Abstract
Recent studies have shown that a number of microRNAs (miRNA or miR) may regulate human breast cancer resistance protein (BCRP/ABCG2), an important efflux transporter responsible for cellular drug disposition, whereas their effects on ABCG2 protein expression are not compared. In this study, we first identified a new proximal miRNA response element (MRE) for hsa-miR-519c within ABCG2 3′-untranslated region (3′UTR) through computational analyses. This miR-519c MRE site was confirmed using dual luciferase reporter assay and site-directed mutagenesis. Immunoblot analyses indicated that ABCG2 protein expression was significantly down-regulated in MCF-7/MX100 cells after transfection with hsa-miR-328- or -519c expression plasmids, and was markedly up-regulated in MCF-7 cells after transfection with miR-328 or -519c antagomir. However, ABCG2 protein expression was unchanged in MCF-7/MX100 cells after transfection with hsa-miR-520h expression plasmids, which was associated with undetectable miR-520h expression. Furthermore, ABCG2 mRNA degradation was accelerated dramatically in cells transfected with miR-519c expression plasmid, suggesting the involvement of mRNA degradation mechanism. Intervention of miR-328 or -519c signaling led to significant change in intracellular mitoxantrone accumulation, as determined by flow cytometry analyses. In addition, we separated RB143 human retinoblastoma cells into stem-like (ABCG2+) and non-stem-like (ABCG2−) populations through immunomagnetic selection, and found that miR-328, -519c and -520h levels were 9-, 15- and 3-fold lower in the ABCG2+ cells, respectively. Our data suggest that miR-519c and -328 have greater impact on ABCG2 expression than miR-520h in MCF-7 human breast cancer cells, and the presence of proximal miR-519c MRE explains the action of miR-519c on shortened ABCG2 3′UTR.
Keywords: miRNA, BCRP/ABCG2, drug disposition, gene regulation, epigenetic, multidrug resistance, stem cell, breast cancer, retinoblastoma
1. Introduction
MicroRNAs (miRNA or miR) are a large group of short, noncoding RNAs that act on their mRNA targets by complementary Watson-Crick base pairings, and control posttranscriptional regulation of target genes through translation inhibition or mRNA cleavage [1, 2]. More than 1000 miRNAs have been identified within human genome, which may regulate thousands of protein-coding genes involved in essentially all critical biological processes [3]. There is also accumulating evidence supporting the hypothesis that miRNAs are involved in the regulation of drug metabolism and disposition [4–8]. Some miRNAs directly target the 3′-untranslated region (3′UTR) of genes encoding drug-metabolizing enzymes and/or drug transporters [9–19]. Other miRNAs act on the 3′UTR of transcriptional regulators (e.g., xenobiotic receptors) of drug-metabolizing enzymes and drug transporters [10, 20–22]. As a result, these miRNAs may determine the final protein expression levels of enzymes or transporters, modulate the capacity of drug metabolism and disposition, and affect the response of cells to xenobiotic drugs.
Breast cancer resistance protein (BCRP/ABCG2) is an ATP-binding cassette membrane transporter that is expressed ubiquitously in human tissues. ABCG2 plays an important role in cellular disposition of a variety of xenobiotic drugs (e.g., mitoxantrone, doxorubicin and topotecan) and metabolites (e.g., estrone-3-sulfate and estradiol-17-β-D-glucuronide) [23, 24]. Variations of ABCG2 gene expression and protein function may result in significant change in pharmacokinetics and drug response. Overexpression of ABCG2 may confer multidrug resistance in cancer cells, which could be caused by gene amplification [25–27] and malfunction of regulatory factors [11]. In addition, ABCG2 is revealed as a critical element of tumorigenic stem cells [28–30], and has been used as one selective biomarker for cancer stem cells.
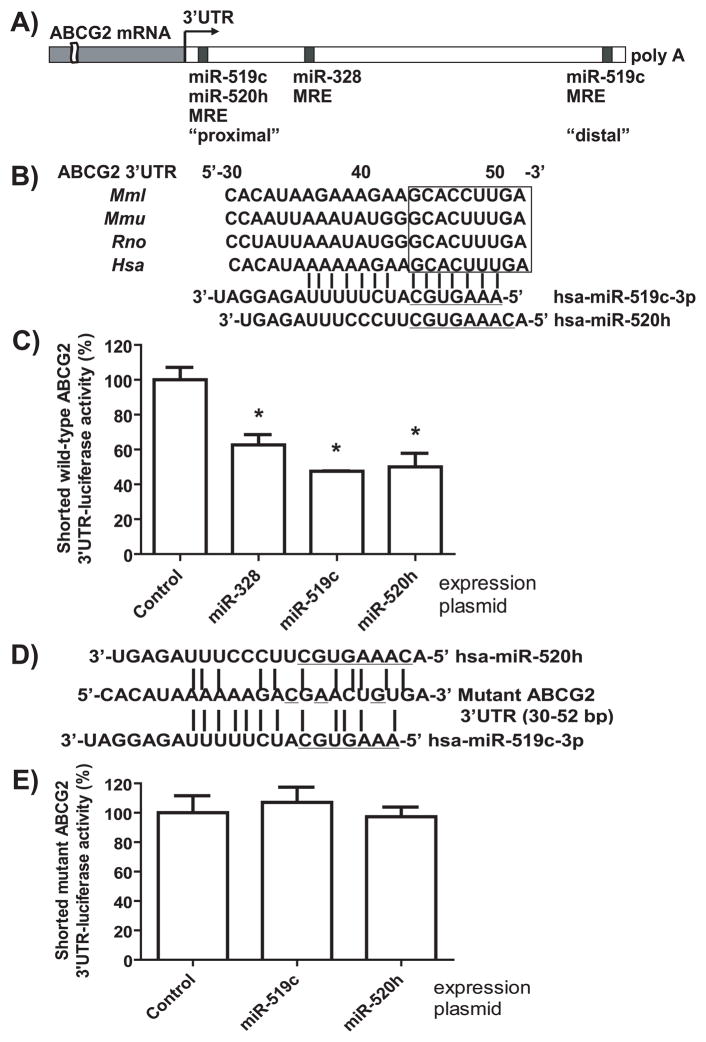
Recent studies have shown that a number of miRNAs may regulate the expression of ABCG2 through their actions on ABCG2 3′UTR. Using a luciferase reporter assay, Liao et al. [31] found that hsa-miR-520h could act on the 3′UTR (30–52 nt from stop codon) of ABCG2 mRNA, and Wang et al. [18] showed that ABCG2 protein expression was indeed down-regulated by miR-520h in pancreatic cancer cells. Meanwhile, To et al. [14, 15] reported a distal miRNA response element (MRE; 1526–1548 nt) for hsa-miR-519c that was close to the poly(A) tail of ABCG2 3′UTR (Fig. 1A), and showed that a miR-519c inhibitor or mimic was able to alter ABCG2 protein expression in A549 human lung adenocarcinoma epithelial cells. Interestingly, the 3′UTR including that of ABCG2 was revealed to be commonly shortened in cancer cells, especially in embryonic stem cells, proliferating cells and drug-resistant cancer cells [14, 32–34]. The absence of this distal miR-519c MRE site in shortened ABCG2 3′UTR might limit the role of miR-519c in ABCG2 regulation. In addition, we found that ABCG2 3′UTR (599–626 nt) could be readily targeted by hsa-miR-328 when considering target-site accessibility or binding energy, and modulation of miR-328 signaling had significant effects on ABCG2 mRNA and protein expression in drug-sensitive MCF-7 and drug-resistant MCF-7/MX100 human breast cancer cells [11]. Nevertheless, it remains unknown which miRNA is more important in posttranscriptional regulation of ABCG2. Therefore, this study aimed to compare the contribution of miR-328, -519c and -520h to the regulation of ABCG2, and delineate the potential action of miR-519c on the shortened 3′UTR of ABCG2 lacking the distal miR-519c MRE site (Fig. 1A).
Figure 1.
A new MRE site for miR-519c proximal to ABCG2 stop codon is identified by bioinformatic analyses, and confirmed by luciferase reporter assay and site-directed mutagenesis. (A) Putative MRE sites for miR-328, -519c and -520h within ABCG2 3′UTR were identified by TargetScan, PITA, Microcosm Targets or RNA22 algorithm. (B) The proximal MRE site at 30–52 nt after stop codon may be targeted by both miR-519c and -520h, which is well conserved in Rhesus (Mml), Mouse (Mmu), Rat (Rno) and Human (Hsa). (C) The luciferase reporter activities (mean ± SD of triplicate cultures) of the shortened wild-type ABCG2 3′UTR (0–1296 nt) were significantly (*P < 0.01) reduced in HEK293 cells after transfection with miR-519c expression plasmid, as well as miR-520h and -328. (D) Base pairings between Mutant ABCG2 3′UTR and miR-519c or -520h are disrupted. (E) Mutant ABCG2 3′UTR-luciferase reporter activity was not affected by miR-519c or -520h.
2. Materials and Methods
2.1. Materials
Mitoxantrone was bought from Sigma-Aldrich (St. Louis, MO). Fumitremorgin C (FTC) was purchased from Alexis (San Diego, CA). Dulbecco’s modified Eagle’s medium (DMEM), RPMI 1640 medium, trypsin, phosphate-buffered saline (PBS) and antibiotics were bought from Mediatech (Manassas, VA). Fetal bovine serum and calf serum were bought from Invitrogen (Carlsbad, AL). Selective miRNA and control antagomirs were purchased from Dharmacon (Lafayette, CO).
2.2. Identification of MRE Sites
Putative MRE sites within the 3′UTR of human ABCG2 (NM_004827) were identified by TargetScan [35], PITA [36], MicroCosm Targets [37] and RNA22 [38].
2.3. Cell Culture
Human embryonic kidney HEK293 cells were cultured in DMEM supplemented with 4.5 g/L glucose, 10 mM HEPES, and 10% fetal bovine serum at 37°C. Human breast cancer MCF-7 cells and mitoxantrone-selected MCF-7/MX100 cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, 100 units/mL penicillin G sulfate and 100 units/ml streptomycin sulfate. Human retinoblastoma cell line RB143 (a gift from Dr. Bruce Ksander) was maintained in DMEM, with 20% calf serum.
2.4. Enrichment of ABCG2+ Cells
The EasySep Human FITC Positive Selection Kit (Stem Cell Technologies, Vancouver, Canada) was used according to instructions, with 2.5 μg/mL of ABCG2-FITC antibody (Stem Cell Technologies). After separation, the ABCG2+ and ABCG2− cells were snap-frozen in liquid nitrogen for nucleic acid extraction.
2.5. Construction of MicroRNA Expression and 3′UTR-Luicferase Reporter Plasmids
Creation of miR-328 expression plasmid (pS-miR-328) and control plasmid (pS-Neg) consisting of a scrambled sequence was described previously [11]. Likewise, miR-519c and -520h precursors were cloned into pSilencer 4.1-CMV-neo vector (Ambion, Austin, TX) using gene-specific primers bearing BamHI and HindIII restriction sites. The resulting plasmids were named pS-miR-519c and pS-miR-520h, respectively. Cloning of a short (0–1296 nt from stop codon) ABCG2 3′UTR-luciferase reporter plasmid was described previously [11]. This 3′UTR fragment does not contain the distal MRE site (1526–1548 nt, close to poly A tail) for miR-519c reported by To et al. [15] but the proximal MRE site (30–52 nt, close to stop codon) for miR-519c identified in this study (Fig. 1A). The QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA) was employed to create an ABCG2 3′UTR mutant that consisted of three nucleotide transitions (A43C, C45A, C49G) at the proximal miR-519c/520h MRE site (Fig. 1A). Primers were shown in Table 1. All clones were confirmed by direct DNA sequencing analyses.
Table 1.
Oligonucleotides used for the construction of plasmids, mutagenesis and qPCR analyses.
| Application | Oligonucleotide | Sequence (5′→3′) |
|---|---|---|
| Cloning | ||
| pS-miR-519c | Forward | CGCGGATCCGCGAAATCAATGGTGCTGGAGCAAGAAGATC |
| Reverse | CCCAAGCTTGGGGGTGGATTACGAGGTCAGGAGATGGAGA | |
| pS-miR-520h | Forward | CGCGGATCCATGCAAACAGGGCCAATAAATGC |
| Reverse | CCAAGCTTAACCCAGAACCCCAACATCATCC | |
| Mutagenesis | ||
| mutant ABCG2 3′UTR | Forward | CTCACATAAAAAAGACGAACTGTGATTGAAGTAT |
| miR-519c and -520h MRE | Reverse | ATACTTCAATCACAGTTCGTCTTTTTTATGTGAGG |
| Stem-loop RT-qPCR | ||
| miR-328 | RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACGGAA |
| Forward | ATATCTGGCCCTCTCTGCCC | |
| miR-519c | RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACATCCTC |
| Forward | GGCGGGAAAGTGCATCTTTTT | |
| miR-520h | RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACTCTA |
| Forward | GGCGACAAAGTGCTTCCCTT | |
| U54 | RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGGCTCA |
| Forward | GCTATTCTGAGCCGTCGTATCCA | |
| U74 | RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAATTGT |
| Forward | CCTGTGGAGTTGATCCTAGTCTGGGTG | |
| Universal reverse primer | GTGCAGGGTCCGAGGT | |
| qPCR | ||
| ABCC1 | Forward | AACCTGGACCCATTCAGCC |
| Reverse | GACTGGATGAGGTCGTCCGT | |
| ABCG2 | Forward | CAGGTGGAGGCAAATCTTCGT |
| Reverse | ACACACCACGGATAAACTGA | |
| ALDH1A1 | Forward | CGCAAGACAGGCTTTTCAG |
| Reverse | TGTATAATAGTCGCCCCCTCTC | |
| CD133 | Forward | TGGATGCAGAACTTGACAACGT |
| Reverse | ATACCTGCTACGACAGTCGTGGT | |
| 18S | Forward | GTAACCCGTTGAACCCCATT |
| Reverse | CCATCCAATCGGTAGTAGCG | |
| GAPDH | Forward | ATCACCATCTTCCAGGAGCGA |
| Reverse | GCTTCACCACCTTCTTGATGT | |
2.6. Luciferase Assay
HEK-293 cells were co-transfected with wild-type or mutant ABCG2 3′UTR-luciferase reporter plasmid, and pS-Neg or miRNA expression plasmid using Lipofectamine 2000 (Invitrogen). Luciferase activities were determined with Dual Luciferase Reporter Assay System (Promega, Madison, WI) at 48 hours after transfection, according to the manufacturer’s protocol. Transfection was conducted in triplicate, and repeated once with separate culture. Renilla luciferase activities were normalized to corresponding firefly luciferase activities, and then compared among different treatments or groups. Additional transfections were used for the evaluation of miRNA expression.
2.7. Quantitative Real-Time Reverse Transcription PCR (qPCR) Analysis
Total RNAs were prepared with Trizol (Invitrogen). Stem-loop reverse transcription (RT) of mature miRNAs and qPCR analyses were conducted as reported [10, 11, 39], using gene-specific primers shown in Table 1. U54 and U74 small nucleolar RNAs were used as internal control for miRNA quantification. To determine ABC transporter (MDR1/ABCB1, MRP1/ABCC1 and BCRP/ABCG2), stem cell biomarker aldehyde dehydrogenase 1A1 (ALDH1A1), glycoprotein CD133, and 18S mRNA levels, total RNAs were reverse transcribed to cDNA using the Superscript II Reverse Transcription-PCR kit (Invitrogen). qPCR analyses of ABC transporters were performed using gene-specific primers (Table 1), and normalized to 18S. In particular, SYBR Green qPCR reactions were conducted on a MyIQ real-time PCR system (Bio-Rad, Hercules, CA). All PCR reactions were conducted in duplicate, and triplicate-cultured cells were tested. All experiments were repeated with separate cultures. The cycle number (CT) at which the amplicon concentration crossed a defined threshold was determined for each miRNA, and CT linear range was established through serial dilutions of control sample. miRNA was defined as undetectable when corresponding CT value was greater than the upper limit or subject to >50% variation. The relative level of each analyte over internal standard was calculated using the formula 2−ΔCT, where ΔCT was the difference in CT values between analyte and internal standard.
2.8. Immunoblot Analysis
MCF-7/MX100 cells were transfected with miRNA expression or control pS-Neg plasmid, and MCF-7 cells were transfected with selective miRNA antagomir or control oligonucleotide. Cells were harvested at 48 hours after transfection. RNA was isolated for qPCR analysis of miRNA expression, as described above. Cell lysates were prepared with RIPA lysis buffer (Rockland Immunologicals, Gilbertsville, PA) supplemented with a complete protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany). Protein concentrations were determined using BCA Protein Assay Kit (Pierce, Rockford, IL). Whole-cell proteins (50 μg) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and electrophoretically transferred onto nitrocellulose membranes (GE Healthcare, Piscataway, NJ). Membranes were then incubated with selective antibody against ABCG2 (Kamiya Biomedical, Thousand Oaks, CA) or GAPDH (Santa Cruz Biotechnology, Santa Cruz, CA), and subsequently with a peroxidase goat anti-mouse IgG (Jackson ImmunoResearch, West Grove, PA). After visualization with an enhanced chemiluminescence detection system (Pierce Biotechnology, Rockford, IL), images were acquired and densitometric analyses were carried out using Kodak Image Station (New Haven, CT).
2.9. RNA Degradation Analysis
Forty eight hours after MCF-7/MX100 cells were transfected with pS-Neg, pS-miR-328 or pS-miR-519c plasmids, actinomycin D (Sigma-Aldrich) was added to a final concentration of 5 μg/mL to block de novo RNA synthesis. Cells were harvested at 0, 2, 4, and 8 hours following the treatment with actinomycin D. ABCG2 mRNA levels were determined by qPCR, and normalized to GAPDH mRNA levels. All treatments were conducted in triplicate.
2.10. Flow Cytometry Analysis
Intracellular drug accumulation was examined with a FACSCalibur flow cytometer (BD Biosciences, Franklin Lakes, NJ), as reported [40–42]. Briefly, MCF-7/MX100 cells were harvested at 48 hours after transfection, and 1×106 cells were incubated for 60 min at 37°C in 800 nM of mitoxantrone in phenol red-free RPMI 1640 medium. Cells incubated in the absence of drug were used as negative controls, and cells incubated with drug and 10 μM FTC were utilized as positive controls. Cells were then washed twice with ice-cold PBS buffer, resuspended, and subjected to flow cytometry analyses with a 635 nm red diode laser and a 670 nm band pass filter. Fluorescence of 10,000 events was collected, and data were processed by CellQuest software (BD Biosciences). D values between stained and unstained cells were calculated by Kolmogorov-Smirnov statistic (CellQuest) [43, 44], and used to assess the difference in drug disposition among cells with different miRNA treatments. All treatments were performed in triplicate, and experiment was repeated at least once.
2.11. Statistical Analysis
All values were expressed as mean ± standard deviation (SD) or standard error of mean (SEM). Depending on the number of groups and variances, values were compared with unpaired Student’s t-test, one-way ANOVA analysis followed by Dunnett’s test, or two-way ANOVA (GraphPad Prism 5, San Diego, CA). Difference was considered statistically significant when probability was less than 0.05 (P < 0.05).
3. Results
3.1. A new, conserved proximal MRE site (30 to 52 nt from stop codon) is identified for miR-519c through computational analysis
Bioinformatic analyses have shown that ABCG2 3′UTR may be targeted by a number of miRNAs [11, 15, 31], including the distal MRE site (1526–1548 nt; Fig 1A) for miR-519c. Herein we show that the putative miR-520 MRE site from 30 to 52 nt within ABCG2 3′UTR [31] may be targeted by miR-519c as well (Fig 1A). Complementary base pairings of miR-519c-3p to this site was supported by computational analyses with PITA (folding energy ΔG, −19.8 Kcal/mol; difference between miRNA-mRNA binding energy and free energy to open the target mRNA site, ΔΔG, −15.6 Kcal/mol) and RNA22 (ΔG, −26.8 Kcal/mol) algorithms, but not identified by TargetScan and MicroCosm Targets algorithms. Interestingly, the miR-328 MRE site (618–626 nt) [11] was identified by PITA, TargetScan and RNA22 (ΔG, −43.0 Kcal/mol) algorithms but not by MicroCosm Targets, whereas the distal miR-519c MRE site [15] was only predicted by TargetScan. Identification of this new proximal MRE site for miR-519c may help explain the apparent effects of miR-519c on ABCG2 protein expression in cancer cells shown in this study and previous studies [14, 15]. Nevertheless, one cannot exclude the possibility that an miRNA may influence protein expression of a gene through its actions on the gene’s regulatory factors [4, 11].
3.2. The proximal miR-519c MRE site is confirmed by site-directed gene mutagenesis and luciferase reporter assay
To test if this proximal MRE site can be targeted by miR-519c, a shortened ABCG2 3′UTR (0–1296 nt) construct [11] lacking the distal miR-519c MRE was utilized in luciferase reporter assay. The shortened wild-type ABCG2 3′UTR-luciferase activity was decreased about 50% in HEK293 cells after transfection with miR-519c expression plasmid, compared with cells transfected with pS-Neg plasmid. By comparison, luciferase activities were decreased around 40% and 50% in cells transfected with miR-328 and -520h expression plasmids, respectively. To further validate the proximal miR-519c MRE site, ABCG2 3′UTR segment (43–50 nt) complementary to the seed sequences of miR-519c and -520h was changed through site-directed mutagenesis. Following the disruption of mRNA-miRNA base pairings (Fig 1C), the mutant ABCG2 3′UTR-luciferase activities remained unchanged in cells after transfection with miR-519c and -520h expression plasmids (Fig 1D). Taken together (Fig 1B and 1D), the segment from 43 to 50 nt is essential for the action of miR-519c, as well as miR-520h, on shortened ABCG2 3′UTR transcripts.
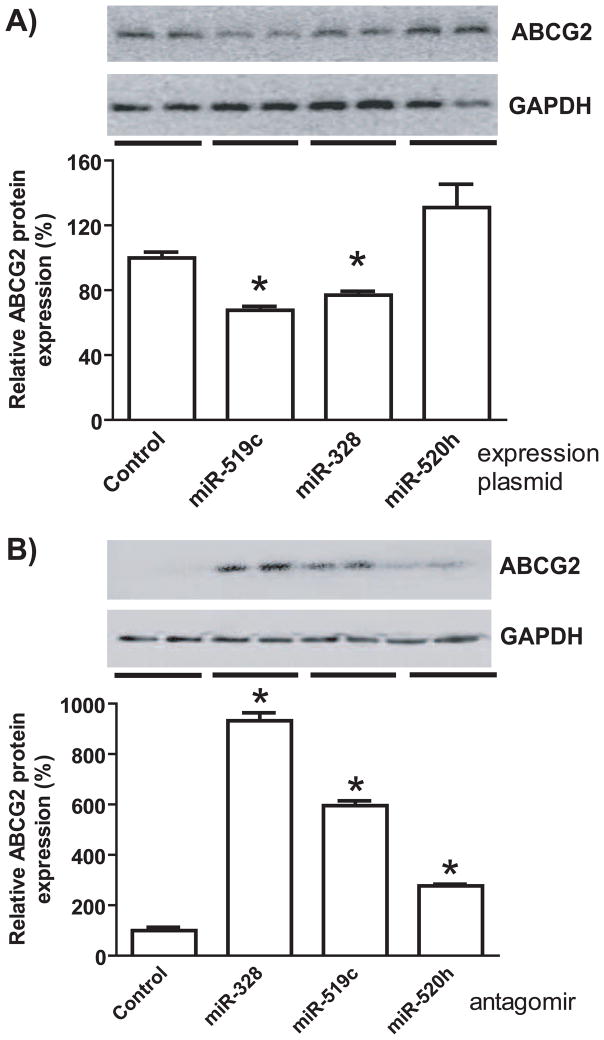
3.3. Modulation of miR-519c or miR-328 signaling significantly alters ABCG2 protein expression in human breast cancer cells, whereas miR-520h has no or minor impact
To directly compare the effects of individual miRNAs on ABCG2 protein expression, we employed both gain- and loss-of-function approaches. After successful creation of miRNA expression plasmids, drug-resistance MCF-7/MX100 cells over-expressing ABCG2 were transfected with control and individual miRNA expression plasmids. Compared with the control, transfection with miR-519c or miR-328 expression plasmids led to 30–35% reduction of ABCG2 protein expression in MCF-7/MX100 cells, whereas miR-520h had no significant impact on ABCG2 protein (Fig. 2A) or mRNA expression (data not shown). Furthermore, selective miRNA antagomirs were utilized to inhibit the function of corresponding miRNAs in drug-sensitive MCF-7 cells with minimal ABCG2 expression. Immunoblot analyses showed that ABCG2 protein levels were sharply elevated in MCF-7 cells transfected with miR-328 or miR-519c antagomir, compared with cells transfected with control oligonucleotide (Fig. 2B). The transfection with miR-520h antagomir resulted in a minor increase in ABCG2 protein expression, and the level was not as dramatic as those altered by miR-328 or miR-519c antagomir. The data indicate that miR-520h has limited impact on ABCG2 protein expression in human breast cancer cells, while miR-519c and miR-328 have much greater contribution to the regulation of ABCG2.
Figure 2.
miR-519c and -328 have greater effects on ABCG2 protein expression than miR-520h in human breast cancer cells. (A) ABCG2 protein levels (mean ± SD of triplicate cultures) were significantly (*P < 0.05) reduced in MCF-7/MX100 cells after transfection with miR-519c- or -328 expression plasmids, whereas ABCG2 remained unchanged in cells transfected with miR-520h plasmid. (B) ABCG2 protein levels were elevated (*P < 0.05) in MCF-7 cells to a much higher degree by miR-328 or -519c antagomir than that by miR-520h antagomir. Whole-cell proteins were separated by SDS-PAGE, and immunoblot analyses were conducted with selective antibody against ABCG2 or GAPDH (loading control).
3.4. The absence of impact of miR-520h on ABCG2 expression in MCF-7/MX100 cells is associated with an undetectable miR-520h expression
To test the hypothesis that the impact of miRNA on target gene expression may be related to the abundance of miRNA within cells, we employed stem-loop RT-qPCR method to compare miRNA expression levels in MCF-7/MX100 cells transfected with control and miRNA expression plasmids. Based on the linear range of quantification (Table 2), miR-328 expression level was revealed to be about 20-fold in MCF-7/MX100 cells after transfection with miR-328 expression plasmid (normalized either to U74 or U54). Interestingly, both miR-519c (CT >34) and miR-520h (CT > 32) in cells transfected with control plasmids were under detection limits (Table 2). However, miR-519c level was sharply increased (CT = 23.6 ± 0.4) in cells following the transfection with miR-519c expression plasmid, whereas miR-520h level remained as undetectable (CT > 32) in cells after transfection with miR-520h expression plasmid. The results suggest that miR-519c and miR-328 precursors are readily processed in human breast cancer cells, and the deficient production of miR-520h (Table 2) may explain the lack of ABCG2 regulation by miR-520h (Fig 2A) in MCF-7/MX100 cells.
Table 2.
Mature miRNA expression in MCF-7/MX100 cells after transfection with control (pS-Neg) or miRNA expression (pS-miR) plasmids, as determined by real-time qPCR analyses. Small RNA U54 and U74 were used as reference genes. CT values represent mean ± SD of quadruplicate cultures.
| CT linear range | CT value | ||
|---|---|---|---|
| pS-Neg | pS-miR | ||
| miR-328 | 22–30 | 27.8 ± 0.3 | 23.3 ± 1.4 |
| U54 | 5–23 | 8.58 ± 0.38 | 8.61 ± 0.33 |
| U74 | 22–34 | 27.7 ± 0.3 | 28.6 ± 0.5 |
| miR-519c | 22–33 | >34 | 23.6 ± 0.4 |
| U54 | 5–23 | 8.58 ± 0.38 | 8.34 ± 0.40 |
| U74 | 22–34 | 27.7 ± 0.3 | 28.3 ± 0.8 |
| miR-520h | 26–32 | >32 | >32 |
| U54 | 5–23 | 8.58 ± 0.38 | 8.60 ± 0.27 |
| U74 | 22–34 | 27.7 ± 0.3 | 28.6 ± 0.3 |
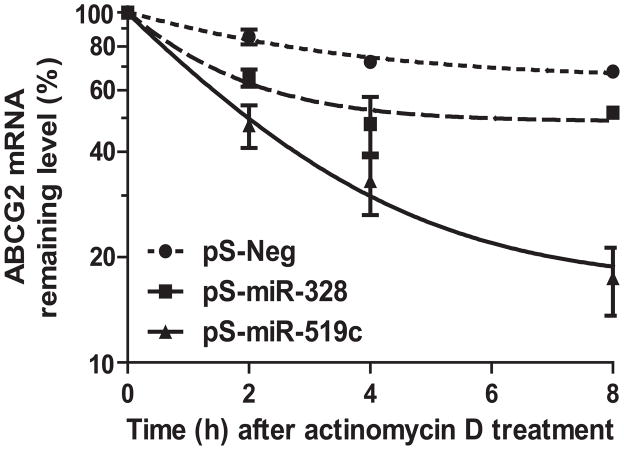
3.5. miR-519c-controlled regulation of ABCG2 likely involves mRNA degradation mechanism
RNA degradation experiments were conducted to assess if miR-328 and miR-519c affect ABCG2 mRNA stability. Actinomycin D was used to block nascent transcription in MCF-7/MX100 cells transfected with control, miR-328 or miR-519c expression plasmids. The data (Fig. 3) showed that miR-519c accelerated the decay (apparent half-life, ~ 2.0 h) of ABCG2 mRNA, compared with the control and miR-328 (half-life, > 8.0 h). The results suggest that mRNA degradation could be an important mechanism underlying miR-519c-mediated posttranscriptional regulation of ABCG2. The higher rate of mRNA decay caused by miR-519c than miR-328 is presumably due to the existence of multiple MRE sites, proximal (this study) and distal [15], for miR-519c.
Figure 3.
ABCG2 mRNA degradation is accelerated significantly (P < 0.001) in MCF-7/MX100 cells after transfection with miR-519c expression plasmid. Actinomycin D was utilized to block nascent RNA synthesis. Data represent mean ± SD (n = 3 in each group) of triplicate cultures. ABCG2 mRNA levels were normalized to GAPDH.
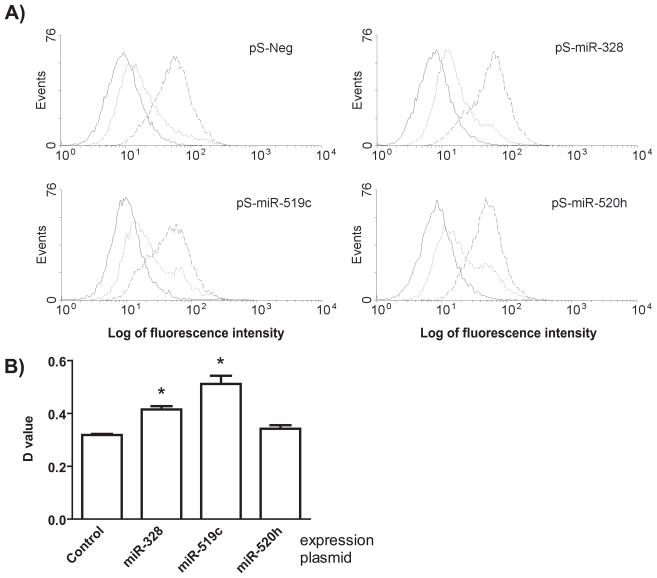
3.6. Intervention of miR-519c or miR-328 signaling affects ABCG2-mediated intracellular drug accumulation
We thus employed flow cytometry analyses to define the impact of miRNAs on ABCG2-mediated cellular drug disposition. Mitoxantrone, a selective substrate of ABCG2, was used as a model drug. Cells in the absence of mitoxantrone showed minimal fluorescence intensity, and fluorescence histogram was shifted to the right when cells were incubated with mitoxantrone (Fig. 4A). Difference between histograms of stained and unstained cells represents the level of drug accumulated within cells, which may be manifested by D value calculated with Kolmogorov-Smirnov statistics [43, 44] (Fig. 4B). As controls, co-incubation of mitoxantrone with the selective inhibitor of ABCG2, FTC, produced the maximal fluorescence intensity, indicating an extensive drug accumulation within MCF-7/MX100 cells when ABCG2 activity was blocked. Compared with the control, MCF-7/MX100 cells transfected with miR-519c or miR-328 expression plasmids had significantly higher D values (Fig. 4B). The results suggest that overexpression of miR-519c or miR-328 could increase intracellular accumulation of mitoxantrone, which is probably due to the decrease of ABCG2 protein expression (Fig. 2A). In contrast, no significant difference in mitoxantrone fluorescence was observed between cells transfected with control and miR-520h expression plasmids, which is consistent with the finding of an unchanged ABCG2 protein expression (Fig. 2A).
Figure 4.
Intervention of miR-519c or -328 signaling alters intracellular mitoxantrone accumulation. (A) Representative fluorescence histograms showing control autofluorescence (solid), and mitoxantrone content after 60 min of incubation in the absence (dotted) or presence (dashed) of 10 μM FTC in MCF-7/MX100 cells transfected with control or individual miRNA expression plasmids. (B) Drug accumulation was significantly (P < 0.05) different between cells transfected with control and miR-519c expression plasmid, as well as control and miR-328 expression plasmid, which was indicated by difference in D value (mean ± SD of triplicate treatments) between stained and unstained cells calculated with Kolmogorov-Smirnov test (CellQuest, BD Biosciences).
3.7. miR-519c, miR-328 and miR-520h are differentially expressed in stem-like ABCG2+ and non-stem-like ABCG- human retinoblastoma cells
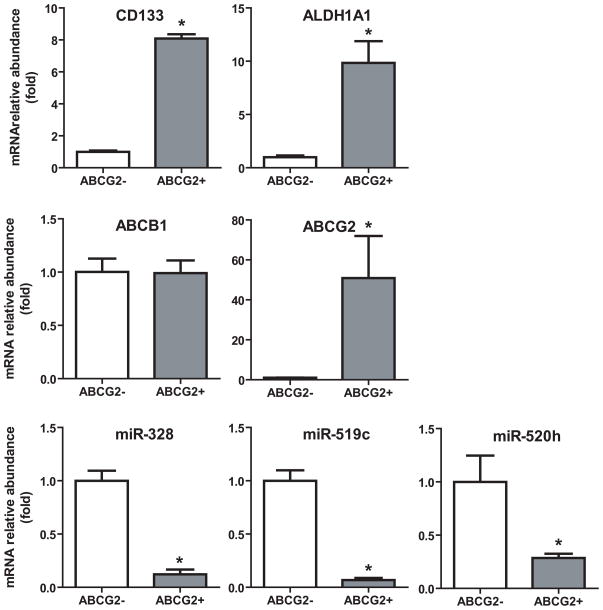
To further evaluate the role of miRNAs in ABCG2 regulation, we investigated the expression of miR-328, -519c and -520h in stem-like cancer cells that are characterized by ABCG2 overexpression. RB143 human retinoblastoma cells were separated into ABCG2+ (stem-like) and ABCG2− (non-stem-like) populations through immunomagnetic positive selection. RT-qPCR assays were used to determine the expression of miRNAs, ABC (ABCB1, ABCC1 and ABCG2) transporters, as well as the stem cell markers CD133 and ALDH1A1 [45]. ABCG2 mRNA expression was approximately 51-fold higher in ABCG2+ cells, a strong indication of cell enrichment by the immunomagnetic selection. As an additional validation of the stem cell phenotype, the stem cell markers ALDH1A1 and CD133 were approximately 10- and 8-fold higher in the ABCG2+ cells, respectively. The ABCC1 transporter was undetectable in both cell populations, whereas ABCB1 expression showed no difference between ABCG2− and ABCG2+ cells. Very interestingly, miR-328, -519c and -520h levels were approximately 9-, 15- and 3-fold lower in ABCG2+ cells, respectively. The degree of difference in miRNA expression between ABCG2+ enriched human retinoblastoma cells and ABCG2− counterparts agrees with the greater contribution from miR-519c and -328 to the regulation of ABCG2 observed in human breast cancer cells (Fig 2 and Fig 4).
4. Discussion
There is emerging evidence supporting the role for noncoding miRNAs in epigenetic regulation of drug-metabolizing enzymes and membrane transporters [4–8], which are molecular determinants of drug absorption, distribution, metabolism and excretion processes. The full length 3′UTR of ABCG2 transcript consists of 1968 bases that may be targeted by regulatory miRNAs [11, 15, 18]. However, in contrast to the existence of both long (~2000 nt) and short (100–400 nt) ABCG2 3′UTR transcripts in drug-sensitive cells, only the short forms were found in drug-resistant cells [14]. Because the short forms are lacking of the putative distal miR-519c MRE (1526–1548 nt) [15], it was suggested that miR-519c might not be able to affect ABCG2 expression in drug-resistant cells [14]. Our current study, however, showed that ABCG2 protein expression was significantly reduced in drug-resistant MCF-7/MX100 cells with a gained miR-519c function (Fig. 2A), in addition to a dramatic elevation of ABCG2 expression in drug-sensitive MCF-7 cells with a reduced miR-519c function (Fig. 2B). Indeed, miR-519c expression level was sharply increased in cells transfected with miR-519c expression plasmid (Table 2), indicating the existence of miR-519c processing machinery in drug-resistant MCF-7/MX100 cells. Furthermore, the suppression of ABCG2 protein expression was associated with a faster degradation of ABCG2 transcripts (Fig. 3), suggesting the involvement of an mRNA decay mechanism in miR-519c-controlled regulation. In addition, our computational analyses and biochemical experiments demonstrated that the proximal MRE site (30–52 nt) assigned to miR-520h was also accessible by miR-519c. The presence of proximal miR-519c MRE site (Fig 1A) may provide a good explanation for the obvious suppression of ABCG2 (Fig 2A) in drug-resistant cells when miR-519c expression/function is restored (Table 2). Rather, the overexpression of ABCG2 in drug-resistant cancer cells could be, at least partially, attributable to the defect of miR-519c expression (Table 2), in addition to the shortening of 3′UTR [14] and gene amplification [25–27].
It is noteworthy that one gene is potentially targeted by a number of miRNAs. Recent studies did show the possible involvement of multiple miRNAs, namely miR-520h, -519c and -328, in posttranscriptional regulation of ABCG2 membrane transporter [11, 15, 18, 31]. In particular, miR-328 modulates intrinsic ABCG2 expression in MCF-7 breast adenocarcinoma cell lines [11], miR-519c regulates endogenous ABCG2 expression in the A549 adenocarcinomic human alveolar basal epithelial cell line [15], and miR-520h changes natural ABCG2 expression in PANC-1 human pancreatic carcinoma cell line [18]. Our current study provided a side-by-side comparison of miR-328, -519c and -520h in regulation of ABCG2 expression in MCF-7 human breast cancer cell lines. Our results (Fig. 2), from both gain- and loss-of-function studies, clearly demonstrated that miR-519c and miR-328 had much greater impact on ABCG2 protein expression than miR-520h in human breast cancer cells. The lack of ABCG2 regulation by miR-520h may be attributed to the deficient processing of miR-520h precursors in MCF-7/MX100 cells (Table 2). As described above and in our previous study [11], the overall effects of an miRNA on ABCG2 expression may be related to the existence and availability of MRE sites, target site accessibility, miRNA abundance and regulatory mechanisms. Due to the difference in one or two factors, e.g., miRNA or mRNA expression profiles, the findings obtained from one type of cell might not be predictable for others.
As shown in other studies [11–13, 16, 19], miRNA-directed change of transporter expression, e.g., ABCB1/MDR1, ABCC1/MRP1 and ABCG2/BCRP, may result in an altered cellular drug disposition and chemosensitivity. Our current study indicated that down-regulation of ABCG2 protein expression (Fig. 2) by miR-519c and -328 led to a modest increase of intracellular accumulation of mitoxantrone (Fig. 4), which could be determined by the degree of change in ABCG2 transporter, selectivity of test drug, and other experimental conditions (e.g., dose, time point and transfection efficiency). These findings suggest that miRNAs could be important factors in regulation of drug disposition, and support the notion that modulation of miRNA expression or function may influence cellular defense against xenobiotic drugs.
Besides the important role in pharmacokinetics, ABCG2 is a suggested marker of stem cells that are capable of self-renewal and differentiation. With the development of the “cancer stem cell” hypothesis [46], high expression of ABCG2 is reported to confer the “side population” cells that efflux Hoechst dye 33342 [28–30], as well as a subpopulation of melanoma cells expressing a high percentage of stem cell marker CD133 [47]. Results from our current study agree with our recent findings that ABCG2 may serve as stem cell marker in human retinoblastoma cells [45]. High expression of other stem cell markers, CD133 and ALDH1A1, was also found in ABCG2+ cells (Fig 5). The most significant finding is the low expression of miR-519c, -328 and -520h in ABCG2+ enriched cells, which is consistent with the reduced expression of ABCG2-regulatory miR-328 found in ABCG2-overexpressing MCF-7/MX100 cells [11] but different from overexpression of miR-520h in CD3+/CD38− human hematopoietic stem cells isolated through fluorescence-activated cell sorting [31]. Our finding of differential expression of ABCG2-regulatory miRNAs in stem-like ABCG2+ cells and their ABCG2− counterparts suggests that further investigation of miRNA signaling in stem cells may provide new insights into ABCG2-mediated chemoresistance and metastasis.
Figure 5.
miR-328, -519c and -520h expression levels are significantly (P < 0.05) lower in stem-like ABCG2+ human retinoblastoma RB143 cells. Note that the stem cell markers ALDH1A1 and CD133 are significantly (P < 0.05) higher in ABCG2+ cells. Data represent mean ± SEM of a total of six measurements in two separate enrichments. ALDH1A1, CD133 and ABC transporter levels were normalized to 18S levels, and miRNA levels were normalized to U74 levels.
In summary, the known miR-520h MRE site from 30 to 52 nt within ABCG2 3′UTR is also accessible by miR-519c-3p. The existence of this proximal and previously-identified distal MRE sites for miR-519c may confer the efficient regulation of ABCG2 by miR-519c, which involves an mRNA degradation mechanism. Our findings also suggest that miR-519c and miR-328 are relatively more important in control of ABCG2 expression in MCF-7 human breast cancer cells, and intervention of miRNA signaling may alter intracellular drug accumulation. In addition, the differential expression of miR-519c, -328 and -520h in ABCG2+ enriched human retinoblastoma cells provides novel insights into understanding miRNA signaling in stem cancer cells.
Acknowledgments
Acknowledgements and disclosure: A-M Yu is supported by grant R01DA021172 from National Institute on Drug Abuse, National Institutes of Health (NIH), and by the Interdisciplinary Research Development Fund from University at Buffalo, The State University of New York. GM Seigel is supported by R21CA127061, Research to Prevent Blindness and U54CA143876 from the National Cancer Institute, NIH. Xin Li was supported by International Program of Project 985 at Sun Yat-Sen University and National Major Projects for science and technology development from Science and Technology Ministry of China, 2009ZX09304-003.
Abbreviations
- miRNA
microRNA
- BCRP/ABCG2
Breast cancer resistance protein/ATP-binding cassette, subfamily G (white), member 2
- hsa-miR-328
human microRNA-328
- miR-519c
microRNA-519c
- miR-520h
microRNA-520h
- MRE
microRNA response element
- 3′UTR
3′-untranslated region
- ALDH1A1
aldehyde dehydrogenase 1A1
- MDR1/ABCB1
P-glycoprotein
- MRP1/ABCC1
multidrug resistance-associated protein 1
- FTC
fumitremorgin C
- PBS
phosphate-buffered saline
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 3.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu AM. Role of microRNAs in the regulation of drug metabolism and disposition. Expert Opin Drug Metab Toxicol. 2009;5(12):1513–28. doi: 10.1517/17425250903307448. [DOI] [PubMed] [Google Scholar]
- 5.Yu AM. Small interfering RNA in drug metabolism and transport. Curr Drug Metab. 2007;8(7):700–8. doi: 10.2174/138920007782109751. [DOI] [PubMed] [Google Scholar]
- 6.Ingelman-Sundberg M, Sim SC, Gomez A, Rodriguez-Antona C. Influence of cytochrome P450 polymorphisms on drug therapies: pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacol Ther. 2007;116(3):496–526. doi: 10.1016/j.pharmthera.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 7.Choudhuri S, Cui Y, Klaassen CD. Molecular targets of epigenetic regulation and effectors of environmental influences. Toxicol Appl Pharmacol. 2010;245(3):378–93. doi: 10.1016/j.taap.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gomez A, Ingelman-Sundberg M. Epigenetic and microRNA-dependent control of cytochrome P450 expression: a gap between DNA and protein. Pharmacogenomics. 2009;10(7):1067–76. doi: 10.2217/pgs.09.56. [DOI] [PubMed] [Google Scholar]
- 9.Tsuchiya Y, Nakajima M, Takagi S, Taniya T, Yokoi T. MicroRNA regulates the expression of human cytochrome P450 1B1. Cancer Res. 2006;66(18):9090–8. doi: 10.1158/0008-5472.CAN-06-1403. [DOI] [PubMed] [Google Scholar]
- 10.Pan YZ, Gao W, Yu AM. MicroRNAs regulate CYP3A4 expression via direct and indirect targeting. Drug Metab Dispos. 2009;37(10):2112–7. doi: 10.1124/dmd.109.027680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan YZ, Morris ME, Yu AM. MicroRNA-328 negatively regulates the expression of breast cancer resistance protein (BCRP/ABCG2) in human cancer cells. Mol Pharmacol. 2009;75(6):1374–9. doi: 10.1124/mol.108.054163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kovalchuk O, Filkowski J, Meservy J, Ilnytskyy Y, Tryndyak VP, Chekhun VF, Pogribny IP. Involvement of microRNA-451 in resistance of the MCF-7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol Cancer Ther. 2008;7(7):2152–9. doi: 10.1158/1535-7163.MCT-08-0021. [DOI] [PubMed] [Google Scholar]
- 13.Zhu H, Wu H, Liu X, Evans BR, Medina DJ, Liu CG, Yang JM. Role of MicroRNA miR-27a and miR-451 in the regulation of MDR1/P-glycoprotein expression in human cancer cells. Biochem Pharmacol. 2008;76(5):582–8. doi: 10.1016/j.bcp.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.To KK, Robey RW, Knutsen T, Zhan Z, Ried T, Bates SE. Escape from hsa-miR-519c enables drug-resistant cells to maintain high expression of ABCG2. Mol Cancer Ther. 2009;8(10):2959–68. doi: 10.1158/1535-7163.MCT-09-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.To KK, Zhan Z, Litman T, Bates SE. Regulation of ABCG2 expression at the 3′ untranslated region of its mRNA through modulation of transcript stability and protein translation by a putative microRNA in the S1 colon cancer cell line. Mol Cell Biol. 2008;28(17):5147–61. doi: 10.1128/MCB.00331-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang Z, Wu H, Xia J, Li Y, Zhang Y, Huang K, Wagar N, Yoon Y, Cho HT, Scala S, Shim H. Involvement of miR-326 in chemotherapy resistance of breast cancer through modulating expression of multidrug resistance-associated protein 1. Biochem Pharmacol. 2010;79(6):817–24. doi: 10.1016/j.bcp.2009.10.017. [DOI] [PubMed] [Google Scholar]
- 17.Mohri T, Nakajima M, Fukami T, Takamiya M, Aoki Y, Yokoi T. Human CYP2E1 is regulated by miR-378. Biochem Pharmacol. 2010;79(7):1045–52. doi: 10.1016/j.bcp.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 18.Wang F, Xue X, Wei J, An Y, Yao J, Cai H, Wu J, Dai C, Qian Z, Xu Z, Miao Y. hsa-miR-520h downregulates ABCG2 in pancreatic cancer cells to inhibit migration, invasion, and side populations. Br J Cancer. 2010;103(4):567–74. doi: 10.1038/sj.bjc.6605724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo L, Liu Y, Bai Y, Sun Y, Xiao F, Guo Y. Gene expression profiling of drug-resistant small cell lung cancer cells by combining microRNA and cDNA expression analysis. Eur J Cancer. 2010;46(9):1692–702. doi: 10.1016/j.ejca.2010.02.043. [DOI] [PubMed] [Google Scholar]
- 20.Takagi S, Nakajima M, Mohri T, Yokoi T. Post-transcriptional regulation of human pregnane X receptor by micro-RNA affects the expression of cytochrome P450 3A4. J Biol Chem. 2008;283(15):9674–80. doi: 10.1074/jbc.M709382200. [DOI] [PubMed] [Google Scholar]
- 21.Ji J, Zhang J, Huang G, Qian J, Wang X, Mei S. Over-expressed microRNA-27a and 27b influence fat accumulation and cell proliferation during rat hepatic stellate cell activation. FEBS Lett. 2009;583(4):759–66. doi: 10.1016/j.febslet.2009.01.034. [DOI] [PubMed] [Google Scholar]
- 22.Takagi S, Nakajima M, Kida K, Yamaura Y, Fukami T, Yokoi T. MicroRNAs regulate human hepatocyte nuclear factor 4alpha, modulating the expression of metabolic enzymes and cell cycle. J Biol Chem. 2010;285(7):4415–22. doi: 10.1074/jbc.M109.085431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krishnamurthy P, Schuetz JD. Role of ABCG2/BCRP in biology and medicine. Annu Rev Pharmacol Toxicol. 2006;46:381–410. doi: 10.1146/annurev.pharmtox.46.120604.141238. [DOI] [PubMed] [Google Scholar]
- 24.van Herwaarden AE, Schinkel AH. The function of breast cancer resistance protein in epithelial barriers, stem cells and milk secretion of drugs and xenotoxins. Trends Pharmacol Sci. 2006;27(1):10–6. doi: 10.1016/j.tips.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 25.Ross DD, Yang W, Abruzzo LV, Dalton WS, Schneider E, Lage H, Dietel M, Greenberger L, Cole SP, Doyle LA. Atypical multidrug resistance: breast cancer resistance protein messenger RNA expression in mitoxantrone-selected cell lines. J Natl Cancer Inst. 1999;91(5):429–33. doi: 10.1093/jnci/91.5.429. [DOI] [PubMed] [Google Scholar]
- 26.Volk EL, Farley KM, Wu Y, Li F, Robey RW, Schneider E. Overexpression of wild-type breast cancer resistance protein mediates methotrexate resistance. Cancer Res. 2002;62(17):5035–40. [PubMed] [Google Scholar]
- 27.Knutsen T, Rao VK, Ried T, Mickley L, Schneider E, Miyake K, Ghadimi BM, Padilla-Nash H, Pack S, Greenberger L, Cowan K, Dean M, Fojo T, Bates S. Amplification of 4q21-q22 and the MXR gene in independently derived mitoxantrone-resistant cell lines. Genes Chromosomes Cancer. 2000;27(1):110–6. [PubMed] [Google Scholar]
- 28.Zhou S, Schuetz JD, Bunting KD, Colapietro AM, Sampath J, Morris JJ, Lagutina I, Grosveld GC, Osawa M, Nakauchi H, Sorrentino BP. The ABC transporter Bcrp1/ABCG2 is expressed in a wide variety of stem cells and is a molecular determinant of the side-population phenotype. Nat Med. 2001;7(9):1028–34. doi: 10.1038/nm0901-1028. [DOI] [PubMed] [Google Scholar]
- 29.Kim M, Turnquist H, Jackson J, Sgagias M, Yan Y, Gong M, Dean M, Sharp JG, Cowan K. The multidrug resistance transporter ABCG2 (breast cancer resistance protein 1) effluxes Hoechst 33342 and is overexpressed in hematopoietic stem cells. Clin Cancer Res. 2002;8(1):22–8. [PubMed] [Google Scholar]
- 30.Scharenberg CW, Harkey MA, Torok-Storb B. The ABCG2 transporter is an efficient Hoechst 33342 efflux pump and is preferentially expressed by immature human hematopoietic progenitors. Blood. 2002;99(2):507–12. doi: 10.1182/blood.v99.2.507. [DOI] [PubMed] [Google Scholar]
- 31.Liao R, Sun J, Zhang L, Lou G, Chen M, Zhou D, Chen Z, Zhang S. MicroRNAs play a role in the development of human hematopoietic stem cells. J Cell Biochem. 2008;104(3):805–17. doi: 10.1002/jcb.21668. [DOI] [PubMed] [Google Scholar]
- 32.Apati A, Orban TI, Varga N, Nemeth A, Schamberger A, Krizsik V, Erdelyi-Belle B, Homolya L, Varady G, Padanyi R, Karaszi E, Kemna EW, Nemet K, Sarkadi B. High level functional expression of the ABCG2 multidrug transporter in undifferentiated human embryonic stem cells. Biochim Biophys Acta. 2008;1778(12):2700–9. doi: 10.1016/j.bbamem.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 33.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320(5883):1643–7. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138(4):673–84. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 36.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39(10):1278–84. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 37.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34(Database issue):D140–4. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126(6):1203–17. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 39.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33(20):e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robey RW, Honjo Y, van de Laar A, Miyake K, Regis JT, Litman T, Bates SE. A functional assay for detection of the mitoxantrone resistance protein, MXR (ABCG2) Biochim Biophys Acta. 2001;1512(2):171–82. doi: 10.1016/s0005-2736(01)00308-x. [DOI] [PubMed] [Google Scholar]
- 41.Zhou XF, Yang X, Wang Q, Coburn RA, Morris ME. Effects of dihydropyridines and pyridines on multidrug resistance mediated by breast cancer resistance protein: in vitro and in vivo studies. Drug Metab Dispos. 2005;33(8):1220–8. doi: 10.1124/dmd.104.003558. [DOI] [PubMed] [Google Scholar]
- 42.Merino G, Real R, Baro MF, Gonzalez-Lobato L, Prieto JG, Alvarez AI, Marques MM. Natural allelic variants of bovine ATP-binding cassette transporter ABCG2: increased activity of the Ser581 variant and development of tools for the discovery of new ABCG2 inhibitors. Drug Metab Dispos. 2009;37(1):5–9. doi: 10.1124/dmd.108.022715. [DOI] [PubMed] [Google Scholar]
- 43.Young IT. Proof without prejudice: use of the Kolmogorov-Smirnov test for the analysis of histograms from flow systems and other sources. J Histochem Cytochem. 1977;25(7):935–41. doi: 10.1177/25.7.894009. [DOI] [PubMed] [Google Scholar]
- 44.Garcia-Escarp M, Martinez-Munoz V, Sales-Pardo I, Barquinero J, Domingo JC, Marin P, Petriz J. Flow cytometry-based approach to ABCG2 function suggests that the transporter differentially handles the influx and efflux of drugs. Cytometry A. 2004;62(2):129–38. doi: 10.1002/cyto.a.20072. [DOI] [PubMed] [Google Scholar]
- 45.Seigel GM, Hackam AS, Ganguly A, Mandell LM, Gonzalez-Fernandez F. Human embryonic and neuronal stem cell markers in retinoblastoma. Mol Vis. 2007;13:823–32. [PMC free article] [PubMed] [Google Scholar]
- 46.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–11. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 47.Monzani E, Facchetti F, Galmozzi E, Corsini E, Benetti A, Cavazzin C, Gritti A, Piccinini A, Porro D, Santinami M, Invernici G, Parati E, Alessandri G, La Porta CA. Melanoma contains CD133 and ABCG2 positive cells with enhanced tumourigenic potential. Eur J Cancer. 2007;43(5):935–46. doi: 10.1016/j.ejca.2007.01.017. [DOI] [PubMed] [Google Scholar]