Dynalign. Therefore, there is more than one predicted structure for the first sequence, and only the structure predicted by the last Dynalign calculation is included in scoring.
2.4 Scoring of prediction accuracy
Predicted structures are evaluated by comparison with known structures. Two scores, sensitivity and positive predictive value (PPV) are calculated. Sensitivity is the fraction of known pairs correctly predicted and PPV is the fraction of predicted pairs in the known structure (Mathews, 2004). When determining whether a predicted pair is consistent with a known base pair, up to one nucleotide rearrangement on one side of the base pair is allowed. Therefore, a predicted base pair (i, j) is correctly predicted if either (i, j), (i−1, j), (i+1, j), (i, j−1) or (i, j−1) appears in the reference structure (Mathews et al., 1999). This scoring scheme reflects the uncertainty of base pair matches in comparative sequence analysis and the conformational dynamics of RNA secondary structures.
3 RESULTS
3.1 Factors that may influence the accuracy of multilign
Multilign uses a series of Dynalign calculations on two sequences to predict the common secondary structure for multiple sequences (Fig. 1). The threshold for allowing pairs in subsequent calculations, the choice of index sequence, the order of the sequences and the number of calculation iterations may impact the Multilign structure prediction. Their influence on prediction accuracy was tested using 5S rRNA sequences.
First, the influence of the threshold was tested. Empirically, it was determined that using both a maxdsvchange of 1% and a MaxPairs equal to the average length of the input sequences provides the best performance. These two cutoffs should be set to allow all the true base pairs in subsequent calculations, and also exclude competing false base pairs. In many cases, false base pairs can occur in structures with comparable or lower free energy changes than true base pairs because of imperfections in the nearest neighbor parameters. As shown in Figure 3A, when MaxPairs is set too small, e.g. 20, the false base pairs of low free energy are removed at the cost of removing true base pairs, therefore PPV is improved at the cost of sensitivity. When MaxPairs is set too large, e.g. 500, the competing false base pairs were not efficiently excluded. False positives were incorporated into the predicted structures and this compromised the prediction accuracy. Setting the MaxPairs to the average length of the input sequences, 120 in this example, is a reasonable choice.
The influence of the choice of index sequence and the order of the other sequences were tested. The order of sequences was randomized and the first one was chosen as index sequence. Overall, it was found that this random ordering does not adversely affect the accuracy. Figure 3B shows an example of this with three different random orderings of 5S rRNA sequences. On average, the performance is the same with the three random orders, although in detail, the accuracy of structure prediction on a single sequence can vary.
The final choice that may influence the accuracy is the number of iterations of the process. For example, after the first iteration, the folding space of index sequence should be well constrained, but those of the other sequences may not be. Figure 3C shows that a second iteration is necessary to improve the prediction of all sequences while extra iterations beyond appear to make no difference in the accuracy.
3.2 Benchmark
The performance of Multilign in structure prediction was evaluated and compared with the nine other methods that predict conserved structures for three or more sequences: FoldalignM (Torarinsson et al., 2007), mLocARNA (Will et al., 2007), MASTR (Lindgreen et al., 2007), Murlet (Kiryu et al., 2007), RNA Alignment and Folding (RAF) (Do et al., 2008), RNASampler (Xu et al., 2007), RNAshapes (Steffen et al., 2006), RNAalifold (Bernhart et al., 2008) and StemLoc (Holmes, 2005). For comparison, the prediction results of single-sequence free energy minimization (Fold; Reuter and Mathews, 2010) and Dynalign on the same dataset are also shown.
The accuracy of structure prediction is illustrated in Figure 4. For each type of RNA, the methods were evaluated over a dataset that is divided into groups of 5, 10 or 20 sequences to show how the number of sequences influences the prediction accuracies.
Fig. 4.
The average structure prediction accuracies (PPV, left and Sensitivity, right) of 12 methods over tRNA, 5S rRNA, SRP RNA, RNase P and small subunit rRNA datasets. There are 400 sequences in tRNA, 100 in 5S rRNA, 20 in SRP RNA, 60 in RNase P and 40 in small subunit rRNA. The dataset for each RNA type is divided into groups of 5, 10 or 20 sequences and predictions were done on each group. The missing bars for some predictions indicate that the methods did not proceed on available hardware. The plotted error bars are 95% confidence intervals.
All multiple sequence methods predict structures of tRNA with high accuracy. Multilign significantly improves the prediction in terms of sensitivity and PPV by 3–4%, as compared with Dynalign. As expected, Fold, the single sequence method, has the lowest prediction accuracies both in sensitivity and PPV. Multilign, FoldalignM, mLocARNA, RAF, RNASampler and StemLoc predict both sensitivity and PPV around or above 90%. The sensitivity of RNAalifold is about the same as Fold, although the PPV is significantly higher.
For the predictions of 5S rRNA, all the multiple sequence methods again perform better than single sequence structure prediction. RNASampler stands out as having a particularly high PPV (> 90%), but at the cost of sensitivity. The accuracy of prediction by single-sequence free energy minimization is the worst, especially for PPV, which has an average of only 62.4%.
The methods were also evaluated on longer sequences, namely the SRP RNA, RNase P RNA and small subunit rRNA. Not all the methods would run for these sequences on the available hardware because of their computational complexity. Surprisingly, for SRP RNA, Dynalign outperformed all the other methods in sensitivity, including Multilign. This is because some of the true base pairs are incorrectly ruled out early in the Multilign process (Supplementary Fig. S2). For RNase P, single-sequence free energy minimization outperforms all the other methods in sensitivity, but not PPV. One possible explanation is that structures of RNase P RNA vary more than those of other RNA types. It is therefore difficult to determine a single consensus structure, causing the algorithms tested here to perform poorly. For small subunit rRNA, only Multilign, Dynalign, RNAalifold, mLocARNA and RAF run successfully on the entire benchmark set. Multilign and mLocARNA have comparable sensitivities and PPVs. The sensitivity of small subunit rRNA prediction by Multilign is much higher than those of RAF (> 10%) and RNAalifold (> 5%), although RAF outperforms Multilign by ∼ 3% in PPV.
It is also notable that the methods demonstrate different patterns of prediction accuracy as a function of number of input sequences. Prediction accuracies of some methods changes remarkably in 5-, 10- and 20-sequence calculations, while scores of others are stable. Multilign, FoldalignM and MASTR tend to have comparable or higher accuracy when using more sequences up to 20, as shown inFigure 4. The prediction accuracies of the other methods show either an opposite pattern or a stochastic pattern in number of sequences utilized.
The CPU time is reported for all the calculations in Table 1. The CPU time requirement for Multilign is roughly equal to the product of Dynalign time requirement, the number of iterations and the number of sequences in the calculation. Table 1 shows that among all the algorithms that work with three or more unaligned sequences, i.e. not including Fold, Dynalign or RNAalifold, the time requirement for Multilign and RNAshapes scale the best in terms of the number of sequences. Although the absolute CPU time of some Multilign calculations is large, Multilign is the only parallelized software and a prediction of five small subunit rRNA sequences can be done in < 1 day with Multilign running in parallel on eight cores.
Table 1.
The average CPU time of 12 methods over the 5S rRNA and small subunit rRNA reported by the Linux time command
| RNA Type | 5S rRNA |
Small subunit rRNA |
||||
|---|---|---|---|---|---|---|
| Sequence Number | 5 | 10 | 20 | 5 | 10 | 20 |
| Multilign | 3 m:39.7 s | 9 m:18.5 s | 27 m:5.8 s | 135 h:26 m:16.9 s | 299 h:30 m:47.3 s | 774 h:9 m14.2 s |
| Fold | 0.16 s | 5 m:17.7 s | ||||
| Dynalign | 34.6 s | 20 h:28 m:9.6 s | ||||
| RNAalifold | 0.05 s | 0.06 s | 0.11 s | 11.92 s | 12.83 s | 26.39 s |
| FoldalignM | 1 m:36.8 s | 6 m:38.2 s | 28 m:11.3 s | N/A | N/A | N/A |
| mLocARNA | 3.63 s | 13.3 s | 49.4 s | 21 h:38 m:22.1 s | 120 h:1 m:4.5 s | 462 h:15 m:7.9 s |
| MASTR | 22.2 s | 1 m:7.64 s | 4 m:9.5 s | N/A | N/A | N/A |
| Murlet | 6.37 s | 23.0 s | 1 m:27.1 s | N/A | N/A | N/A |
| RAF | 0.58 s | 1.86 s | 7.75 s | 4 m:6.6 s | 11 m:40.2 s | 37 m:40.3 s |
| RNASampler | 12.9 s | 1 m:39.1 s | 7 m:13.1 s | N/A | N/A | N/A |
| RNAshapes | 50.3 s | 1 m:42.7 s | 3 m:32.2 s | N/A | N/A | N/A |
| StemLoc | 20 m:41.2 s | 1 h:23 m:6.9 s | 6 h:7 m:19.5 s | N/A | N/A | N/A |
For Fold and Dynalign, the average of each calculation is reported here. Times are reported in hours (h), minutes (m) and seconds (s). N/A, ‘Not Applicable,’ is reported for programs that did not complete the calculation for small subunit rRNA on the available hardware.
4 DISCUSSION
Multilign combines free energy minimization and comparative sequence analysis to predict RNA secondary structures of multiple sequences with higher or comparable accuracy to Dynalign. It has among the best accuracy as compared with other algorithms that predict conserved structures for multiple sequences.
4.1 Strengths and limitations of Multilign
Multilign inherits the strengths of Dynalign. It uses a dynamic programming algorithm to guarantee that the lowest free energy conserved structure will be found for two sequences. The total free energy score optimized by Dynalign does not depend on sequence identity, which allows the algorithm to predict consensus secondary structures for homologous sequences with little sequence identity, e.g. as low as 20% in previous benchmarks (Harmanci et al., 2007). Suboptimal structures can be predicted to provide alternative solutions and show the well-definedness of prediction by creating dot plots.
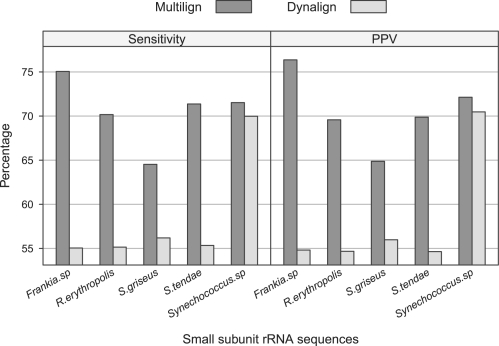
Multilign extends Dynalign to multiple sequence prediction and improves the prediction accuracy as shown in Figure 4 for all types of RNA tested except for SRP RNA. Although the average improvement of Multilign over Dynalign does not appear large, this is because the improvement on sequences is averaged. The improvement of prediction accuracy for a single sequence can be large. Dynalign predicts structures well for a majority of the sequences, but poorly for a subset of sequences. Multilign, however, tends to improve the average accuracy of sequences for which Dynalign does well, but additionally it also dramatically improves the accuracy for structures poorly predicted by Dynalign (Fig. 5 and Supplementary Table S2). In other words, Multilign does well by ensuring that the structure prediction accuracy will be uniform and generally good for all sequences in a set.
Fig. 5.
The comparison of Multilign and Dynalign prediction on the level of single sequences. The plot shows Multilign and Dynalign prediction of five small subunit rRNA sequences from species listed along x axis.
Additionally, Multilign is able predict structures of an arbitrary number of long sequences. To our knowledge, this work is the first report of the prediction of multiple unaligned sequences as long as small subunit rRNA, which have a mean length of 1526 nt in this test set. Most algorithms failed to predict structures for long sequences or large number of homologous sequences on the hardware used for this study. Multilign has computation complexity linear to the number of sequences in time. In memory, the requirement does not increase with increasing number of sequences.
Overall, Multilign improves prediction accuracy as compared with Dynalign. This is not, however, guaranteed in all cases, such as SRP RNA. Dynalign is accelerated by two steps of prefiltering that restrict the space of solutions that needs to be considered. One prefilter predicts structures for each sequence by free energy minimization and only base pairs that are in secondary structures within 30% of the lowest free energy are then allowed in Dynalign (Uzilov et al., 2006). The other constrains the allowed alignment space with a probabilistic model predicted using a Hidden Markov Model (Harmanci et al., 2007). It is known that these two steps exclude few genuine base pairs from consideration. Therefore, they influence a Dynalign structure prediction little, but they turn out to be a hidden problem for Multilign. If a genuine base pair is prohibited by a particular pairwise Dynalign calculation, it is permanently prohibited in all the following calculations (Supplementary Fig. S2). This effect is cumulative and can be a problem for sequence families that have diverse structures.
4.2 Prospectus
Great efforts have been placed on improving RNA secondary structure prediction. One way is to mimic comparative sequence analysis by predicting a structure for multiple sequences simultaneously. Some RNA types demonstrate great structural diversity, however, and are hard for current existing algorithms to predict their structures accurately. As reported in this work, structure prediction of SRP RNA and RNase P RNA remains a difficult problem because of structural heterogeneity.
Another point worth noting is that not all methods predicting conserved structures for multiple sequences are guaranteed to perform better on average than single sequence folding. Furthermore, many of the available algorithms do not necessarily improve in prediction accuracy when more homologous sequences are utilized by the prediction. Clearly, no method yet replaces human expertise for comparative sequence analysis. The predictions from these programs need to be considered as hypotheses. Users may, for example, want to use multiple programs to develop hypotheses for subsequent study.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank J.M. Keegan for work on an early prototype of Multilign. Computer time was provided by the University of Rochester Center for Research Computing.
Funding: National Institutes of Health (R01HG004002 to D.H.M.).
Conflict of Interest: none declared.
REFERENCES
- Aguirre-Hernandez R, et al. Computational RNA secondary structure design: empirical complexity and improved methods. BMC Bioinformatics. 2007;8:34–34. doi: 10.1186/1471-2105-8-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey RT. Structures of regulatory elements in mRNAs. Curr. Opin. Struct. Biol. 2006;16:299–306. doi: 10.1016/j.sbi.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Bellamy-Royds AB, Turcotte M. Can Clustal-style progressive pairwise alignment of multiple sequences be used in RNA secondary structure prediction? BMC Bioinformatics. 2007;8:190–190. doi: 10.1186/1471-2105-8-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhart SH, et al. RNAalifold: improved consensus structure prediction for RNA alignments. BMC Bioinformatics. 2008;9:474–484. doi: 10.1186/1471-2105-9-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhart SH, Hofacker IL. From consensus structure prediction to RNA gene finding. Brief. Funct. Genomics Proteomics. 2009;8:461–461. doi: 10.1093/bfgp/elp043. [DOI] [PubMed] [Google Scholar]
- Brown JW. The ribonuclease P database. Nucleic Acids Res. 1999;27:314–316. doi: 10.1093/nar/27.1.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond JM, et al. Thermodynamics of three-way multibranch loops in RNA. Biochemistry. 2001;40:6971–6981. doi: 10.1021/bi0029548. [DOI] [PubMed] [Google Scholar]
- Dirks RM, et al. Paradigms for computational nucleic acid design. Nucleic Acids Res. 2004;32:1392–1403. doi: 10.1093/nar/gkh291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do CB, et al. A max-margin model for efficient simultaneous alignment and folding of RNA sequences. Bioinformatics. 2008;24:i68–i76. doi: 10.1093/bioinformatics/btn177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowell RD, Eddy SR. Evaluation of several lightweight stochastic context-free grammars for RNA secondary structure prediction. BMC Bioinformatics. 2004;5:71–79. doi: 10.1186/1471-2105-5-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedor MJ, Williamson JR. The catalytic diversity of RNAs. Nat. Rev. Mol. Cell Biol. 2005;6:399–412. doi: 10.1038/nrm1647. [DOI] [PubMed] [Google Scholar]
- Gutell RR. Collection of small subunit (16S- and 16S-like) ribosomal RNA structures. Nucleic Acids Res. 1993;21:3051–3054. doi: 10.1093/nar/21.13.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell RR, et al. The accuracy of ribosomal RNA comparative structure models. Curr. Opin. Struct. Biol. 2002;12:301–310. doi: 10.1016/s0959-440x(02)00339-1. [DOI] [PubMed] [Google Scholar]
- Harmanci A, et al. Efficient pairwise RNA structure prediction using probabilistic alignment constraints in Dynalign. BMC Bioinformatics. 2007;8:130–130. doi: 10.1186/1471-2105-8-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmanci AO, et al. PARTS: Probabilistic Alignment for RNA joinT Secondary structure prediction. Nucleic Acids Res. 2008;36:2406–2416. doi: 10.1093/nar/gkn043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes I. Accelerated probabilistic inference of RNA structure evolution. BMC Bioinformatics. 2005;6:73–77. doi: 10.1186/1471-2105-6-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiryu H, et al. Murlet: a practical multiple alignment tool for structural RNA sequences. Bioinformatics. 2007;23:1588–1598. doi: 10.1093/bioinformatics/btm146. [DOI] [PubMed] [Google Scholar]
- Kiss-Laszlo Z. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Larsen N, et al. The signal recognition particle Database (SRPDB) Nucleic Acids Res. 1998;26:177–178. doi: 10.1093/nar/26.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Li PTX, et al. Real-time control of the energy landscape by force directs the folding of RNA molecules. Proc. Natl Acad. Sci. USA. 2007;104:7039–7044. doi: 10.1073/pnas.0702137104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgreen S, et al. MASTR: multiple alignment and structure prediction of non-coding RNAs using simulated annealing. Bioinformatics. 2007;23:3304–3311. doi: 10.1093/bioinformatics/btm525. [DOI] [PubMed] [Google Scholar]
- Long D, et al. Potent effect of target structure on microRNA function. Nat. Struct. Mol. Biol. 2007;14:287–294. doi: 10.1038/nsmb1226. [DOI] [PubMed] [Google Scholar]
- Lu ZJ, Mathews DH. Efficient siRNA selection using hybridization thermodynamics. Nucleic Acids Res. 2008;36:640–647. doi: 10.1093/nar/gkm920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoumi B, Turcotte M. Simultaneous alignment and structure prediction of three RNA sequences. Int. J. Bioinform. Res. Appl. 2005;1:230–245. doi: 10.1504/IJBRA.2005.007581. [DOI] [PubMed] [Google Scholar]
- Mathews DH. Using an RNA secondary structure partition function to determine confidence in base pairs predicted by free energy minimization. RNA. 2004;10:1178–1190. doi: 10.1261/rna.7650904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH. Predicting a set of minimal free energy RNA secondary structures common to two sequences. Bioinformatics. 2005;21:2246–2253. doi: 10.1093/bioinformatics/bti349. [DOI] [PubMed] [Google Scholar]
- Mathews DH. Predicting RNA secondary structure by free energy minimization. Theor. Chem. Acc. 2006;116:160–168. [Google Scholar]
- Mathews DH, Turner DH. Dynalign: an algorithm for finding the secondary structure common to two RNA sequences. J. Mol. Biol. 2002;317:191–203. doi: 10.1006/jmbi.2001.5351. [DOI] [PubMed] [Google Scholar]
- Mathews DH, Turner DH. Prediction of RNA secondary structure by free energy minimization. Curr. Opin. Struct. Biol. 2006;16:270–278. doi: 10.1016/j.sbi.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Mathews DH, et al. Secondary structure model of the RNA recognized by the reverse transcriptase from the R2 retrotransposable element. RNA. 1997;3:1–16. [PMC free article] [PubMed] [Google Scholar]
- Mathews DH, et al. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- Mathews DH, et al. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl Acad. Sci. USA. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen P, et al. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- Ravasi T, et al. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 2006;16:11–19. doi: 10.1101/gr.4200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter JS, Mathews DH. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010;11:129–129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankoff D. Simultaneous solution of the RNA folding, alignment and protosequence problems. SIAM J.Appl. Math. 1985;45:810–825. [Google Scholar]
- Sharma CM, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- Sprinzl M, Vassilenko KS. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 2005;33:D139–140. doi: 10.1093/nar/gki012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen P, et al. RNAshapes: an integrated RNA analysis package based on abstract shapes. Bioinformatics. 2006;22:500–503. doi: 10.1093/bioinformatics/btk010. [DOI] [PubMed] [Google Scholar]
- Szymanski M, et al. 5S ribosomal RNA data bank. Nucleic Acids Res. 1999;27:158–160. doi: 10.1093/nar/27.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafer H, et al. The impact of target site accessibility on the design of effective siRNAs. Nat. Biotechnol. 2008;26:578–583. doi: 10.1038/nbt1404. [DOI] [PubMed] [Google Scholar]
- The Encode Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Fantom Consortium the Riken Genome Exploration Research Group Phase I & II Team. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- Torarinsson E, et al. Thousands of corresponding human and mouse genomic regions unalignable in primary sequence contain common RNA structure. Genome Res. 2006;16:885–889. doi: 10.1101/gr.5226606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torarinsson E, et al. Multiple structural alignment and clustering of RNA sequences. Bioinformatics. 2007;23:926–932. doi: 10.1093/bioinformatics/btm049. [DOI] [PubMed] [Google Scholar]
- Turner DH, Mathews DH. NNDB: the nearest neighbor parameter database for predicting stability of nucleic acid secondary structure. Nucleic Acids Res. 2010;38:D280–282. doi: 10.1093/nar/gkp892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzilov AV, et al. Detection of non-coding RNAs on the basis of predicted secondary structure formation free energy change. BMC Bioinformatics. 2006;7:173–183. doi: 10.1186/1471-2105-7-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendeix FAP, et al. Anticodon domain modifications contribute order to tRNA for ribosome-mediated codon binding. Biochemistry. 2008;47:6117–6129. doi: 10.1021/bi702356j. [DOI] [PubMed] [Google Scholar]
- Washietl S, et al. Mapping of conserved RNA secondary structures predicts thousands of functional noncoding RNAs in the human genome. Nat. Biotechnol. 2005a;23:1383–1390. doi: 10.1038/nbt1144. [DOI] [PubMed] [Google Scholar]
- Washietl S, et al. Fast and reliable prediction of noncoding RNAs. Proc. Natl Acad. Sci. USA. 2005b;102:2454–2459. doi: 10.1073/pnas.0409169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will S, et al. Inferring noncoding RNA families and classes by means of genome-scale structure-based clustering. PLoS Comput. Biol. 2007;3:e65–e65. doi: 10.1371/journal.pcbi.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia T, et al. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- Xu X, et al. RNA Sampler: a new sampling based algorithm for common RNA secondary structure prediction and structural alignment. Bioinformatics. 2007;23:1883–1891. doi: 10.1093/bioinformatics/btm272. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.