Summary
Apoptosis occurs in the nervous system during normal development, but can also be induced by disease or after exogenous insults such as DNA damage. Depending on the magnitude and timing of the stimulus, apoptosis can be sporadic or widespread. Because of the highly ordered structure of the nervous system, immunohistochemical detection approaches provide a wealth of information about the spatiotemporal nature of apoptosis in this tissue. Therefore, immunohistochemistry offers valuable insights into the neuropathology of disease processes and the identification of specific cell populations that are susceptible to apoptosis. In this chapter we outline standard approaches for the immunohistochemical analysis of apoptosis in the nervous system, with an emphasis on methodology useful for studies involving DNA damage-induced apoptosis.
Keywords: Immunohistochemistry, DNA damage, p53, TUNEL, caspase-3, γH2AX, 53BP1
1. Introduction
Studies of apoptosis in the nervous system are particularly informative during neural development because of the susceptibility of proliferative and differentiating neural cells to apoptosis. The development of the nervous system occurs in a reiterated manner involving widespread areas of proliferation adjacent to regions of differentiation and migration (1–4). Neuroanatomically, this laminar arrangement is tremendously informative when immunohistochemistry is combined with mammalian model systems such as the mouse to analyze apoptosis. These approaches have proven valuable in deciphering tissue and cell-type specificity in diseases and after various insults. For example, studies of genotoxicity have utilized apoptotic analyses as a major means of determining the consequences of this type of stress in the nervous system (5–7). Organisms have developed efficient DNA repair mechanisms to maintain the integrity of genomic information. However, while repair of DNA lesions is an option, the tremendous proliferative capacity of germinal zones in the nervous system makes apoptosis and subsequent cellular replacement a more common strategy to deal with genomically compromised cells (8–10).
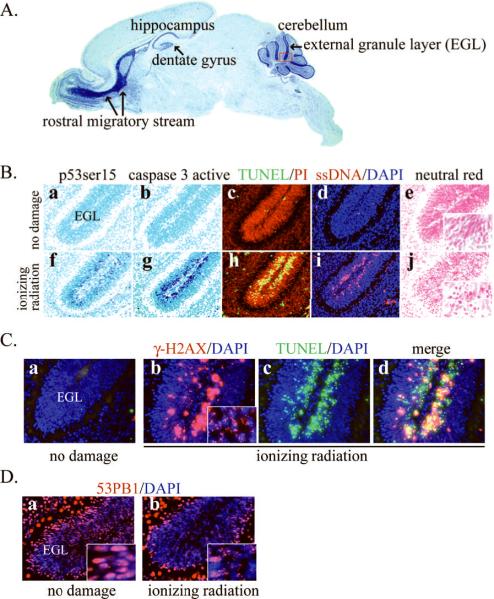
Similar to the embryo, postnatal brain development also contains proliferating regions such as the dentate gyrus of the hippocampus or the cerebellar external granule layer (EGL) which are also vulnerable to DNA damage-induced apoptosis (8) (Figure 1A). While immature neurons are quite susceptible to DNA induced apoptosis, fully mature neurons are relatively resistant to this particular insult, and they do not typically undergo apoptosis (8). The signaling pathway triggered by DNA damage in the nervous system generally results in p53 stabilization and activation to initiate transcription-dependent apoptosis (5, 8–10). The response of p53 to DNA damage can be readily detected using p53 and phospho-p53 (serine 15) immunohistochemistry (Figure 1B a, f). A major outcome of p53 signaling is the induction of PUMA (p53 upregulated mediator of apoptosis) a pro-apoptotic Bcl2 family member, which is critical for DNA damage-mediated apoptosis in the nervous system (11). Caspases becomes activated to effect apoptosis, and in the case of caspase-3, visualization by immunohistochemistry is relatively straightforward (5) (Figure 1B b, g). Finally, the end-stage of programmed cell death can be detected by enzymatic labeling such as TUNEL, single strand DNA (ssDNA) immunoreactivity or simple Neutral Red staining to identify pyknotic cells (Figure 1B, c–e, h–j). With regard to DNA damage, immunohistochemistry is also valuable for detecting DNA double strand breaks (DSBs), and available reagents such as antibodies against phosphorylated H2AX (γH2AX) and 53BP1which form punctate nuclear focal staining in response to DSBs, are particularly useful in this regard (8, 12, 13) (Figure 1C–D; these foci generally persist until DNA damage is resolved. TUNEL and γH2AX foci can often overlap in proliferating areas of the developing brain (figure 1C).
Figure 1. Immunohistochemical analysis of apoptosis after DNA damage in the postnatal mouse brain.
(A) Susceptible areas to DNA damage-induced apoptosis during postnatal brain development include the rostral migratory system, the hippocampus including the dentate gyrus, and the cerebellum particularly the external granular layer (EGL). A sagittal section of a postnatal day 7 mouse brain with Nissl staining is shown. The red box indicates an example of the cerebellar EGL shown in B–D.
(B) Immunohistochemical detection of apoptosis process in the EGL of postnatal day 5 brain induced by ionizing radiation. The lower panel shows activation of p53 (f) and caspase 3 (g), TUNEL positive signal (h), single strand DNA immunopositive signal (i), and pyknosis visualized by Neutral Red staining (j) induced by DNA damage. Inset panels in e and j are magnified views to illustrate the morphology of pyknotic cells. Apoptotic cells are not evident without any insult to the developing brain (a–e). p53 and caspase 3 immunoreactivity were visualized with the VIP substrate kit and the brains were counterstained with Methyl Green (a–b, f–g). TUNEL was done using ApopTag Fluorescein in situ Apoptosis Detection Kit with DAPI counterstaining (c, h). ssDNA antibody was detected with Cy3 conjugated secondary antibody with PI counterstaining (d, i).
(C) Phosphorylated H2AX (γH2AX) is a measure of DNA double strand breaks, and are visualized as nuclear punctate staining (inset panel in b), called foci. In the absence of DNA damage no γH2AX signal is found (a). γ-H2AX staining is detected after DNA damage using Cy3 secondary antibodies (b) and apoptosis is detected by TUNEL using fluorescein (c); panel d is a merge of panels b and c.
(D) Foci formation of 53BP1 after IR induced DNA breaks. Similar to γH2AX, 53BP1 is another early responder to DNA double strand breaks. While 53BP1 is distributed evenly in the nucleus without any DNA damage, 53BP1 foci form in the nucleus after ionizing radiation (b) and were visualized using Cy3-coupled secondary antibodies.
In the following, we describe methods suitable for the immunohistochemical analysis of apoptosis, with some additional details regarding analysis of DNA damage-induced apoptosis.
2. Materials
2.1 Neutral Red staining
1% Neutral Red (Cat# 861251, Sigma-Aldrich) in 0.1 M acetic acid (pH 4.8).
Shake or stir Neutral Red solution overnight to completely dissolve Neutral Red power, and filter the solution. Store Neutral Red solution in an amber bottle at room temperature. It is stable for several months. Maintain an acidic pH since solution color changes from red to yellow between pH 6.8 to 8.0. Care: Neutral red powder is possibly mutagenic.
2.2 Methyl Green staining
0.5% Methyl Green (Cat# 323829, Sigma-Aldrich) in 0.1 M sodium acetate solution (pH 4.2 adjusted with glacial acetic acid).
Dissolve Methyl Green overnight and then filter. It is stable for several months in an amber bottle at room temperature.
2.3 Paraformaldehyde
4% paraformaldehyde (Cat#P6148, Sigma-Aldrich) in PBS (pH 7.4).
After adding paraformaldehyde power, heat the solution to 65 ~ 70°C while stirring the solution continuously. Once paraformaldehyde has dissolved, add several drops of 5N NaOH until the solution becomes clear. Cool the solution and filter. Before using the paraformaldehyde solution, adjust pH back to pH 7.4. This entire process should be done in a fume hood. Buffered paraformaldehyde solution is stable for about a month at 4 °C. Alternatively make aliquots and freeze them down for long-term storage. Paraformaldehyde is toxic and needs appropriate disposal by neutralization with a formalin neutralizer such as Vytac (Cat#5401, Richard-Allan Scientific).
2.4 Antigen-Retrieval solution
Solution A – 0.1 M citric acid.
Solution B – 0.1 M sodium citrate.
Working Solution – 9 ml of Solution A / 41 ml of Solution B in 450 ml of distilled water.
Make the working solutions just before use and adjust pH to 6.0. Autoclaving Solutions A and B helps to extend shelf life.
2.5 Primary and secondary antibodies for apoptosis studies
2.5.1 Primary antibodies
p53 (CM5); rabbit polyclonal antibody, 1:1000, Antigen-retrieval technique applied, Cat# VP-P956, Vector Laboratories.
Phospho-p53 (serine 15); rabbit polyclonal antibody, 1:250 (adult brain), 1:150 (embryo), Antigen-retrieval technique applied, Cat# 9284, Cell Signaling Technology.
Active caspase 3; rabbit monoclonal antibody, 1:500, Antigen-retrieval technique applied, Cat# 559565, BD PharMingen.
ssDNA; rabbit polyclonal antibody, 1:300, Cat# 18731, IBL Co
γH2AX (serine 139); rabbit polyclonal antibody, 1:500, Antigen-retrieval technique applied, Cat# ab11174, Abcam.
53BP1; rabbit polyclonal antibody, 1:500, Antigen-retrieval technique applied, Cat# A300-272A, Bethyl.
2.5.2 Secondary antibodies
Biotinylated Goat anti-Rabbit IgG (H+L), 1:500, Jackson ImmunoResearch.
Cy3 (cyanine) conjugated Goat (or Donkey) anti-Rabbit IgG (H+L), 1:400, Jackson ImmunoResearch, alternatively Alexa Fluor 555 Goat (or Donkey) anti-Rabbit IgG (H+L), 1:1000, Invitrogen.
FITC (fluorescein) conjugated Goat (or Donkey) anti-Rabbit IgG (H+L), 1:200, Jackson ImmunoResearch, alternatively Alexa Fluor 488 Goat (or Donkey) anti-Rabbit IgG (H+L), 1:1000, Invitrogen.
2.5.3 Blocking solution
5% normal serum (Goat serum, Vector Laboratories or Donkey serum, Jackson ImmunoResearch) / 1% bovine serum albumin (crystallized, Cat# A4378, Sigma) in PBST (0.4% Triton X-100, Sigma in PBS), and add thimerosal (Cat# T-5125, Sigma) to make 0.01% solution for longer storage.
2.6 Available kits
Biotin-Avidin System reagent; Vectastain Elite ABC kit, Vector Laboratories.
Substrate kits for colorimetric detection; Vector VIP (for violet color) or DAB (brown or dark blue color) substrate kit, Vector Laboratories. 4 mg of diaminobenzidine (DAB, Sigma) in 10 ml of 0.05 M Tris buffer (pH 7.2) and add 16.7 μl of 3% H2O2 before use for brown color development. Alternatively, 2 mg of DAB/250 mg Nickel Sulfate (Sigma) in 10 ml of 0.175 M sodium acetate (pH 6.5 adjusted with glacial acetic acid) and add 8.3 μl of 3% H2O2 before use for dark blue color development.
Mounting medium for fluorescence with DAPI (blue nucleus staining) or Propidium iodide (generates a red nuclear staining), Vector Laboratories.
Terminal uridine deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) kit; ApopTag in situ Apoptosis Detection Kit, Millipore.
3. Methods
The methods described below correspond to the apoptotic analyses shown in Figure 1. As an example of the general approaches using these methods, DNA damage-induced apoptosis in the postnatal immature cerebellum is shown. However, the methods below are applicable to apoptotic analysis in all regions of the brain. It should be noted that the effects of DNA damage from ionizing radiation are widespread and can simultaneously affect large areas of immature neural cells. Depending on the type of insult or lesion being studied, the relative abundance of apoptotic cells will vary. Moreover, in some cases apoptosis may be sporadic, and so multiple approaches using different assays will ensure greater confidence in the evaluation of cell death. Due to the cellular heterogeneity and three-dimensional structure of the brain, it is also very important to compare carefully matched and defined neuroanatomical areas between control and experimental groups when performing apoptotic analyses via immunohistochemistry.
3.1 Neutral Red staining (or Methyl Green) staining to detect apoptotic cells
Apoptotic cells will appear as fragmented and condensed individual cells or in groups alongside unaffected cells with normal morphology upon histological evaluation (e.g. Figure 1B e,j).
Wash slides containing tissue sections with PBS for 10 minutes (2 times).
Before staining, briefly rinse slides with tap water.
Staining slides with neutral red solution for 1 to 5 minutes (Neutral Red) (note 4.1) or 5 to 10 minutes (Methyl Green) (note 4.2).
Wash off extra staining solution in running tap water.
-
Dehydration step:
50% alcohol ~1 minute 70% alcohol ~1 minute (note 4.3) 95% alcohol ~1 minute 100% alcohol ~1 minute, 2 times Xylene ~5 minute, 3 times Mount slides with non-aqueous mounting medium such as DPX for histology (Cat# 44581, Fluka).
3.2 Antigen Retrieval Treatment (note 4.4)
Antigen retrieval is a method that is often used to reveal the epitopes of an antigen that facilitates antibody binding and therefore a clear immunohistochemical signal (14).
Incubate slides with Antigen-Retrieval working solution (see 2.4) in Coplin jar made of polypropylene with screw cap for 10 minutes.
Tightly close screw cap and place 4 Coplin jars symmetrically on the microwave tray, and microwave for 10 minutes (total 420 kJoules, 70% power of 1000 watt microwave) (note 4.5). Check the level of working solution to cover all the slides and add more solution as necessary. Handle with caution since the solution will be very hot.
After microwaving, remove screw tops, and cool slides down in Antigen Retrieval working solution slowly to room temperature. It usually takes up to 1 hour or longer.
3.3 Immunohistochemistry – Colorimetric detection
Brain tissue fixation is done by transcardial perfusion with 4% buffered paraformaldehyde (see 2.3) followed by routine cryopreparation and cryosectioning procedures.
Perform Antigen Retrieval Treatment (see 3.2), if it is necessary (note 4.6)
Wash slides with PBS for 10 minutes (2 times).
Incubate slides with H2O2 solution (2 ml of 30% H2O2 in 100 ml of Methanol) for 30 minutes to quench endogenous peroxidase.
Rinse slides in PBS for 5 minutes (2 times).
Place slides in humid containers.
Gently spread blocking solution (see 2.5.3) (note 4.7) to cover entire sections and incubate for 1 to 2 hours on an orbital shaker at room temperature.
Decant blocking solution on absorbents.
Apply primary antibody solution (see 2.5.1) diluted in blocking solution (see 2.5.3) to slides in humid containers and incubate overnight on an orbital shaker at room temperature.
Rinse slides in PBS for 10 minutes (2 times).
Incubate slides with biotinylated IgG (see 2.5.2) diluted in blocking solution (see 2.5.3) for 1 to 2 hours on an orbital shaker at room temperature.
Rinse slides in PBS for 10 minutes (2 times).
Add the ABC complex solution diluted in PBS (note 4.8) on the slides and incubate for 1 to 2 hours on an orbital shaker at room temperature.
Rinse in PBS for 10 minutes (2 times).
Visualize positive signals with substrate kits (see 2.6); monitor the development of color under the microscope (note 4.9).
Stop enzyme reaction by rinsing slides with running tap water.
Counter staining with Methyl Green (see 3.1).
Dehydration and mounting (see 3.1).
3.4 Immunohistochemistry – Fluorescence detection
Perform Antigen Retrieval Treatment (see 3.2) if it is necessary.
Wash slides with PBS for 10 minutes (1 time).
Rinse slides in PBST for 15 minutes (2 times).
Place slides in humid containers.
Gently spread blocking solution (see 2.5.3) (note 4.7) to cover entire sections and incubate for 1 to 2 hours on an orbital shaker at room temperature.
Decant blocking solution on absorbents.
Apply primary antibody solution (see 2.5.1) diluted in blocking solution (see 2.5.3) to slides in humid containers and incubate overnight on an orbital shaker at room temperature.
Rinse slides in PBST for 15 minutes (2 times).
Add fluorescent secondary antibody (see 2.5.2) diluted in blocking solution (see 2.5.3) on the slides and incubate for 1 to 2 hours in the dark.
Rinse slides in PBST for 15 minutes (2 times) in the dark.
Wash slides with PBS briefly.
Mount slides with mounting medium for fluorescence (see 2.6) (note 4.10).
3.4. TUNEL or double staining with TUNEL
Perform Antigen Retrieval Treatment (see 3.2) for double staining. This is not required for TUNEL method itself (note 4.11).
Wash slides with PBS for 10 minutes (2 times).
Post-fix sections in pre-cooled fixative (2 ethanol : 1 acetic acid) for 5 minutes at −20 °C
Rinse slides in PBS for 5 minutes (2 times).
Apply equilibration buffer to slides provided in the kit in a humid chamber for at least 10 seconds at room temperature.
Decant equilibration buffer.
Apply working strength terminal deoxynucleotidyl transferase (TdT) on the slides and incubate in a humid chamber for 1 hour at 37 °C (note 4.12).
Stop enzyme reaction in working strength stop/wash buffer provided in the kit for 10 minutes.
Rinse slides in PBS 10 minutes (2 times).
For double staining with TUNEL, perform 3.4 steps 3 to 8.
Add working strength anti-digoxigenin conjugate (fluorophore tagged) diluted in the blocking solution provided in the kit, for double staining add fluorescent secondary antibody (see 2.5.2) in the same blocking solution together (note 4.13).
Incubate for 1 hour at room temperature in the dark (note 4.12).
Wash slides in PBS 15 minutes (2 times) in the dark.
Mount slides with mounting medium for fluorescence (see 2.6) (note 4.10).
Acknowledgement
The authors thank McKinnon lab members for their input and advice. Support of this work was from the NIH and the ALSAC charities of SJCRH.
Footnotes
One of the hallmarks of apoptosis is condensation of chromatin, called pyknosis, which can be visualized by Neutral Red staining (figure 1B e and j). Round dense staining of nucleus or fragmented nuclear staining is the sign of cell death. However, some brain areas contain granule cells that have small round nucleus, so careful reading of Neutral Red staining requires not to mislabel this type of cells as pyknotic cells.
Proliferating cells and differentiating neurons tend to stain much stronger than fully mature neurons do in Methyl Green nuclear staining.
50~70% alcohol remove partially Neutral Red or Methyl Green staining.
Antigen Retrieval was originally developed for formalin-fixed, paraffin-embedded tissues (14, 15); this method can also be applied to paraformaldehyde-fixed frozen tissues for better immunohistochemistry signals.
Using a pressure cooker or a steamer is an alternative method for heating.
Even though all exemplified primary antibodies in this protocol except ssDNA antibody (see 2.5.1) require Antigen Retrieval treatment for better/specific signals, this treatment is not always applicable to all primary antibodies. It is necessary to test the applicability of Antigen Retrieval treatment to new antibodies.
The choice of species for normal serum (see 2.5.2) is dependent on the species in which secondary antibody is raised. For example, use goat normal serum for the blocking solution, when biotinylated or fluorophore conjugated secondary antibody is raised in goat.
The ABC complex in PBS should be made at least 30 minutes prior to use.
It is important to cover the entire sections with substrate solution quickly for even development of positive signals. Adding Triton X-100 (0.04%) to substrate solution helps to break the surface tension of the solution.
Fluorophore Cy3 or/and FITC (see 2.5.2) are compatible with DAPI counterstaining. The combination of FITC and PI is good, yet Cy3 signal is not distinguishable from PI counterstaining (Figure 1B and C).
This TUNEL protocol is based on ApopTag fluorescence in Situ Apoptosis Detection Kit (see 2.6), such as Fluorescein Detection Kit (Millipore) (Figure 1C).
Keep the timing of incubation. Longer incubation gives high nonspecific background staining.
The TUNEL kit provides fluorophore tagged Anti-Digoxigenin that is affinity purified sheep polyclonal antibody, so double staining is possible with antibodies raised in other species.
References
- 1.Dehay C, Kennedy H. Nat Rev Neurosci. 2007;8:438–50. doi: 10.1038/nrn2097. [DOI] [PubMed] [Google Scholar]
- 2.Huang ZJ, Di Cristo G, Ango F. Nat Rev Neurosci. 2007;8:673–86. doi: 10.1038/nrn2188. [DOI] [PubMed] [Google Scholar]
- 3.Jacobsen M. Developmental Neurobiology. Plenum Press; New York: 1991. [Google Scholar]
- 4.Wang VY, Zoghbi HY. Nat Rev Neurosci. 2001;2:484–91. doi: 10.1038/35081558. [DOI] [PubMed] [Google Scholar]
- 5.Chong MJ, Murray MR, Gosink EC, Russell HR, Srinivasan A, Kapsetaki M, Korsmeyer SJ, McKinnon PJ. Proc Natl Acad Sci U S A. 2000;97:889–94. doi: 10.1073/pnas.97.2.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee Y, Chong MJ, McKinnon PJ. J Neurosci. 2001;21:6687–93. doi: 10.1523/JNEUROSCI.21-17-06687.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orii KE, Lee Y, Kondo N, McKinnon PJ. Proc Natl Acad Sci U S A. 2006;103:10017–22. doi: 10.1073/pnas.0602436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee Y, McKinnon PJ. Neuroscience. 2007;145:1365–74. doi: 10.1016/j.neuroscience.2006.07.026. [DOI] [PubMed] [Google Scholar]
- 9.Norbury CJ, Zhivotovsky B. Oncogene. 2004;23:2797–808. doi: 10.1038/sj.onc.1207532. [DOI] [PubMed] [Google Scholar]
- 10.Roos WP, Kaina B. Trends Mol Med. 2006;12:440–50. doi: 10.1016/j.molmed.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 11.Jeffers JR, Parganas E, Lee Y, Yang C, Wang J, Brennan J, MacLean KH, Han J, Chittenden T, Ihle JN, McKinnon PJ, Cleveland JL, Zambetti GP. Cancer Cell. 2003;4:321–8. doi: 10.1016/s1535-6108(03)00244-7. [DOI] [PubMed] [Google Scholar]
- 12.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. J Biol Chem. 1998;273:5858–68. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 13.Ward I, Chen J. Curr Top Dev Biol. 2004;63:1–35. doi: 10.1016/S0070-2153(04)63001-8. [DOI] [PubMed] [Google Scholar]
- 14.Shi SR, Cote RJ, Taylor CR. J Histochem Cytochem. 2001;49:931–7. doi: 10.1177/002215540104900801. [DOI] [PubMed] [Google Scholar]
- 15.Shi SR, Key ME, Kalra KL. J Histochem Cytochem. 1991;39:741–8. doi: 10.1177/39.6.1709656. [DOI] [PubMed] [Google Scholar]