Abstract
Pathogens that cause chronic infections often employ antigenic variation to evade the immune response and persist in the host. In Treponema pallidum (T. pallidum), the causative agent of syphilis, the TprK antigen undergoes variation of seven variable regions (V1-V7) by nonreciprocal recombination of silent donor cassettes with the tprK expression site. These V regions are the targets of the host humoral immune response during experimental infection. The present study addresses the causal role of the acquired immune response in the selection of TprK variants in two ways: 1) by investigating TprK variants arising in immunocompetent vs immunosuppressed hosts, and 2) by investigating the effect of prior specific immunization on selection of TprK variants during infection. V region diversity, particularly in V6, accumulates more rapidly in immunocompetent rabbits than in pharmacologically immunosuppressed rabbits (treated with weekly injections of methylprednisolone acetate). In a complementary experiment, rabbits pre-immunized with V6 region synthetic peptides had more rapid accumulation of V6 variant treponemes than control rabbits. These studies demonstrate that the host immune response selects against specific TprK epitopes expressed on T. pallidum, resulting in immune selection of new TprK variants during infection, confirming a role for antigenic variation in syphilis.
Keywords: Syphilis, TprK, antigenic variation, immune selection
INTRODUCTION
Pathogens frequently use antigenic variation mechanisms to evade the adaptive immune response, resulting in persistent infection. Syphilis is a lifelong infection in the absence of antibiotic treatment, and the mechanisms of persistence have been the subject of speculation for decades. Syphilis manifests in distinct clinical stages, of which the primary (ulcerative lesion) and secondary (skin rash, mucosal lesions) stages are infectious. The infectious lesions contain large numbers of treponemes but, after weeks to months, heal spontaneously after local immune clearance of the bacteria by opsonophagocytosis (1-3). The subsequent years to decades of persistent syphilis infection are usually asymptomatic, but approximately 30% of untreated individuals with latent infection develop tertiary syphilis (4). The manifestations of tertiary syphilis include gummas, cardiovascular syphilis, and late neurosyphilis, which can cause insanity, blindness, paralysis or death.
Suggested mechanisms of T. pallidum's persistence despite the host's efforts to eradicate the infection include residence within intracellular or immune privileged niches to hide from the immune effectors (5-9), T. pallidum‘s ability to cloak its surface with a coat of host serum proteins or mucopolysaccharides to avoid immune recognition (10-12), and immunosuppression of the host caused by syphilis infection (13, 14). All of these theories have lost favor in light of subsequent investigations. Freeze-fracture electron microscopy of T. pallidum revealed a paucity of integral membrane proteins in the T. pallidum outer membrane (OM), perhaps accounting for the relatively poor antigenicity of this spirochete's surface (15, 16). However, because T. pallidum can be phagocytized in the presence of opsonic antibody, antibody targets must be present on the surface of the bacterium. Furthermore, the finding that treponemes harvested from infected tissues later in infection, following the clearance of the majority of treponemes from the early lesions, are resistant to opsonophagocytosis, raised the possibility that antigenic variation might occur in T. pallidum, but no specific variable antigen was identified (17). The subsequent identification and investigation of TprK provided the first candidate antigen of T. pallidum that might function in immune evasion. TprK is highly heterogeneous among and within T. pallidum strains, with sequence diversity localized in seven discrete regions (V1-V7) flanked by conserved domains (18, 19). Centurion-Lara et al. (20) recently proposed a model whereby sequence diversity is generated by non reciprocal recombination (gene conversion) between multiple silent donor cassette segments and the single tprK expression site, similar to other bacterial variable antigens (21-23). This mechanism could theoretically generate millions of chimeric TprK variants that, if surface-exposed, could continuously alter T. pallidum's surface antigenic profile.
During infection, TprK is targeted by the host immune response, with T cell responses directed at epitopes located primarily in the conserved regions of the antigen, while the humoral response targets the V regions (24). LaFond and colleagues demonstrated that, while antibodies against the V region sequences of the infecting inoculum develop during infection, little or no antibody reactivity against newly arising V region variants is detectable (25).
The present study addresses the direct causal role of acquired immunity in variant selection in two complementary ways: 1) by investigating the T. pallidum TprK variants arising in immunocompetent vs. immunosuppressed hosts, and 2) by investigating the effect of prior specific immunization on selection of T. pallidum variants during infection. These studies provide evidence for the role of the acquired immune response in the selection of TprK variants during the course of infection and confirm that TprK variation is central to immune evasion during syphilis.
MATERIALS AND METHODS
T. pallidum Chicago strain propagation and derivation of the clonal Chicago C isolates
T. pallidum subsp. pallidum, Chicago strain, originally obtained from Paul Hardy and Ellen Nell (Johns Hopkins University, Baltimore, MD), was propagated intratesticularly (IT) in New Zealand white rabbits as previously reported (17). Because the Chicago strain treponemes have diverse sequences in the tprK locus, two clonal isolates (for our purposes, a clonal isolate is defined as an isolate with very limited or no detectable tprK sequence diversity) were derived in our laboratory as previously described (25). The resulting isolates, obtained after two IT expansions of the clonal population in the initial skin biopsy (25) were called Chicago C1 and Chicago C2, and used as inocula for the experimental infections (described below) without any further passage. Aliquots of Chicago C1 and C2 treponemes were stored as viable frozen stocks in liquid nitrogen and in lysis buffer for DNA sequence analysis. The level of sequence homogeneity of the tprK locus in both isolates was assessed by fragment length analysis (described below) and DNA sequencing. Approval of the protocols involving animal use was obtained in advance from the University of Washington Institutional Animal Care and Use Committee (IACUC).
Experimental infection of pharmacologically immunosuppressed and control rabbits with the Chicago C1 strain
The Chicago C1 strain was used to intradermally (ID) infect five pharmacologically immunosuppressed rabbits and five control rabbits at ten sites each on their clipped backs; 106 treponemes were injected per site. Pharmacological immunosuppression was achieved by weekly intramuscular (IM) injections of 20 mg of methylprednisolone acetate (Sicor, Irvine, CA). Optimal dosage was determined in a pilot experiment. Treatment was started three days before experimental infection and doses were administrated weekly for a total of six injections. A lesion biopsy (4-mm punch biopsies taken under local lidocaine anesthesia) was harvested from each rabbit weekly for a period of 5 weeks and used for both DNA and RNA extraction. Blood samples were also collected weekly to measure antibodies in the two groups. Venereal Disease Research laboratory (VDRL) titers were compared between treated and control rabbits as a measure of the effectiveness of the immunosuppressive treatment. Immunosuppression was further evaluated by quantification of rabbit IFNγ mRNA levels (normally the predominant cytokine in syphilis lesions) and T. pallidum burden in skin lesions by quantitative real-time PCR on reverse-transcribed RNA samples (qRT-PCR). tprK mRNA levels were also evaluated by qRT-PCR to determine whether there were any effects of immunosuppression on tprK expression by T. pallidum. Generation of sequence diversity in the tprK locus during the course of infection was determined using fragment length analysis (FLA; described below) of individual TprK V regions and by sequence analysis.
Experimental infection of V region-immunized and control rabbits with the Chicago C2 strain
Synthetic peptides based on the Chicago C2 V5 and V6 sequences were purchased from GenScript Corporation (Piscataway, NJ) and conjugated to the Keyhole Limpet Hemocyanin (KLH) carrier protein using the Imject Maleimide Activated mcKLH Kit (Pierce, Rockford IL) according to the manufacturer's protocol. Because maleimides reacts with sulfhydryl groups, a cysteine was added at the COOH-terminal of the Chicago C2 V6 peptide during the synthesis process, while the V5 peptides naturally ends with a cysteine and no further modification was required. Peptide-carrier conjugates were separated from free carrier molecules using desalting columns (provided with the kit), and conjugate concentration was determined using the BCA Protein Assay Kit (Pierce). Peptide-carrier complexes were stored at −20°C until use. Two groups of three adult male rabbits each were immunized with the Chicago C2 V5-KLH and V6-KLH conjugates, respectively, while a third group was immunized with the KLH carrier alone. Prior to injection, antigens (200 μg/dose) were emulsified in Freund's Incomplete Adjuvant (Sigma, St. Louis, MO). A total of six doses were administrated IM at 20-day intervals; efficacy of immunization was measured by ELISA detection of specific antibodies. At the end of the immunization cycle, the three groups of immunized rabbits, plus three unimmunized control rabbits, were infected intradermally at ten sites per rabbit with 105 Chicago C2 treponemes per site. Following ID infection, tissue biopsies from the leading edge of one lesion in each rabbit were taken at days 12 and 20 using a four mm biopsy punch (Miltex, Inc, York, PA) under local lidocaine anesthesia.
Specific procedures
Antibody testing
VDRL titers were determined weekly on sera obtained from immunosuppressed and control rabbits; reagents (VDRL antigen and buffered saline) were purchased from Becton Dickinson (Sparks, MD), and used according to the manufacturer's instructions. The development of antibodies to Chicago C1 V6 was determined by ELISA using synthetic peptides representing the predominant Chicago C1 V6 sequence, as previously described (25). Sera obtained from rabbits immunized with Chicago C2 V5-KLH, V6-KLH, and KLH alone were similarly tested by ELISA for their ability to recognize the respective immunogens (synthetic V5 and V6 peptides and unconjugated KLH). All ELISA results represent the mean OD ± the standard error of results for all animals in the group; individual sera were tested in triplicate wells per serum sample.
Nucleic acid extraction and manipulation
Immediately upon harvest, each biopsy was minced with a sterile blade. Half of the lesion tissue was resuspended in 400 μl of 1X lysis buffer (10mM Tris, pH 8.0; 0.1M EDTA; 0.5% sodium dodecyl sulfate) for DNA extraction, and the other half in 400 μl of Ultraspec buffer (Biotecx Laboratories Inc, Houston, TX) for total RNA isolation. DNA extraction was performed as previously described (26) using the QIAamp DNA Mini Kit (Qiagen Inc., Chatsworth, CA), according to the manufacturer's protocol. Extracted samples were stored at −80°C until use. RNA isolation was performed following the Ultraspec manufacturer's instructions as already described (27); after extraction, DNaseI treatment was performed as reported (28). Reverse transcription (RT) of DNA-free RNA was performed using the Superscript II First Strand Synthesis Kit (Invitrogen, Carlsbad, CA) with random hexamers according to the provided protocol. cDNA sample preparation and storage for real-time amplification were also previously reported (28).
Real-time quantification of T. pallidum and rabbit mRNA
A relative quantification protocol using external standards was chosen to analyze mRNA levels at the time of biopsy harvest. This approach normalizes the amount of message from one or more target genes to the mRNA of a reference gene. TP0574 (the 47 kDa lipoprotein) was used as reference when the target to be measured was tprK mRNA; rabbit HPRT was chosen to normalize levels of rabbit interferon-gamma (IFNγ) mRNA. The quantity of T. pallidum in skin lesions was determined as the ratio between TP0574 and rabbit HPRT mRNA levels. The rationale behind the use of the TP0574 (over several other candidates) as a reference gene for T. pallidum has been discussed in detail (28), and real-time amplification protocols for TP0574, tprK (28) (primers in Table I), rabbit HPRT and IFN-γ (29) have previously been described. Amplification reactions and data collection were carried out using the LightCycler 1.0 (Roche, Basel, Switzerland) instrument. All reactions were performed following the manufacturer's instructions with the Roche FastStart DNA Master plus SYBR Green Kit (Roche). Triplicate amplifications were performed for each gene per sample using three microliters of the cDNA preparations; a known concentration of a linear plasmid DNA containing all of the targets in its polylinker was amplified concurrently in each run as an internal standard and amplification control. Results were analyzed using the LightCycler 3.5 software (Roche). Differences in levels of gene expression between groups were compared using Students t-test, with significance set at p<0.05.
Table I.
Primers and synthetic peptides used in this study
| Primers | |||
|---|---|---|---|
| Purpose | Name | 5′ to 3′ sequence | Size (bp) |
| Full-length tprK amplification and sequencing |
tprK-S1 | ACCGGGCATGAATTTTCTTT | 1593 |
| tprK-As1 | CCATACATCCCTACCAAATCA | ||
|
| |||
| tprK-int-As2 | CCTACCCGCTGATACACCAC | ||
| M13rev2 | CAGGAAACAGCTATGAC | ||
| M13for2 | GTAAAACGACGGCCAG | ||
|
| |||
|
tprK fluorescent fragment length analysis3 |
FAM-V1-S | GTGGGCTCAGGTTTCGTTC | 187 |
| V1-As | CGCATAGACATTCCCCTCAC | ||
|
| |||
| FAM-V2-S | GGGGCTCACGTTTGATATTG | 183 | |
| V2-As | CCGGTGAGCTCCACTTTAAT | ||
|
| |||
| FAM-V3-S | GAGCGTACGCGTGAAGATG | 166 | |
| V3-As | TAGCAGCCAGAGCACACAGA | ||
|
| |||
| FAM-V4-S | CTTTGGGGTCTGTGTGCTCT | 112 | |
| V4-As | AACGATACCCCAACGTCAAC | ||
|
| |||
| FAM-V5-S | TTGGGGTATCGTTGGTTCTC | 173 | |
| V5-As | CCCAAATCAAGACCCTCAAG | ||
|
| |||
| FAM-V6-S | AAACCAAGGGGTCTGATCCT | 188 | |
| V6-As | TAGACGATACGAACCCCAGA | ||
|
| |||
| FAM-V7-S | TGGGTGAGTATGGTTGGGTTA | 159 | |
| V7-As | GCCGAATCTCCACCTTCTCT | ||
|
| |||
| Real-time qPCR |
TP0574-S | CGTGTGGTATCAACTATGG | 313 |
| TP0574-As | TCAACCGTGTACTCAGTGC | ||
|
| |||
| RT-tprK-S | AGTTTGCGTCTAACACCGACTG | 410 | |
| RT-tprK-S | TCGCATGGCCATGTTGAGAAAT | ||
|
| |||
| rHPRT-S | TGATAGATCCATTCCTATGACTGTAGA | 265 | |
| rHPRT-As | GGGTCCTTTTCACCAGCAG | ||
|
| |||
| rIFN-γ-S | TTCTTCAGCCTCACTCTCTCC | 224 | |
| rIFN-γ-As | TGTTGTCACTCTCCTCTTTCC | ||
|
| |||
|
Synthetic peptides
| |||
| Purpose | Name | NH2- to COOH- terminus sequence | Size (aa) |
|
| |||
| ELISA | ChicC1-V6 | VHYKVLKARAQAPAAVPAAADDIYF | 25 |
|
| |||
| Immunization | ChicC2-V5 | ASQASNVFQGVFLTTPMQKDDC4 | 22 |
| ChicC2-V6 | MPVHYKVLKARARAGAAVPAAADDIYFPV | 29 | |
tprK-S/As primers were used for both full-length ORF amplification and sequencing after cloning.
Sequencing primer. M13for and M13rev primers are PCR-II TOPO vector primers flanking the cloning site.
All sense primers are 5′-fluoresceine (6-FAM) labeled.
The final C residue was added during synthesis to allow conjugation with the KLH carrier.
tprK amplification, fragment length analysis (FLA), cloning and sequencing
DNA extracted from lesion biopsies was used to amplify the tprK gene to evaluate presence of sequence diversity by 1) fragment length analysis of amplicons of individual V regions and 2) sequencing of the full-length tprK ORF. To perform FLA analysis, each of the seven tprK V regions was amplified using a fluorescent (6-FAM-labeled) sense primer and an unlabelled antisense primer (primer sequences and amplicon sizes are shown in Table I) complementary to the unique conserved sequences flanking each tprK V region. Amplifications were performed using the AccuPrime Pfx DNA Polymerase (Invitrogen) with approximately 100 ng of DNA template in each reaction and primers at a final concentration of 400 nM. Pfx Polymerase is supplied with a 10 X reaction mixture containing 10 mM MgSO4, and 3 mM dNTPs. A touch-down amplification protocol was adopted to minimize nonspecific amplification products: during the first 10 cycles; the annealing temperature was decreased by 1°C per cycle until the optimal temperature of 55°C was reached. Initial denaturation and final extension steps were of 10 min each; denaturation, annealing and extension times were 1 min, 30 sec, and 30 sec, respectively, for a total of 45 cycles. Amplification and amplicon sizes were checked on 2% agarose gels, and products purified using the QIAquick PCR purification kit (Qiagen). Concentrations were measured with a ND-1000 instrument (NanoDrop Technologies, Wilmington, DE), and all samples diluted to 0.2 ng/μl final concentration. One microliter of each sample was mixed with 15.4 μl of Hi-Di Formamide (Perkin Elmer/Applied Biosystems, Foster City, CA) and 0.1 μl of MapMarker400 Rox-labeled DNA ladder (Bioventures Inc., Murfreesboro, TN); samples were transferred to a 96-well plate and denatured by incubation at 95°C for 2 min, briefly chilled on ice and loaded on a ABI3730xl DNA analyzer (Perkin Elmer/Applied Biosystems). Graphically, the resulting electropherograms contain red peaks, generated by the ROX-labeled DNA ladder, and blue peaks, representing amplification products for a V region. Because changes in length of the V regions do not modify the tprK reading frame, the blue peaks will necessarily be three nucleotides apart from each other when length diversity is present within the V region. Electropherograms were analyzed using the GeneMapper 4.0 software package (Perkin Elmer/Applied Biosystems); data relative to V region fragment length (determined by comparison to the ROX-labeled marker) and intensity (measured by area under the peak) were collected. For the inoculum and for each rabbit lesion at each time point, V region diversity observed by FLA was calculated as the reciprocal of Simpson's Diversity Index (RSDI) (30). The RSDI value takes into account not only the number of different V region sizes represented, but also their relative proportions; therefore it is the most appropriate to interpret the FLA results. In this context, RSDI =1/∑px2, where px = (area underneath the peak with size x / sum of the areas underneath all peaks). Using this interpolation, a RSDI value of 1 indicates the presence of a single V region size, while values >1 indicate increasing diversity in V region sizes, with higher values seen when higher proportions of different sizes are present.
Sequencing of the tprK ORF from the expression site was conducted as previously described (31), with the PCR amplicons being cloned into E. coli to permit sequencing of genes from individual T. pallidum cells. Plasmid extraction was carried on using either the Plasmid Mini Kit (Qiagen) or the Montage Plasmid Miniprep96 Kit (Millipore, Billerica, MA). Approximately 10 tprK clones from each amplification were sequenced. Sequencing primers are listed in Table I. Nucleotide sequences were translated and analyzed using the BioEdit Sequence Alignment Editor program, available at (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). A V region was considered diverse if it differed from any sequence seen in the original “founder” Chicago C1 inoculum. Sequence diversity was calculated using the predicted amino acid sequence for each V region compared to the inoculum (founder) sequences as follows: Diversity Score = number of clones with a V region sequence not seen in the inoculum / number of clones sequenced. Thus a diversity of 0 would result from a sample in which no new sequences were identified compared to the inoculum; a diversity score of 1 would indicate that all of the sequences obtained were different from those seen in the inoculum.
RESULTS
Derivation of the Chicago C1 and C2 clonal isolates
Both FLA and sequence analysis showed that the tprK locus in the Chicago C1 and C2 clonal isolates used as the inocula for the studies reported here are characterized by very low diversity. V region sequences and RSDI values for these isolates are provided in Table SI. For each V region, the inoculum sequences were designated as the founder sequences and used to calculate the Diversity Score for each V region from samples obtained during experimental infection.
Immunosuppression reduces selection for tprK sequence variants during syphilis infection
Effectiveness of immunosuppressive treatment
The effectiveness of methylprednisolone treatment was measured by comparison of serum VDRL antibody titers and lesion IFNγ mRNA levels in treated rabbits compared to controls. As shown in Fig.1A, treated animals had a significant delay in development of VDRL antibody, with lower overall antibody titers after infection with the Chicago C1 strain, confirming immunosuppression in these rabbits. It should be noted, however, that the VDRL titer in the treated rabbits was modestly increased at week 5 suggesting that a mild immune response might have developed at that point despite the methylprednisolone treatment. Rabbit IFNγ expression, which is robust during syphilis infection in rabbits and humans, was found to be significantly reduced in immunosuppressed rabbits compared to controls at each time point (Fig.1B), further confirming the effectiveness of immunosuppression in the treated group. Also, because immunocompetent subjects are able to resolve early syphilis lesions by clearance of T. pallidum, we quantified viable T. pallidum (measured by mRNA), showing that T. pallidum could be detected in lesion biopsies from suppressed rabbits for at least two weeks longer than in controls (Fig.1C). T. pallidum 47 kd lipoprotein (TP0574) mRNA (normalized to the rabbit HPRT message) could be detected up to week 3 after experimental infection in controls, compared to at least week 5 in treated rabbits. These data confirm the efficacy of methylprednisolone for inducing an immunosuppressed state in these rabbits.
Figure 1. Effectiveness of pharmacological treatment and tprK expression.
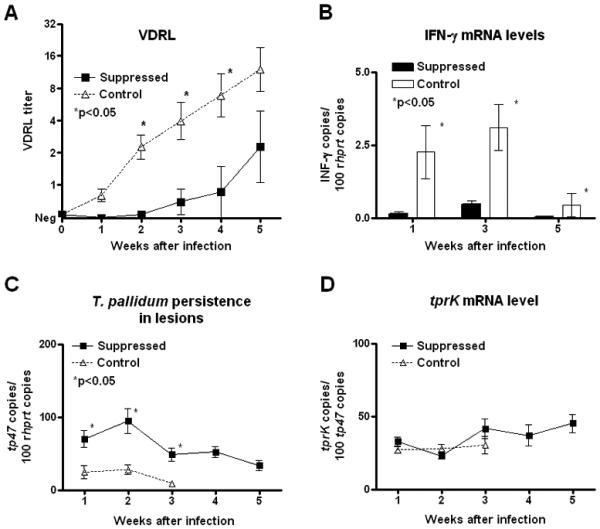
Significant delay in development of VDRL antibody titers (Panel A), reduced IFN-γ expression (Panel B), and longer persistence of T. pallidum cells in lesions from suppressed rabbits (Panel C) confirmed the effectiveness of pharmacological immunosuppression. Pharmacological treatment did not alter tprK mRNA levels in treponemes from control and treated rabbits (Panel D).
To be certain that any observed reduction in proportion of TprK variants in immunosuppressed rabbits was not due to down-regulation of tprK expression in those animals, we measured tprK mRNA by qRT-PCR. As shown in Fig 1D, immunosuppressive treatment did not alter the level of tprK expression per treponeme, with no significant difference between treated and control groups of rabbits during the time that T. pallidum mRNA could be detected by amplification.
Pharmacological immunosuppression reduces selection for TprK variants during syphilis infection
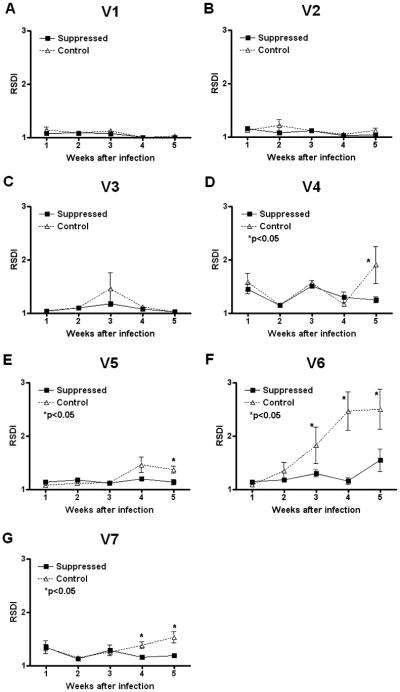
The accumulation of diversity in each tprK V region during the course of infection was investigated by both FLA and full-length tprK DNA sequencing. FLA results, reported as RSDI values, show that V region diversity increases with time in immunocompetent rabbits in V4, V5, V6, and V7, compared to immunosuppressed animals (Fig. 2D-G). In contrast, no difference in sequence diversity was seen for V1, V2, and V3 (Fig. 2A-C) between the two groups of rabbits. V6 RSDI values start to differ significantly (p<0.05) at week 3 postinfection, while a significant difference was seen for V7 at week 4, and at week 5 postinfection for V4 and V5. Very modest increases in V6 sequence diversity were seen at week 5 of infection in immunosuppressed animals. This may correspond to developing breakthrough specific immunity in the methylprednisolone-treated rabbits at week 5, as suggested by a slight increase in VDRL (Fig. 1A) and anti-V6 (Fig. 4) antibody titer at that time.
Figure 2. Accumulation of diversity in tprK V regions determined by FLA analysis.
RSDI (reciprocal of Simpson's diversity index) values show that, in immunocompetent rabbits infected with the Chicago C1 strain, sequence diversity tends to accumulate more rapidly in V6, V4, V5, and V7 than in treated rabbits (Panels D-G). No difference in diversity was seen for V1, V2, and V3 (Panels A-C) between the two groups of rabbits.
Figure 4. Correlation of development of anti-V6 antibody with presence of V6 variants.
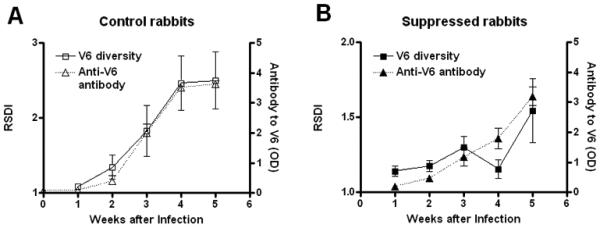
In immunocompetent rabbits, development of antibodies against the predominant Chicago C1 V6 variant steadily increases during experimental infection (panel A, Δ Symbol). Anti-V6 antibody however, fails to develop until later in infection and is significantly lower in titer in treated rabbits than in controls (p<0.05) at week 3 and 4 after infection (Panel B, ▲ Symbol). In both groups, developing anti-V6 antibody parallels the accumulation of new V6 variants seen by FLA analysis (reported in Fig. 2F and here again for comparison purposes) and sequencing (Fig. 3F), suggesting a role for antibody in selection of new TprK variants in vivo.
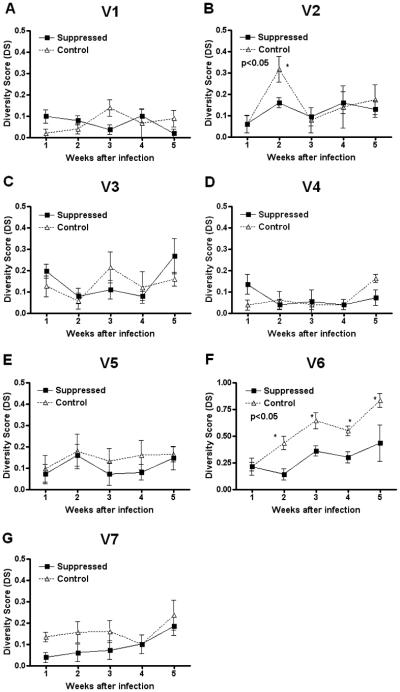
These same lesion samples were also analyzed by gene sequencing to determine whether any V region had diversified from the founder amino acid sequences for this isolate. Diversity score (DS) was calculated, as described above for each V region at weeks 1-5 postinfection (Fig. 3A-G). Similar to the FLA results, sequencing data showed absence of significant differences in DS values for V1, V2, and V3 during the course of infection (Fig. 3A-C) while V6 was found to be significantly more diverse by week 2 after infection in control compared to treated rabbits (Fig. 3F). In contrast to FLA analysis results, DS values for V4, V5, and V7 did not show significant differences in sequence diversity between treated and control rabbits (Fig. 3D, E, and G), likely reflecting the limited sampling that is practicable with sequencing.
Figure 3. Accumulation of diversity in tprK V regions determined by sequence analysis.
Diversity Score (DS) values for TprK V1-V7 (Panels A-G) at weeks 1-5 after infection with the Chicago C2 strain. The sequences of at least 10 TprK variants were determined for each lesion per time point. For each group of sequences, DS is calculated as the number of new V regions detected divided by the total number of sequences determined. When DS = 0, no new variants are detected compared to the inoculum, while a value of 1 indicates that none of the sequences is identical to the known inoculum sequences. DS values were found to be significantly higher (p<0.05) in controls with respect to suppressed rabbits for V6 (Panel F) at week 2 of infection.
Specific antibody titer correlates with immune selection for TprK V6 variants
In control rabbits infected with the Chicago C1 strain, measurable antibodies against the predominant Chicago C V6 peptide steadily increased postinfection (Fig. 4A); however, anti-V6 antibody titer was essentially unchanged through week 4 of infection in the immunosuppressed rabbits (Fig. 4B), with the titer increasing only at week 5. In both groups, antibody titer paralleled RSDI (reported in Fig. 2F and again, separately, in Fig. 4A and B for comparison purposes) and full-length tprK sequencing, suggesting a role for specific anti-V6 antibody in selection of TprK variants in vivo.
Prior immunization with tprK V6 selects for V6 variants in rabbits infected with Chicago C2
To complement the results described above, we asked whether prior immunization with V5 and V6 peptides (conjugated to KLH) would increase selection for TprK variants following infection with the Chicago C2 isolate. Efficacy of immunization was demonstrated by ELISA, showing that specific antibodies to V5 and V6 were induced by the immunization protocol (data not shown).
FLA analysis of tprK V5 and V6 in lesions from immunized rabbits infected with Chicago C2 showed that accumulation of diversity in V6, but not in V5, is influenced by the presence of antibody against the original V region sequence (Fig. 5A and B). V6 diversity significantly increased only in V6-immunized rabbits, but not in unimmunized, KLH-immunized or V5-immunized controls (Fig. 5B), confirming the specificity of the selective effect on V6. In contrast, V5 diversity was not shown to be significantly affected by the presence of pre-existing anti-V5 antibody (Fig. 5A).
Figure 5. Accumulation of sequence diversity in tprK V5 and V6 regions in pre-immunized rabbits.
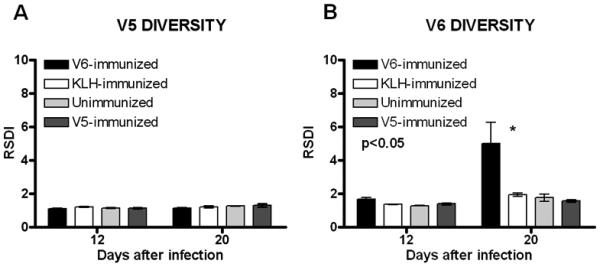
In Chicago C2-infected rabbits, V5 diversity is not significantly different in rabbits previously immunized with V5-KLH, V6-KLH, or KHL alone, or in unimmunized controls (Panel A). In contrast, V6 variants more readily are seen in rabbits previously immunized with V6-KLH, compared to V5-KLH- or KLH-immunized rabbits or in unimmunized controls (Panel B).
DISCUSSION
Antigenic variation of the TprK antigen of T. pallidum is hypothesized to explain the persistence of T. pallidum in the host despite a robust immune response (18, 19, 25, 31, 32). An essential role for TprK in syphilis pathogenesis is strongly supported by the fact that its seven variable regions are targeted by the humoral immune response during experimental infection (24), and also by the fact that, despite the high recombination rate between over 50 donor sequences and the tprK coding sequence, such rearrangements always result in an intact tprK ORF.
The involvement of TprK in immune evasion has not been directly addressed until now. A role in immune evasion implies that the target antigen is accessible to immune components and that an immunological function affects the survival of the individual bacterial cell expressing the antigen. While the function of the TprK protein is unknown, computer prediction (pSORTb; http://www.psort.org/psortb/) suggests that the protein is located in the outer membrane. Three-dimensional structural predictions of TprK (not shown) analysis yield a tertiary structure typical of gram-negative porins that reside in the bacterial outer membrane. However, given the very fragile nature of the T. pallidum cell, surface exposure of TprK has yet to be experimentally demonstrated by biochemical analysis. We previously reported (18) that antibodies raised against the T. pallidum Nichols strain TprK are opsonic, which strongly supports surface exposure. Recent experiments (unpublished) conducted in our laboratory have confirmed these results using a number of different antisera raised against recombinant TprK or TprK fragments; analogous opsonization studies with the Chicago strain ongoing in our laboratory confirm this finding for this strain. These studies support the surface exposure of TprK and a potential role for variation of this antigen as a means of immune evasion in syphilis. In complementary studies reported here, we demonstrated the direct effect of acquired immunity in selection of TprK variants during the course of infection. The naturally developing immune response during infection, and pre-existing specific anti-V6 immunity, both resulted in the accumulation of a higher proportion of TprK variants than seen in comparison rabbits.
We used two methods for quantitating V region sequence change in these studies; fragment length analysis (FLA) reported as reciprocal of the Simpson's Diversity Index (RSDI) and direct sequencing of ~10 clones per TprK amplicon. Each of these methods has advantages and disadvantages. Sequence analysis is useful in that it provides the actual sequence data, which are essential for analyses beyond the scope of diversity evaluation (i.e. epitope identification and donor site usage). However, the number of sequences that can practically be obtained per sample is limited by cost and time, and thus the results do not fully reflect the extent of the sequence diversity of the original sample. FLA analysis potentially compensates for these limitations by providing analysis of all DNA species in the V region amplicons, thus giving a more comprehensive snapshot of the entire V region population in the sample at a given time. It is likely that FLA analysis would identify variants that are infrequently represented in the sample and are thus not likely to be identified by limited sequencing. This may account for the observed higher sensitivity of FLA, compared to sequencing, for detecting variation in V4, V5, and V7 in the Chicago C1-infected rabbits (Figs. 2 and 3). However, two sequence variants with the same V region length would not be distinguished by FLA and thus even this method likely underestimates the true magnitude of V region diversity in a given sample.
Using both methods for measuring diversity, our analyses showed a remarkable accumulation of diversity in V6 during the course of experimental infection with the Chicago C1 strain in immunocompetent rabbits, with a significantly higher number of variants generated compared to immunologically suppressed rabbits. FLA analysis results for V4, V5, and V7 suggests that these V regions might also be involved in immune evasion, even though differences between the control and treated rabbits becomes significant much later than for V6. In contrast, both analytical methods showed no difference between groups for V1, V2, and V3. It could be hypothesized that V4-V7 may be more accessible to the host's antibodies, which could therefore more easily facilitate the clearance of T. pallidum cells carrying the original V region sequences. We have noted in the past that V1, V2, and V3 are less likely to vary in sequence than the other V regions, and also are less likely to induce specific antibodies than are other V regions (25).
Although pre-existing immunity to V6 selected for V6 variants following infection with Chicago C2, immunity to V5 had no such effect. This was puzzling in light of our finding that V5 variants appeared during the course of Chicago C1 infection, as described above. It is possible that the anti-V5 antibodies evoked by immunization with synthetic V5 peptide-KLH conjugate did not reflect antibodies induced during infection by the natural conformation of the V5 region in context of the mature TprK protein in T. pallidum cells and were thus not functional in selection of V5 variants in our studies.
In all T. pallidum strains examined to date, V6 is the most variable of the V regions. Although no experimental evidence is available on TprK protein structure, V6 could occupy a key location that would be highly susceptible to antibody-binding, consistent with the seemingly requisite high level of diversity exhibited by this variable region.
Because T. pallidum appears to go to great length to preserve the ability to express TprK, one could postulate other biological roles for TprK besides altering T. pallidum surface antigenicity. Because of the nature of syphilis infection, sequence diversity could favor adaptation to changing microenvironmental conditions that T. pallidum encounters in the dissemination from the site of primary infection to distant body locations. LaFond et al. (19), using clustering analysis, demonstrated that tprK sequences from treponemes in primary chancres are more likely to cluster within a patient than among patients, and therefore that tprK sequence variability is more limited within a T. pallidum isolate than among isolates. This suggests that different strains might express disparate repertoires of tprK genes and could help to explain the biological basis for the pathogenetic differences exhibited by T. pallidum strains during experimental infection as, for instance, propensity to invade the host central nervous system (33), or ability to cause more or less severe lesions in the rabbit model (34). If TprK functions as a porin, sequence diversity could influence the specificity with which metabolic intermediates are translocated across the OM, potentially affecting the survival of variants in particular anatomical niches.
TprK clearly undergoes antigenic variation, with immune selection being evident as variants are able to evade the host immune response. We postulate that immune selection occurs via opsonophagocytosis, mediated by anti-TprK antibodies, but it is unclear which specific epitopes are functional in this setting. It is important to remember as well that there are undoubtedly other surface-exposed antigens on T. pallidum, and the role of these molecules in pathogenesis and persistence also requires further study.
Supplementary Material
ACKNOWLEGEMENTS
The authors are grateful to Dr. Rebecca E. LaFond for her suggestions to improve the manuscript, Prof. Ram Samudrala of the Department of Microbiology at the University of Washington for his help with the TprK structural predictions (not shown), Bruce Godfrey of the Comparative Genomics Center at the University of Washington for assistance with the FLA experiments, and Elizabeth Miko Robertson for manuscript preparation.
Abbreviations used in this manuscript
- IMPs
integral membrane proteins
- OM
outer membrane
- ID
intradermal
- IT
intratesticular
- IV
intravenous
- VDRL
venereal disease research laboratory
- FLA
fragment length analysis
- RT
reverse transcriptase
- RSDI
reciprocal of Simpson's diversity index
- IFN-γ
interferon-γ
- DS
diversity score
- KLH
Keyhole Limpet Hemocyanin
Footnotes
This work was supported by NIH grants AI42143 and AI63940 (to S.A.L. and A.C.L.).
BIBLIOGRAPHY
- 1.Lukehart SA, Miller JN. Demonstration of the in vitro phagocytosis of Treponema pallidum by rabbit peritoneal macrophages. J Immunol. 1978;121:2014–2024. [PubMed] [Google Scholar]
- 2.Shaffer JM, Baker-Zander SA, Lukehart SA. Opsonization of Treponema pallidum is mediated by immunoglobulin G antibodies induced only by pathogenic treponemes. Infect Immun. 1993;61:781–784. doi: 10.1128/iai.61.2.781-784.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker-Zander SA, Lukehart SA. Macrophage-mediated killing of opsonized Treponema pallidum. J Infect Dis. 1992;165:69–74. doi: 10.1093/infdis/165.1.69. [DOI] [PubMed] [Google Scholar]
- 4.Gjestland T. The Oslo study of untreated syphilis. Acta Dermato Venereol. 1955;35:11–368. doi: 10.2340/00015555343368. [DOI] [PubMed] [Google Scholar]
- 5.Azar HA, Pham TD, Kurban AK. An electron microscopic study of a syphilitic chancre. Engulfment of Treponema pallidum by plasma cells. Arch Pathol. 1970;90:143–150. [PubMed] [Google Scholar]
- 6.Sykes JA, Miller JN, Kalan AJ. Treponema pallidum within cells of a primary chancre from a human female. Br J Vener Dis. 1974;50:40–44. doi: 10.1136/sti.50.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sykes JA, Miller JN. Intracellular location of Treponema pallidum (Nichols strain) in the rabbit testis. Infect Immun. 1971;4:307–314. doi: 10.1128/iai.4.3.307-314.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Medici MA. The immunoprotective niche-a new pathogenic mechanism for syphilis, the systemic mycoses and other infectious diseases. J Theor Biol. 1972;36:617–625. doi: 10.1016/0022-5193(72)90012-4. [DOI] [PubMed] [Google Scholar]
- 9.Sell S, Salman J, Norris SJ. Reinfection of chancre-immune rabbits with Treponema pallidum. I. Light and immunofluorescence studies. Am J Pathol. 1985;118:248–255. [PMC free article] [PubMed] [Google Scholar]
- 10.Christiansen S. Protective layer covering pathogenic treponematosis. Lancet. 1963;1:423–425. doi: 10.1016/s0140-6736(63)92309-2. [DOI] [PubMed] [Google Scholar]
- 11.Alderete JF, Baseman JB. Surface characterization of virulent Treponema pallidum. Infect Immun. 1980;30:814–823. doi: 10.1128/iai.30.3.814-823.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fitzgerald TJ, Johnson RC. Surface mucopolysaccharides of Treponema pallidum. Infect Immun. 1979;24:244–251. doi: 10.1128/iai.24.1.244-251.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wicher V, Wicher K. In vitro cell response of Treponema pallidum-infected rabbits. II. Inhibition of lymphocyte response to phytohaemagglutinin by serum of T. pallidum-infected rabbits. Clin Exp Immunol. 1977;29:487–495. [PMC free article] [PubMed] [Google Scholar]
- 14.Wicher V, Wicher K. In vitro cell response of Treponema pallidum-infected rabbits. III. Impairment in production of lymphocyte mitogenic factor. Clin Exp Immunol. 1977;29:496–500. [PMC free article] [PubMed] [Google Scholar]
- 15.Radolf JD, Norgard MV, Schulz WW. Outer membrane ultrastructure explains the limited antigenicity of virulent Treponema pallidum. Proc Natl Acad Sci USA. 1989;86:2051–2055. doi: 10.1073/pnas.86.6.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walker EM, Borenstein LA, Blanco DR, Miller JN, Lovett MA. Analysis of outer membrane ultrastructure of pathogenic Treponema and Borrelia species by freeze-fracture electron microscopy. J Bacteriol. 1991;173:5585–5588. doi: 10.1128/jb.173.17.5585-5588.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lukehart SA, Shaffer JM, Baker-Zander SA. A subpopulation of Treponema pallidum is resistant to phagocytosis: possible mechanism of persistence. J Infect Dis. 1992;166:1449–1453. doi: 10.1093/infdis/166.6.1449. [DOI] [PubMed] [Google Scholar]
- 18.Centurion-Lara A, Godornes C, Castro C, Van Voorhis WC, Lukehart SA. The tprK gene is heterogeneous among Treponema pallidum strains and has multiple alleles. Infect Immun. 2000;68:824–831. doi: 10.1128/iai.68.2.824-831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LaFond RE, Centurion-Lara A, Godornes C, Rompalo AM, Van Voorhis WC, Lukehart SA. Sequence diversity of Treponema pallidum subsp. pallidum tprK in human syphilis lesions and rabbit-propagated isolates. J Bacteriol. 2003;185:6262–6268. doi: 10.1128/JB.185.21.6262-6268.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Centurion-Lara A, LaFond RE, Hevner K, Godornes C, Molini BJ, Van Voorhis WC, Lukehart SA. Gene conversion: a mechanism for generation of heterogeneity in the tprK gene of Treponema pallidum during infection. Molecular Microbiology. 2004;52:1579–1596. doi: 10.1111/j.1365-2958.2004.04086.x. [DOI] [PubMed] [Google Scholar]
- 21.Zhang JR, Norris SJ. Genetic variation of the Borrelia burgdorferi gene vlsE involves cassette-specific, segmental gene conversion. Infect Immun. 1998;66:3698–3704. doi: 10.1128/iai.66.8.3698-3704.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barbour AG. Antigenic variation of a relapsing fever Borrelia species. Annu Rev Microbiol. 1990;44:155–171. doi: 10.1146/annurev.mi.44.100190.001103. [DOI] [PubMed] [Google Scholar]
- 23.Barbet AF, Lundgren A, Yi J, Rurangirwa FR, Palmer GH. Antigenic variation of Anaplasma marginale by expression of MSP2 mosaics. Infect Immun. 2000;68:6133–6138. doi: 10.1128/iai.68.11.6133-6138.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morgan CA, Molini BJ, Lukehart SA, Van Voorhis WC. Segregation of B and T cell epitopes of Treponema pallidum repeat protein K to variable and conserved regions during experimental syphilis infection. J Immunol. 2002;169:952–957. doi: 10.4049/jimmunol.169.2.952. [DOI] [PubMed] [Google Scholar]
- 25.LaFond RE, Molini BJ, Van Voorhis WC, Lukehart SA. Antigenic variation of TprK V regions abrogates specific antibody binding in syphilis. Infect Immun. 2006;74:6244–6251. doi: 10.1128/IAI.00827-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giacani L, Sun ES, Hevner K, Molini BJ, Van Voorhis WC, Lukehart SA, Centurion-Lara A. Tpr homologs in Treponema paraluiscuniculi Cuniculi A strain. Infect Immun. 2004;72:6561–6576. doi: 10.1128/IAI.72.11.6561-6576.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giacani L, Hevner K, Centurion-Lara A. Gene organization and transcriptional analysis of the tprJ, tprI, tprG and tprF loci in the Nichols and Sea 81-4 Treponema pallidum isolates. J Bacteriol. 2005;187:6084–6093. doi: 10.1128/JB.187.17.6084-6093.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giacani L, Molini B, Godornes C, Barrett L, Van Voorhis WC, Centurion-Lara A, Lukehart SA. Quantitative analysis of tpr gene expression in Treponema pallidum isolates: differences among isolates and correlation with T-cell responsiveness in experimental syphilis. Infect Immun. 2007;75:104–112. doi: 10.1128/IAI.01124-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Godornes C, Leader BT, Molini BJ, Centurion-Lara A, Lukehart SA. Quantitation of rabbit cytokine mRNA by real-time RT-PCR. Cytokine. 2007;38:1–7. doi: 10.1016/j.cyto.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunter PR, M G. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. Journal of Clinical Microbiology. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.LaFond RE, Centurion-Lara A, Godornes C, Van Voorhis WC, Lukehart SA. TprK sequence diversity accumulates during infection of rabbits with Treponema pallidum subsp. pallidum Nichols strain. Infect Immun. 2006;74:1896–1906. doi: 10.1128/IAI.74.3.1896-1906.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morgan CA, Lukehart SA, Van Voorhis WC. Protection against syphilis correlates with specificity of antibodies to the variable regions of Treponema pallidum repeat protein K. Infect Immun. 2003;71:5605–5612. doi: 10.1128/IAI.71.10.5605-5612.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tantalo LC, Lukehart SA, Marra CM. Treponema pallidum strain-specific differences in neuroinvasion and clinical phenotype in a rabbit model. J Infect Dis. 2005;191:75–80. doi: 10.1086/426510. [DOI] [PubMed] [Google Scholar]
- 34.Turner TB, Hollander DH. Biology of the Treponematoses. World Health Organization; Geneva: 1957. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.