Abstract
Recent studies in vitro have shown that the CREB co-activator, Transducer of Regulated CREB activity (TORC), is required for transcriptional activation of the corticotropin releasing hormone (CRH) gene. To determine the physiological importance of TORC 2 regulating CRH transcription during stress, we examined the localization of TORC 2 in CRH neurons, and the relationship between changes in CRH hnRNA, nuclear translocation of TORC 2, and binding of TORC 2 to the CRH promoter. Immunohistochemistry revealed TORC 2 immunoreactivity (irTORC 2) in the dorsolateral (magnocellular) and dorsomedial (parvocellular) regions of the hypothalamic paraventricular nucleus (PVN). While staining was mostly cytosolic in basal conditions, there was a marked increase in nuclear irTORC 2 in the dorsomedial region at 30 min restraint, concomitant with increases in CRH hnRNA levels. Levels of nuclear irTORC 2 and CRH hnRNA had returned to basal 4h after stress. Double staining immunohistochemistry showed TORC 2 co-staining in 100% of detected CRH neurons, and nuclear translocation following 30 min restraint in 61%. Cellular distribution of TORC 2 in the dorsolateral PVN was unaffected by restraint. Chromatin immunoprecipitation (ChIP) experiments showed recruitment of TORC 2 and phospho-CREB by the CRH promoter following 30min restraint, but 4h after stress only phospho-CREB was associated with the CRH promoter. The demonstration that TORC 2 translocates to the nucleus of hypothalamic CRH neurons and interacts with the CRH promoter in conjunction with the activation of CRH transcription during restraint stress, provides strong evidence for the involvement of TORC 2 in the physiological regulation of CRH transcription.
Keywords: TORC 2, hypothalamus, corticotropin releasing hormone, transcription
INTRODUCTION
The regulation of CRH transcription depends on cAMP/protein kinase A (PKA) dependent pathways (1-5), leading to recruitment of phosphorylated cAMP response element binding protein (pCREB) by a cAMP response element (CRE) located at position −230 in the CRH promoter (4,6-8). This CRE is essential for cyclic AMP dependent and independent regulation of CRH transcription (7,9,10). However, more recently evidence has indicated that phospho-CREB alone is not sufficient to support CRH transcription. While a number of signaling pathways lead to CREB phosphorylation (11,12), cAMP signaling is required for stimulating CRH expression (2,3,6,9,13,14). In studies using the neuronal cell line, 4B, or hypothalamic primary neuronal cultures, treatment with either the phorbol ester phorbol-12-myristate-13-acetate (PMA) a protein kinase C activator, or forskolin (adenylate cyclase activator) both led to CREB phosphorylation. Nevertheless, only forskolin treatment was able to stimulate CRH transcription, as shown by reporter gene assays in 4B cells, or the induction of CRH primary transcript in primary cultures of hypothalamic neurons (6). A similar situation has been suggested in experiments in rats in which microinjection of both 8-bromo-cAMP and the phorbol ester, TPA in the PVN increase plasma ACTH and pituitary POMC mRNA levels, but only cAMP augments CRH mRNA levels (15). Although not sufficient to drive CRH transcription, CREB is required for CRH promoter activation since the CREB dominant negative, A-CREB, blunts the stimulatory effect of forskolin on CRH promoter activity (6). The above evidence suggested that activation of CRH transcription requires the presence of a CREB co-activator which requires cyclic AMP for its activation. Recent studies have confirmed this premise and provide strong evidence that such a co-activator is Transducer Of Regulated CREB activity (TORC) (16).
The TORC family, comprising TORC 1, 2 and 3, was discovered and characterized in 2003 by two groups while independently searching for proteins that interacted with CREB (17,18). TORC potentiates the transcriptional activity of CREB by binding to its dimerization domain and facilitating the recruitment of the transcriptional complex. In basal conditions, TORC is in the cytoplasm in a phosphorylated inactive state and bound to the scaffolding protein, 14-3-3. Activation of cyclic AMP/PKA signaling causes rapid TORC shuttling to the nucleus (17-19). In the nucleus, TORC 2 interacts with the dimerized leucine zipper of the CREB DNA-binding and dimerization domain (bZip). TORC recruitment does not influence CREB DNA binding activity, but following its recruitment by the promoter enhances the interaction of CREB with the TATA binding protein associated factor (TAF4) component of Transcription Factor II D (TFIID) (17). Studies in the hypothalamic cell line 4B show TORC translocation to the nucleus upon stimulation of the cells with forskolin. In the same study, TORC overexpression potentiates the stimulatory effect of forskolin on CRH promoter activity. Conversely, in 4B cells and primary cultures of hypothalamic neurons, knockout of TORC (especially TORC 2) using siRNA blunts CRH transcription (16).
The aim of the present study is to determine whether TORC 2 is involved in the regulation of CRH transcription during stress in the rat. Immunohistochemical studies were conducted to investigate the presence of TORC 2 in CRH neurons of the PVN. In addition, the effects of restraint stress on nuclear translocation of TORC 2 and the recruitment of phospho-CREB and TORC 2 by the CRH promoter were examined.
MATERIALS AND METHODS
Animals and in vivo procedures
Adult male Sprague-Dawley rats weighing 275-325g were housed three per cage on a 14-10h light-dark cycle with food and water available ad libitum, for at least one week prior to experimentation. To determine the distribution of TORC 2 in the PVN and the time course of the changes in TORC 2, rats were subjected to restraint stress by placing them into plastic restrainers (2.5 × 6 inches) for 1h. Groups of 4 rats were killed by decapitation at 0.5 or at 4 hours (3h after initiation of 1h restraint). Control rats were removed from the cages and killed within 30 seconds. Brains were immediately removed and either frozen in isopentane at −40 °C and stored at −80 °C until sectioned in a cryostat for in situ hybridization. For immunohistrochemistry, coronal brain pieces (or blocks) in between the optic chiasm and the mammillary bodies were fixed by immersion in 4% paraforamaldehyde for 8h at 4° C before transferring to 30% sucrose and kept at 4° C until sectioning. For western blot and chromatin immunoprecipitation assay (ChIP), the hypothalamic region microdissected from a coronal section in between the optic chiasma and 1 mm rostral from the mammillary bodies. Sections were placed flat on a chilled rubber cork and cut at the top and 1mm lateral at each side of the 3rd ventricle with a scalpel blade. After removing an additional 1mm from the ventral side, in order to exclude the suprachiasmatic, supraoptic and arcuate nuclei, hypothalamic blocks weighting about 25 mg, containing the PVN were frozen in 1.5 ml microtubes on dry ice. The whole procedure from decapitation to freezing of the tissue was performed in about 3 min.
Trunk blood was collected in plastic tubes and serum was separated by centrifugation and stored at −80 °C for ACTH and corticosterone determination. All experiments were performed in the morning with rats killed between 9 and 11 AM. All procedures and experimental protocols were performed according to NIH guidelines and approved by the NICHD Animal Care and Use Committee.
ACTH and Corticosterone Assays
Blood samples for ACTH and corticosterone measurement were collected in ice-chilled 50 ml tubes containing 100 μl of 500 mM EDTA and 500 TIU of aprotinin. After centrifugation for 10 min, 1500 x g, plasma was stored at −80°C until time of assay. Plasma levels of ACTH were measured using kit reagents from ACTH IRMA Immunoradiometric Assay, USA DiaSorin (Stillwater, Minnesota) according to the manufacturer’s instructions. Corticosterone levels were measured using the Rat Corticosterone Coat-A-Count kit (Diagnostic Products Corporation (DPC), Los Angeles,CA) according to the manufacturer’s instructions.
In Situ Hybridization Histochemistry
Serial coronal sections (12 μm) were cut through the medial parvocellular subdivision of the PVN in a cryostat (−15°C), thaw mounted onto poly-L-lysine coated slides (4 sections per slide), and stored at −80°C until processed for in situ hybridization. Transcription of antisense 35S-labeled probes was performed as previously described after linearization with XbaI of intronic CRH plasmid, kindly provided by Dr Robert Thompson (Ann Arbor, MI).
Prehybridization and hybridization procedures were performed as previously described (20). Briefly, prior to hybridization, sections were thawed at room temperature, fixed in 4% formaldehyde for 5 min at room temperature, washed 3 times with PBS, and acetylated for 10 min, at room temperature in 0.25% acetic anhydride in 0.1M triethanolamine/0.9% NaCl (pH 8.0). Sections were dehydrated and delipidated by sequential transfers through ethanol and chloroform and air dried before hybridization. Sections were hybridized at 55°C, overnight, with 2 × 106 cpm 35-S labeled probe, and then non-specifically bound probe was removed by washing in 50% formamide/250 mM NaCl at 60°C for 10-15 min, followed by ribonuclease A treatment for 30 min at 37°C and 3 washes in 0.1 × SSC at 50°C for 15 min. Following the final wash, sections were dried at room temperature and opposed to Biomax MR film for two weeks, developed, and fixed (Eastman Kodak, Rochester, NY). The optical density of film autoradiographic images of parvocellular CRH hnRNA was measured in a computerized image analysis system (Imaging Research, St. Catherine, Ontario, Canada), using the public domain ImageJ program (developed at the National Institutes of Health, and available at http://rsb.info.nih.gov/NIH-Image). Optical densities obtained in two consecutive sections containing the medial parvocellular subdivision per rat were averaged and used to calculate group means. The results are presented as mean and SE of the fold-change over basal levels.
Western blot
Cytosolic and nuclear proteins from the PVN region were prepared using kit reagents from NE-PER™ Nuclear and Cytoplasmic Extraction Reagent (Pierce, Rockford, IL) as previously described (16). Protein concentration was quantified by spectrophotometry using the BCA Protein Assay (Pierce, Thermo Fisher Scientific Inc, Rockford, IL). For Western Blot, 15μg of cytoplasmic or nuclear extract was loaded and separated in a 10% Tris-Glycine gel (Invitrogen) for beta actin and histone deacetylase 1 (HDAC1), or a 6% Tris-Glycine gel (Invitrogen) for TORC 2. Proteins were transferred to a PVDF membrane (GE Amersham Biosciences), incubated with 5% non-fat milk in 1 x TBST (TBS plus 0.05% Tween-20) for 1h and incubated overnight at 4°C with TORC 2 (Calbiochem/EDM Chemicals, Gibbstown, NJ), at 1:6000 dilution. After washing in 1 x TBST, membranes were incubated for 1h at room temperature with a horseradish peroxidase-conjugated donkey anti-rabbit IgG at a dilution of 1:10,000. Detection of immunoreactive bands was performed using ECL Plus TM reagents (GE Amersham Biosciences) followed by exposure to BioMax MR film (Eastman Kodak, Rochester, NY, USA). Blots for cytoplasmic and nuclear proteins were exposed in the same film. After film exposure, blots were stripped and assayed for HDAC1 in the nucleus and β-actin in cytoplasm as a loading control. The intensity of the bands was quantified using the computer image analysis system, ImageJ. Results are expressed as fold-change over the values in control rats after correction for protein loading using HDAC1 for the nucleus and beta-actin for cytoplasm.
Immunostaining for TORC 2 in the rat brain sections
Hypothalamic blocks of three rats per each experimental group were embedded in 3% agarose and sectioned on vibratome Leica, Germany (thickness = 40 μm). After briefly washing with PBS, sections were incubated in 0.5% H2O2 in PBS for 1 hr at room temperature, washed in PBS (6 × 10 min) and incubated overnight in PBS containing 1% Triton X100 and 10% normal goat serum (NGS) at 4°C to block non-specific binding of antibodies. Sections were then incubated for 24 hrs at 4°C with rabbit anti TORC 2 antibodies (Calbiochem, 1:10.000. Lot # D00062801 and Lot # D00069636) diluted in PBS containing 1% NGS and 0.5% Triton X100. Next day sections were incubated with biotinylated goat anti-rabbit IgG (Vector Elite kit, Vector Laboratories, Inc., Burlingame, CA; 1:500) in PBS containing 0,05% Triton X100 and 5% NGS, for 2 hours at room temperature. After washing in PBS for 1 h, ABC complex (Vector Laboratories, CA) in PBS was applied for 1 h at RT. Finally, after 6 × 10 min washing in PBS, the TORC 2 immunoproduct was visualized with freshly prepared diaminobenzidine (Sigma) and H2O2 in PBS for 10 min. After rinsing in PBS, sections were dehydrated through ascending concentrations of ethanol, cleared in xylol and mounted with embedding medium (Entellan, Merck, Rahway, NJ). Sections processed without application of the primary TORC 2 antibodies were served as a control of specificity of immunostaining.
For semi-quantitative analysis of number of TORC 2 immunoreactive cells in parvo- and magnocellular subdivisions of the PVN, sections containing distinct parvo- and magnocellular subdivisions of the PVN were selected for the analysis. Images taken at 20x on Zeiss Axioskop 2 plus microscope coupled with Zeiss AxioCam HRc camera were then analyzed in Adobe Photoshop (Adobe, Mountain View, CA) manually by placing a frame 200 × 300 μm over the parvocellular subdivision of the PVN and a frame 150 × 150 μm over magnocellular PVN subdivision. Cell counts were made on two or three sections per each region, bilaterally, in each animal and performed by an operator blind to the experimental treatment. Results represent the number of cells containing either nuclear or cytoplasmic TORC 2 staining per 100 neurons. One Way ANOVA-based statistic was performed using Webpage: http://faculty.vassar.edu.
To evaluate colocalization of TORC 2 with CRH, double fluorescent immunostaining was performed. Sections were preincubated in PBS containing 1% Triton X100 and 10% NGS overnight at 4°C, prior to incubation for 24 hrs at 4°C with primary rabbit antibodies against TORC 2 (Calbiochem, 1:1,000, the same lot as for DAB-based staining), and guinea pig antibodies against CRH (Peninsula Labs, 1:10,000) diluted in PBS containing 1% NGS and 0.5% Triton X100 (21). Sections were then washed 6 × 10 min with PBS before addition of FITC-conjugated goat anti-rabbit and a Cy3-conjugated donkey anti-guinea pig IgGs (Jackson ImmunoResearch Labs, Inc., 1:500), for visualization of TORC 2 and CRH immunosignals, respectively. Sections were washed with PBS (6 × 10 min at RT) before mounting with Mowiol (Sigma).
To further demonstrate nuclear localization of TORC2 in CRH cells of rats subjected to 30min restraint, double stained sections were counterstained with 4′,6-diamidino-2-phenylindole (DAPI, Sigma-Aldrich). DAPI was added in PBS (1μg/ml) during the last washing of sections prior to mounting.
Sections processed without application of the primary TORC 2 or CRH antibodies served as a control of specificity of immunostaining. All images were taken as a single plane scan on a confocal microscope Leica TCS NT microscope.
For semi-quantitative analysis of the number of CRH-IR neurons containing nuclear TORC 2 immunostaining, one hundred immunoreactive CRH neurons in the PVN from each animal of the 30 min restraint group were analyzed in Adobe Photoshop on single confocal images taken at 63x. CRH immunoreactive neurons with clearly visible TORC 2 immunostaining in their nuclei were manually counted. The results represent the mean ± SE of the percent of CRH neurons displaying nuclear TORC 2-immunoreactivity in 3 rats per experimental group.
Chromatin immunoprecipitation assay
To determine whether TORC 2 is recruited by the CRH promoter during activation of CRH transcription during stress, we performed chromatin immunoprecipitation (ChIP) assays. ChIP assays were performed using kit reagents from the ChIP-IT Express kit (Active Motif, Carlsbad, CA), according to the manufacturers protocol with some modifications. Briefly, pooled hypothalamic tissue from 2 or 3 rats (about 50 to 75 mg) was homogenized in 500 μl of 1% formaldehyde using a hand held motorized homogenizer Bio-Gen Pro200 (Pro Scientific, Oxford, CT), for 10sec, setting 2, and incubated for 10 min at room temperature in order to cross-link the DNA–protein complexes. After terminating the cross-linking reaction with glycine, homogenates were centrifuged and washed three times in cold PBS containing protease inhibitors and phosphatase inhibitors. Pellets were then resuspended in 300 μl of SDS lysis buffer (Millipore/Upstate, Temecula, CA) with all the inhibitors and incubated on ice for 10 minutes, and sonicated 8 cycles of 30 seconds on and 30 seconds off at high level output (Bioruptor from Diagenode, Belgium) to generate 0.2-1 kb DNA fragments of chromatin.
After preclearing using Protein A/G plus beads (Santa Cruz Biotech, Santa Cruz, CA), immunoprecipitation was performed on 15 μg of chromatin with 3 μg of either anti-TORC 2 antibody (Bethyl, Montgomery, TX) or phospho-CREB antibody, Millipore/Upstate, Temecula, CA), or rabbit IgG for negative control in the presence of ChIP-IT Protein G Magnetic beads (Active Motif) at 4 °C under rotation, in a total volume of 200μl following the manufacturer’s protocol (Active Motif). Prior to addition, Protein G Magnetic beads were preincubated with one μg of herring sperm DNA per immunoprecipitation reaction in order to reduce the background. After overnight incubation DNA-protein immuno-complexes were collected by Protein G Magnetic beads and followed by washing, elution and reversing cross-links. After treatment with protinase K, The immunoprecipitated DNA fragments was quantified using real-time PCR with the following primers designed to amplify the CRH promoter region encompassing the 112 bp containing the CRE (forward, 5′- tcagtatgttttccacacttggat-3′ and reverse, 5′-tttatcgcctccttggtgac-3′). Calibration curves to assess the efficiency of the PCR reaction were performed using 0.025 to 250ng of rat genomic DNA were linear between 23 and 39 Ct. The amplification efficiency of the primers was usually about 100%, ranging from 90 to 110%. In immunoprecipitated chromatin for animals in basal conditions or 3h after termination of stress, CRH promoter detection was in the low end of the standard curve (33 to 37 cycles). In immunoprecipitated chromatin from rats subjected to 30 min restraint, CRH promoter was detected at 26 to 29 cycles. The amount of CRH promoter calculated from the standard curve was normalized by the total input (amount of CRH promoter in chromatin not subjected to immunoprecipitation).
Analysis of results and statistical analysis
Statistical analysis was performed by one-way analysis of variance (ANOVA) followed by Fisher’s least significant difference procedure (PLSD) test to assess statistical significance between control and experimental groups. Data are presented as the mean and SE of the values obtained in the number of experiments indicated in results or legends to the figures. For ChIP experiments, data were normalized by log transformation prior to statistic analysis.
RESULTS
Plasma hormone and hypothalamic CRH hnRNA responses to restraint stress
Plasma ACTH and corticosterone levels in basal conditions were 64.8±6.3 pg/ml, and 82.1±10.1 ng/ml, respectively. Values increased significantly at 30 min restraint, (407.6±28.0 pg/ml for ACTH, and 494.2±39.2 ng/ml for corticosterone), and had returned to basal at 4h, 3h after the termination of the stress (37.6±3.9 pg/ml and 26.6±4.0 ng/ml for ACTH and corticosterone respectively), n=8, p<0.001 for both ACTH and corticosterone, F= 19.2 and 16.4, respectively).
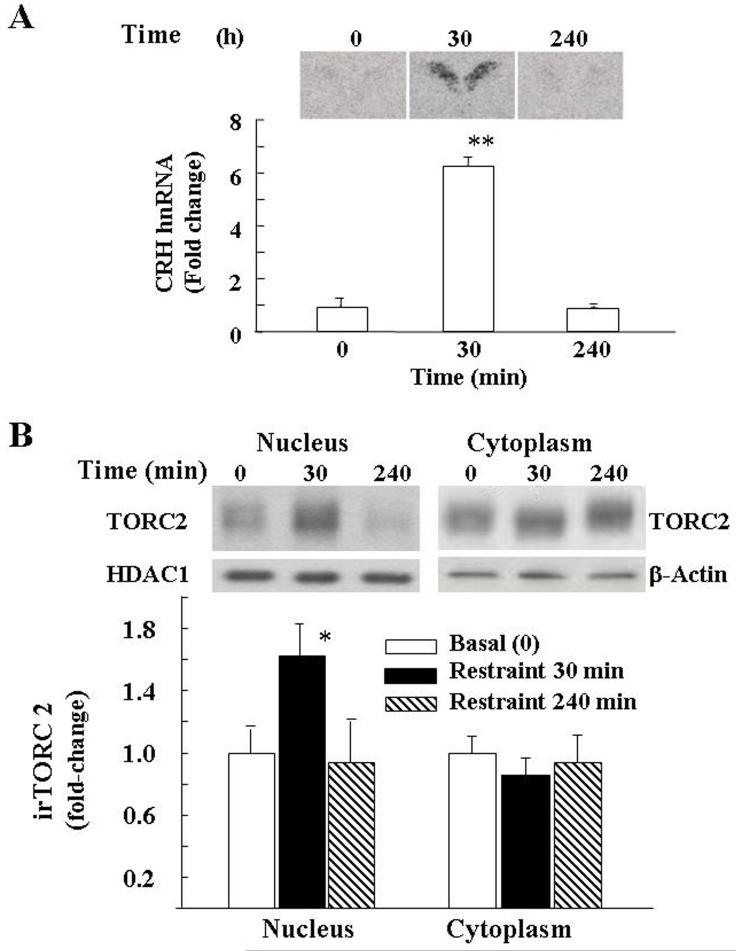
In situ hybridization analysis of CRH hnRNA in the PVN showed very low levels in basal conditions. Levels increased significantly after 30 min restraint (p<0.001, F=21.3) and had returned to basal values by 4h (Fig 1–A).
Fig 1.
Effect of restraint stress on CRH hnRNA, measured by in situ hybridization (A), and immunoreactive TORC 2 (irTORC 2), measured by western blot in nuclear and cytoplamic proteins in the microdissected dorsal hypothalamic region from individual rats (B). Rats were subjected to restraint stress for 1h and killed at 30 min or 4h after termination of the stress (240 min). Data are the mean and SE of data obtained from 5 rats for CRH hnRNA or 6 rats for irTORC 2. The intensity of the TORC 2 bands were corrected for β-actin in the cytoplasm and for HDAC1 in the nucleus. Images from representative rats are shown above the bars. *, p< 0.05; **, p< 0.001
Effect of restraint stress on nuclear and cytosolic TORC 2 in the hypothalamus
Western blot analysis of TORC 2 levels in cytosolic and nuclear proteins from whole hypothalamic homogenates evidenced a band of about 85 kDa corresponding to the molecular size of TORC 2. Overall analysis of the results from hypothalamic tissue from 4 to 5 rats per group shows a significant effect of stress (p<0.04, F=4.6). Although, there was variability between animals, there was a significant increase of the nuclear TORC 2 band at 30 min during restraint stress (65±19 over the basal values, p<0.05). Nuclear levels of TORC 2 3h after release from the restrainers, were no different from basal. No changes in intensity of the TORC 2 band were observed in the cytoplasm though the migration of the band was slightly accelerated at 30 min restraint (Fig 1-B).
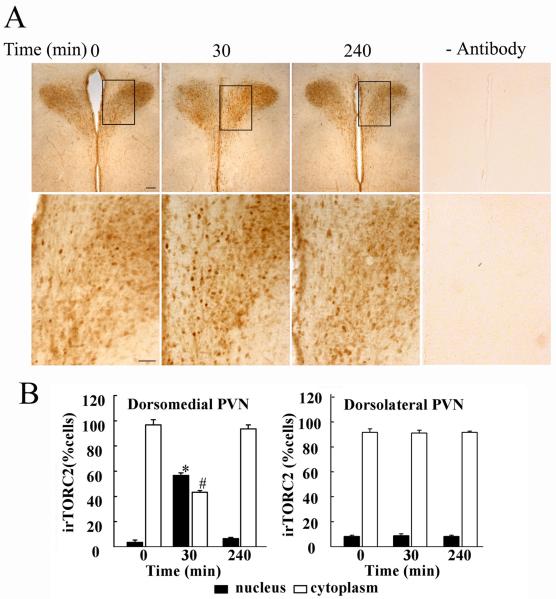
To determine whether the effects of stress on TORC 2 translocation occur in the PVN, immunohistochemical studies were performed in hypothalamic sections of control and stressed rats. As shown in Fig 2-A, TORC 2 staining was evident in both the dorsomedial and dorsolateral regions of the PVN, with the highest staining in the dorsolateral region corresponding to the topographic location of magnocellular neurons. No staining was observed in the absence of the primary antibody. As shown in the higher magnification images, there was an increase in nuclear staining at 30 min restraint (p<0.001 compared with controls), which returned to control values at 4h (Fig 1-A and B). This was accompanied by a decrease in the number of cells with cytoplasmic staining (p<0.002). Overall analysis of the average of TORC 2-stained cells in the cytoplasm and nucleus in 3 rats per group showed significant effects of restraint stress only in the dorsomedial region of the PVN, corresponding to the distribution of CRH neurons (p<0.001, F=265.6, and p<0.03, F=16.3, for nuclear and cytoplasmic staining, respectively).
Fig 2.
Immunohistochemical staining for TORC 2 in hypothalamic sections from control rats (basal), 30 min restraint and 3h after 1h restraint (240 min) (A). As shown by the higher magnification of representative images in the lower panels, there is an increase in nuclear staining for TORC 2 following 30 min restraint. Replacing the TORC 2 antibody by non-immune globulin yielded no staining over the background. The quantitative analysis of the number of cells showing nuclear and cytoplasmic staining in the dorsomedial and dorsolateral regions of the PVN is shown in B. Bars represent the mean and SE of measurements expressed as percent of cytosolic and nuclear location per total number of cells were counted in each animal in 3 rats. *, p<0.01 higher than nuclear staining in basal (time 0), or 3h after termination of 1h stress (240 min). #, p<0.01 lower than cytoplasmic staining in basal, or 3h after termination of 1h stress (240 min).
In contrast to the parvocellular region, there were no significant effects in the dorsolateral region, containing magnocellular vasopressin and oxytocin neurons (p<0.4, F=0.8, and p<0.6, F=0.5, for nuclear and cytoplasmic staining, respectively).
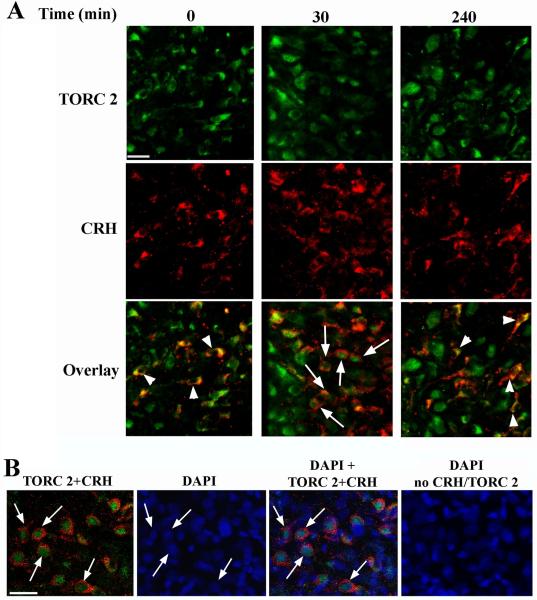
To confirm that nuclear translocation of TORC 2 during restraint stress occurred in CRH neurons of the PVN, double staining immunohistrochemistry for CRH and TORC 2 was performed in hypothalamic sections of controls and rats subjected to restraint. As shown in Fig 3A, in basal conditions there was clear co-localization of irTORC 2 in the cytoplasm of all detected CRH expressing neurons. Consistent with the observations with DAB staining, at 30 min restraint there was an increase in cells with nuclear TORC 2 staining. Co-staining with the nuclear marker DAPI performed in double stained sections of rats subjected to restraint for 30 min confirmed the nuclear localization of irTORC2 in this group (Fig 3-B). Omission of TORC 2 and CRH antibodies in sections stained with DAPI showed no immunoreactive signals (Fig 3-B). Quantitative analysis of the number of double-labeled cells after 30 min restraint indicated that only 61±3.5% of CRH neurons show nuclear TORC 2 staining. Consistent with the data in Fig 2, irTORC 2 was seen mostly in the cytoplasm of CRH neurons (Fig 3).
Fig 3.
Co-localization of immunoreactive CRH and immunoreactive TORC 2 in the dorsomedial region of the PVN Rats were subjected to restraint stress for up to 1h and killed at 30 min or 3h after termination of the stress (240 min) (A). Hypothalamic sections were stained for TORC 2 and CRH as described in Methods. CRH neurons with nuclear localization of irTORC2 indicated by arrows, and neurons with cytoplasmic irTORC 2 are shown by arrowheads. Nuclear localization of TORC2 in rats subjected to 30 min restraint was confirmed by DAPI staining as shown in B. Arrows point to the corresponding DAPI staining in 4 neurons. No staining was observed in the absence of TORC and CRH antibodies. The magnification bars correspond to 20μm.
Stress induces TORC 2 recruitment by the CRH promoter in the hypothalamus
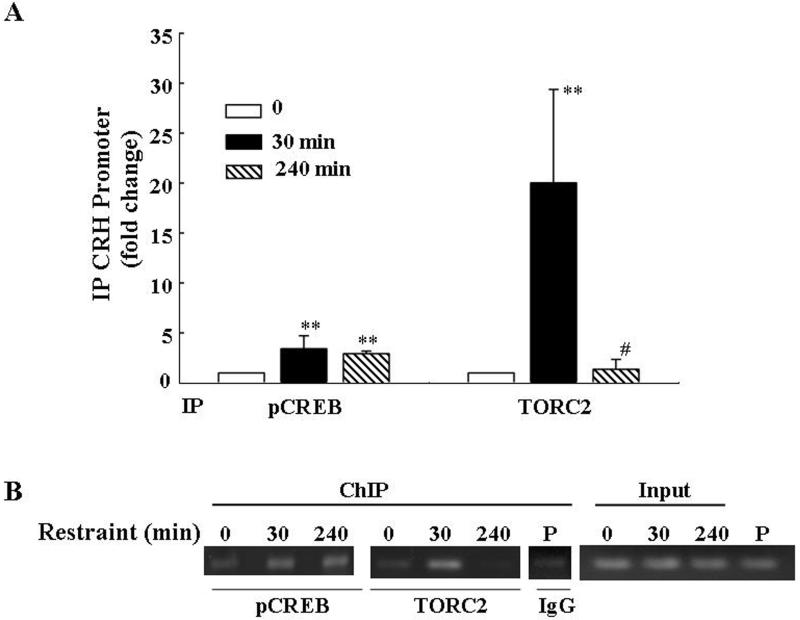
The ability of TORC 2 and phospho-CREB to interact with the CRH promoter during stress in conjunction with the activation of CRH transcription was examined by ChIP assays in hypothalamic chromatin. Overall analysis of the results in 4 experiments showed a significant effects of stress on phospho-CREB and TORC 2 recruitment by the CRH promoter (p<0.01, F=7.2 and p< 0.01, F=7.3, respectively). As shown in Fig. 4, CRH promoter immunoprecipitation with the phospho-CREB antibody increased by 30 min of stress (p<0.01 compared to basal), in correspondence with the increase in CRH hnRNA, but it was still significantly higher than controls by 4h (p<0.01), when CRH hnRNA had returned to basal values. In contrast, the amount of CRH promoter immunoprecipitated with the TORC 2 antibody markedly increased by 30min (p<0.02), but returned to basal values by 4h parallel to the changes in CRH hnRNA shown in Fig 1-A. Confirming the specificity of the protein binding to the CRH promoter, a lower intensity PCR band was observed in the pooled remaining cross-linked DNA after immunoprecipitation with non-immune globulin. Gel analysis of the PCR products of the immunoprecipitated CRH promoter in rat hypothalamic chromatin subjected to restraint for 30 min and 4h in a representative experiment is shown in Fig 4-B)
Fig 4.
Restraint stress induces recruitment of phospho-CREB and TORC2 by the CRH promoter in the hypothalamic PVN region. ChIP assays using phospho-CREB and TORC 2 antibodies and cross-linked DNA from microdissected hypothalamic PVN region of control rats and rats subjected to restraint stress for 30min or 3h after 1h restraint (240 min) (A). Amounts of CRH promoter DNA in the immunoprecipitant was measured by qRT-PCR. Bars represent the mean and SE of the results obtained in 4 experiments using pooled tissue from 2 or 3 rats per experimental point. Gel shows the results obtained in pools of hypothalamic tissue from 3 rats per group in a representative experiment (B). **, p< 0.01 higher than basal; **, p<0.02 higher than basal; #, p<0.02 lower than 30 min.
DISCUSSION
It has been long accepted that activation of CRH transcription depends on cAMP/protein kinase A (PKA) dependent pathways and binding of phosphorylated cAMP response element binding protein (pCREB) to CRE in the CRH promoter (4,6,8). While the requirement of cyclic AMP and CREB is still a fact, recent in vitro studies suggest that activation of CRH transcription involves nuclear translocation and recruitment of the CREB co-activator, TORC, by the CRH promoter (16). The present study shows that TORC 2 protein is expressed in CRH neurons of the hypothalamic PVN in rats and that all detected CRH neurons co-express TORC 2. Moreover, restraint stress induces nuclear translocation and recruitment of TORC 2 by the CRH promoter in conjunction with activation of CRH transcription. It should be noted that while all detected CRH neurons express TORC 2, only a proportion of them (about 60%) showed TORC 2 translocation to the nucleus. Since siRNA blockade of TORC 2 expression in hypothalamic neurons markedly reduces forskolin-induced CRH transcription (16), it could be assumed that transcriptional activation occurs only in neurons showing nuclear translocation of TORC. The partial number of CRH neurons showing nuclear translocation of TORC 2 raises the possibility that some CRH neurons do not respond (at least transcriptionally) to stress, or more likely that the proportion of responsive neurons could depend on the intensity of the stimulus. On the other hand, since nuclear translocation of TORC 2 was evaluated only at the peak of CRH transcription activation (22-24), it is possible that additional neurons showed nuclear TORC 2 staining at earlier time points. Although unlikely, it is not possible to rule out that TORC 1 or 3, which were not examined in this study, may mediate CRH transcription in the CRH neurons without TORC 2 staining in the nucleus.
In addition to TORC 2, the other members of the gene family, TORC 1 and 3 are expressed in the brain. In situ hybridization studies have shown that TORC 1 is the most abundant and widely distributed in the brain (25), but all three TORC subtypes are present in the PVN (26). While the present immunohistochemical data confirms the participation of TORC 2 in the regulation of CRH during stress, the question of whether TORC 1 and 3 have a role will require further investigation. In contrast to the high specificity of the TORC 2 antibody (Calbiochem) used in this study, none of the TORC 1 or TORC 3 antibodies available were suitable for immunohistochemistry. Previous studies using the cell line 4B or primary cultures of hypothalamic neurons and siRNA blockade for the different TORC subtypes had suggested that the predominant subtype required for CRH transcription is TORC 2, though TORC 3 may also be involved (16).
TORC potentiates the transcriptional activity by interacting with the bZip domain of CREB and subsequently facilitating the recruitment of the transcriptional complex (17,27). Consistent with this interaction, previous studies in 4B cells transfected with the CRH promoter have shown co-immunoprecipitation of TORC 2 and CREB (16). In the same study, stimulation of cells with forskolin induces recruitment of TORC 2 to the transfected CRH promoter. The present chromatin immunoprecipitation experiments using hypothalamic chromatin from controls and stressed rats clearly show that the endogenous CRH promoter is indeed able to recruit TORC 2. Moreover, TORC 2 was associated with the CRH promoter only at the time when CRH transcription is activated. It is noteworthy that in the present experiments and previous studies (24) phospho-CREB remains bound to the CRH promoter after CRH transcription has returned to basal values. Although a more detailed analysis of the temporal interaction of TORC 2 and phospho-CREB with the CRH promoter remain to be elucidated, the data suggest that termination of transcription does not depend on the removal of CREB from the CRE in the CRH promoter. The fact that TORC 2 is no longer present in the CRH promoter, or in the nucleus, when CRH transcription has returned to basal, suggests that removal of TORC 2 from the chromatin and its export from the nucleus is part of the mechanism of termination of CRH transcription during stress.
The mechanisms leading to TORC translocation are under investigation. In basal conditions, TORC is in the cytoplasm in a phosphorylated state and bound to the scaffolding protein, 14-3-3. TORC phosphorylation is mediated by members of the AMP-activated protein kinase (AMPK) family of Ser/Thr protein kinases, including salt inducible kinase 1, (SIK1) (28). Protein kinase A inactivates these kinases, thus preventing TORC phosphorylation and allowing its release from 14-3-3 and translocation to the nucleus, where it interacts with CREB. Studies in progress suggest that SIK1 plays a role regulating TORC translocation in hypothalamic neurons and we have evidence that there is marked induction of SIK1 in the parvocellular region of the PVN during stress (29).
In addition to CRH-stained neurons, other cells in the dorsomedial region of the PVN showed TORC 2 labeling suggesting the involvement of TORC 2 in the function of other parvocellular and periventricular neuroendocrine neurons. Also, the presence of prominent TORC 2 staining in the dorso-lateral region of the PVN, containing magnocellular vasopressin and oxytocin neurons implicates possible roles of the co-activator in the transcriptional regulation of these hormones. While this suggests that TORC 2 is involved in the function of other cell types in the hypothalamus, it is clear from the topographic distribution and co-staining data that restraint stress did not cause overt TORC 2 redistribution in neurons other than CRH neurons. The neuron type specificity of the redistribution (translocation of TORC 2 into the nucleus), in conjunction with the recruitment of TORC by the CRH promoter at the time when transcription is activated, provide strong support for a role of TORC 2 in the physiological activation of CRH transcription during stress.
In summary, this study in rats demonstrates firstly that TORC 2 is present in CRH neurons of the hypothalamic PVN, and secondly that stress causes transient TORC 2 translocation to the nucleus and its recruitment by the CRH promoter concomitantly with the activation of CRH transcription. These data support of the hypothesis that TORC 2 acts as the CREB co-activator required for activation of CRH transcription during stress.
ACKNOWLEDGEMENTS
This work was supported by the Intramural Research Program, National Institute of Child Health and Human Development and the Max Planck Society. The authors thank Drs. Sergey Khrulev and Günter Giese for technical help with triple staining and confocal microscopy.
REFERENCES
- 1.Adler GK, Smas CM, Fiandaca M, Frim DM, Majzoub JA. Regulated expression of the human corticotropin releasing hormone gene by cyclic AMP. Mol Cell Endocrinol. 1990;70:165–174. doi: 10.1016/0303-7207(90)90156-3. [DOI] [PubMed] [Google Scholar]
- 2.Cheng YH, Nicholson RC, King B, Chan EC, Fitter JT, Smith R. Corticotropin-Releasing Hormone Gene Expression in Primary Placental Cells Is Modulated by Cyclic Adenosine 3′,5′-Monophosphate. J Clin Endocrinol Metab. 2000;85:1239–1244. doi: 10.1210/jcem.85.3.6420. [DOI] [PubMed] [Google Scholar]
- 3.Nikodemova M, Kasckow J, Liu H, Manganiello V, Aguilera G. Cyclic adenosine 3′,5′-monophosphate regulation of corticotropin-releasing hormone promoter activity in AtT-20 cells and in a transformed hypothalamic cell line. Endocrinology. 2003;144:1292–1300. doi: 10.1210/en.2002-220990. [DOI] [PubMed] [Google Scholar]
- 4.Seasholtz AF, Thompson RC, Douglass JO. Identification of a cyclic adenosine monophosphate-responsive element in the rat corticotropin-releasing hormone gene. Mol Endocrinol. 1988;2:1311–1319. doi: 10.1210/mend-2-12-1311. [DOI] [PubMed] [Google Scholar]
- 5.McEvoy A, Bresnihan B, Fitzgerald O, Murphy EP. Corticotropin-Releasing Hormone Signaling in Synovial Tissue Vascular Endothelium Is Mediated through the cAMP/CREB Pathway. Ann N Y Acad Sci. 2002;966:119–130. doi: 10.1111/j.1749-6632.2002.tb04209.x. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y, Kamitakahara A, Kim A, Aguilera G. Cyclic Adenosine 3′,5′-Monophosphate Responsive Element Binding Protein Phosphorylation Is Required But Not Sufficient for Activation of Corticotropin-Releasing Hormone Transcription. Endocrinology. 2008;149:3512–3520. doi: 10.1210/en.2008-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nicholson RC, King BR, Smith R. Complex regulatory interactions control CRH gene expression. Front Biosci. 2004;9:32–39. doi: 10.2741/1204. [DOI] [PubMed] [Google Scholar]
- 8.Wolfl S, Martinez C, Majzoub JA. Inducible Binding of Cyclic Adenosine 3′,5′-Monophosphate (cAMP)-Responsive Element Binding Protein (CREB) to a cAMP-Responsive Promoter in Vivo. Mol Endocrinol. 1999;13:659–669. doi: 10.1210/mend.13.5.0282. [DOI] [PubMed] [Google Scholar]
- 9.Guardiola-Diaz HM, Boswell C, Seasholtz AF. The cAMP-responsive element in the corticotropin-releasing hormone gene mediates transcriptional regulation by depolarization. J Biol Chem. 1994;269:14784–14791. [PubMed] [Google Scholar]
- 10.Liu Y, Kalintchenko N, Sassone-Corsi P, Aguilera G. Inhibition of Corticotrophin-Releasing Hormone Transcription by Inducible cAMP-Early Repressor in the Hypothalamic Cell Line, 4B. J Neuroendocrinol. 2006;18:42–49. doi: 10.1111/j.1365-2826.2005.01383.x. [DOI] [PubMed] [Google Scholar]
- 11.Deisseroth K, Tsien RW. Dynamic Multiphosphorylation Passwords for Activity-Dependent Gene Expression. Neuron. 2002;34:179–182. doi: 10.1016/s0896-6273(02)00664-5. [DOI] [PubMed] [Google Scholar]
- 12.Shaywitz AJ, Greenberg ME. CREB: A Stimulus-Induced Transcription Factor Activated by A Diverse Array of Extracellular Signals. An Rev Biochem. 1999;68:821–861. doi: 10.1146/annurev.biochem.68.1.821. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y, Hatalski CG, Brunson KL, Baram TZ. Rapid phosphorylation of the CRE binding protein precedes stress-induced activation of the corticotropin releasing hormone gene in medial parvocellular hypothalamic neurons of the immature rat. Mol Brain Res. 2001;96:39–49. doi: 10.1016/s0169-328x(01)00265-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emanuel RL, Girard DM, Thull DL, Majzoub JA. Second Messengers Involved in the Regulation of Corticotropin-Releasing Hormone mRNA and Peptide in Cultured Rat Fetal Hypothalamic Primary Cultures. Endocrinology. 1990;126:3016–3021. doi: 10.1210/endo-126-6-3016. [DOI] [PubMed] [Google Scholar]
- 15.Itoi K, Horiba N, Tozawa F, Sakai Y, Sakai K, Abe K, Demura H, Suda T. Major role of 3′,5′-cyclic adenosine monophosphate-dependent protein kinase A pathway in corticotropin-releasing factor gene expression in the rat hypothalamus in vivo. Endocrinology. 1996;137:2389–2396. doi: 10.1210/endo.137.6.8641191. [DOI] [PubMed] [Google Scholar]
- 16.Liu Y, Coello AG, Grinevich V, Aguilera G. Involvement of Transducer of Regulated cAMP Response Element-Binding Protein Activity on Corticotropin Releasing Hormone Transcription. Endocrinology. 2010;151:1109–1118. doi: 10.1210/en.2009-0963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conkright MD, Canettieri G, Screaton R, Guzman E, Miraglia L, Hogenesch JB, Montminy M. TORCs: Transducers of Regulated CREB Activity. Molecular Cell. 2003;12:413–423. doi: 10.1016/j.molcel.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 18.Iourgenko V, Zhang W, Mickanin C, Daly I, Jiang C, Hexham JM, Orth AP, Miraglia L, Meltzer J, Garza D, Chirn GW, McWhinnie E, Cohen D, Skelton J, Terry R, Yu Y, Bodian D, Buxton FP, Zhu J, Song C, Labow MA. Identification of a family of cAMP response element-binding protein coactivators by genome-scale functional analysis in mammalian cells. Proc Natl Acad Sci USA. 2003;100:12147–12152. doi: 10.1073/pnas.1932773100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takemori H, Kajimura J, Okamoto M. TORC-SIK cascade regulates CREB activity through the basic leucine zipper domain. FEBS J. 2007;274:3202–3209. doi: 10.1111/j.1742-4658.2007.05889.x. [DOI] [PubMed] [Google Scholar]
- 20.Ma XM, Aguilera G. Differential regulation of corticotropin-releasing hormone and vasopressin transcription by glucocorticoids. Endocrinology. 1999;140:5642–5650. doi: 10.1210/endo.140.12.7214. [DOI] [PubMed] [Google Scholar]
- 21.Sawada K, Fukui Y, Hawkes R. Spatial distribution of corticotropin-releasing factor immunopositive climbing fibers in the mouse cerebellum: Analysis by whole mount immunohistochemistry. Brain Res. 2008;1222:106–117. doi: 10.1016/j.brainres.2008.05.029. [DOI] [PubMed] [Google Scholar]
- 22.Kovacs KJ, Sawchenko PE. Regulation of stress-induced transcriptional changes in the hypothalamic neurosecretory neurons. J Mol Neurosci. 1996;7:125–133. doi: 10.1007/BF02736792. [DOI] [PubMed] [Google Scholar]
- 23.Ma XM, Levy A, Lightman SL. Rapid changes in heteronuclear RNA for corticotrophin-releasing hormone and arginine vasopressin in response to acute stress. J Endocrinol. 1997;152:81–89. doi: 10.1677/joe.0.1520081. [DOI] [PubMed] [Google Scholar]
- 24.Shepard JD, Liu Y, Sassone-Corsi P, Aguilera G. Role of glucocorticoids and cAMP-mediated repression in limiting corticotropin-releasing hormone transcription during stress. J Neurosci. 2005;25:4073–4081. doi: 10.1523/JNEUROSCI.0122-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li S, Zhang C, Takemori H, Zhou Y, Xiong Z. TORC1 Regulates Activity-Dependent CREB-Target Gene Transcription and Dendritic Growth of Developing Cortical Neurons. J Neurosci. 2009;29:2334–2343. doi: 10.1523/JNEUROSCI.2296-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanchez-Watts G, Liu Y, Aguilera G, Watts A. The rat forebrain distribution of mRNAs encoding the three isoforms of the transducer of regulated CREB activity (TORC); Soc Neurosci Annual Meeting; San Diego, CA. 2010; Abst 792.11. [Google Scholar]
- 27.Ravnskjaer K, Kester H, Liu Y, Zhang X, Lee D, Yates JR, 3, Montminy M. Cooperative interactions between CBP and TORC2 confer selectivity to CREB target gene expression. EMBO J. 2007;26:2880–2889. doi: 10.1038/sj.emboj.7601715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takemori H, Okamoto M. Regulation of CREB-mediated gene expression by salt inducible kinase. J Steroid Biochem Mol Biol. 2008;108:287–291. doi: 10.1016/j.jsbmb.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y, Watts AG, Sanchez-Watts G, Aguilera G. Stimulus- and brain region-specific regulation of salt inducible kinase and AMP activated protein kinase in the hypothalamus of the rat; Soc Neurosci Ann Meeting; San Diego, CA. 2010; Abst 792.1. [Google Scholar]