Abstract
Neurotrophins (NTs) represent a family of proteins that play an important role in the survival, development, and function of neurons. Extensive efforts are currently being made to develop small molecules endowed with agonist or antagonist NT activity. The structurally versatile N-termini of these proteins are considered regions of interest for the design of new molecules. By combining experimental and computational approaches, we analyzed the intrinsic conformational preferences of the N-termini of two of the most important NTs: NGF (NGF-Nter) and NT4 (NT4-Nter). Circular dichroism spectra clearly indicate that both peptides show a preference for random coil states. Because this finding does not preclude the possibility that structured forms may occur in solution as minor conformational states, we performed molecular-dynamics simulations to gain insights into the structural features of populated species. In line with the circular dichroism analysis, the simulations show a preference for unstructured states for both peptides. However, the simulations also show that for NT4-Nter, and to a lesser extent for NGF-Nter, helical conformations, which are required for binding to the Trk receptor, are present in the repertoire of structures that are intrinsically accessible to these peptides. Accordingly, molecular recognition of NTs by the Trk receptor is accomplished by the general mechanism known as population shift. These findings provide a structural rationale for the observed activity of synthetic peptides based on these NT regions. They also suggest strategies for the development of biologically active peptide-based compounds.
Introduction
Neurotrophins (NTs) play a crucial role in the differentiation, survival, and maintenance of nerve cells (1). This protein family includes nerve growth factor (NGF), brain-derived neurotrophic factor (BDNF), neurotrophin 3 (NT3), neurotrophin 4/5 (NT4), and neurotrophin 6 (NT6) (2,3). Although the fine details of their function have yet to be fully elucidated, it is generally accepted that these proteins exert their effects by interacting with two types of transmembrane receptors belonging to either the tyrosine kinase (Trk) or tumor necrosis factor (p75) families. The Trk receptors (TrkA, TrkB, and TrkC) show specificity in their preference for NT binding, whereas the p75 receptor interacts with all NTs, albeit with slightly different binding affinities.
The involvement of NTs in biological processes linked to the emergence of severe and widespread pathologies has suggested their use as promising therapeutic agents. Although several independent investigations have indeed provided support for this idea (4,5), the use of NTs as drugs has been hampered by their poor pharmaceutical properties (6). Therefore, particular attention is being given to the development of mimetics endowed with NT-like activities. Over the last 15 years, investigators have used different approaches to synthesize and characterize a number of NT agonists or competitive antagonists (7–13), some of which have also proven to be active in vivo. Indeed, experiments conducted with synthetic NGF agonists have shown improvements in memory and cholinergic phenotype in cognitively impaired aged rats (14), and the ability to restore neuronal function and reduce reactive gliosis in a rat model of chronic constriction injury (7).
The rational design of effective NT mimetics relies strongly on detailed information about their interaction with receptors. Extensive crystallographic investigations have provided a detailed picture of NTs structures in their unliganded states and in complex with receptors (Trk and p75) (15–24). Comparative analyses of different crystallographic structures have unveiled an intriguing dynamic behavior of NT N-terminal regions. There is clear evidence that these fragments, which are likely unfolded in unliganded NTs, become ordered only upon binding to Trk receptors (24). Moreover, N-terminal regions of different NTs exhibit different structural properties. Indeed, the N-terminus of NGF mediates the interactions with TrkA by assuming an α-helical structure (24). On the other hand, the N-terminal region of NT4 forms a 3–10 helix upon binding to TrkB (20).
To date, computational studies aimed at unveiling the dynamic properties of NTs and their receptors remain rather limited (25,26). Previous molecular-dynamics (MD) simulations focused on analyzing the dynamic properties of the complex between NGF and TrkA, as well as the energetic factors involved in the NGF N-terminal recognition by the TrkA receptor (25,26).
Because the N-terminal regions of NTs are considered hot spots for the design of NT agonist/antagonists (27), we performed a combined circular dichroism (CD) and MD investigation aimed at identifying the intrinsic conformational preferences of the N-terminal regions of both NGF and NT4. The results of our investigation provide insights into the recognition process between NTs and Trk receptors, and guidelines for the design of new NT mimetics.
Materials and Methods
Peptide synthesis
Peptides corresponding to the N-terminal region of NT4 and NGF (see Table 1 for the sequence) were purchased from Inbios Laboratories (Pozzuoli, Italy) and used without further purification.
Table 1.
Sequences of the peptides characterized in this study
| NT4-Nter | |
| NGF-Nter |
The region highlighted in gray represents the α-helical region detected in the structure of the complex NGF-TrkA (PDB code: 1WWW), and the 3–10 helix found in the complex NT4-TrkB (PDB code: 1HCF).
CD spectroscopy
CD spectra on the peptides were recorded with a Jasco J-810 spectropolarimeter equipped with a Peltier temperature control system (model PTC-423-S). Molar ellipticity per mean residue, [θ] in deg cm2 × dmol−1, was calculated from the equation [θ] = [θ]obs × mrw × (10 × l × C)−1, where [θ]obs is the ellipticity measured in degrees, mrw is the mean residue molecular mass, C is the protein concentration in g × l−1, and l is the optical path length of the cell in centimeters. Far-UV measurements (190–230 nm) were carried out with a 0.1 cm optical path length cell. The concentration of NGF-Nter and NT4-Nter was 0.030 and 0.027 mM, respectively. The CD spectra reported in Fig. 1 were signal-averaged over three scans. Other parameters of the CD spectra registration are identical to those adopted in previous studies (28).
Figure 1.
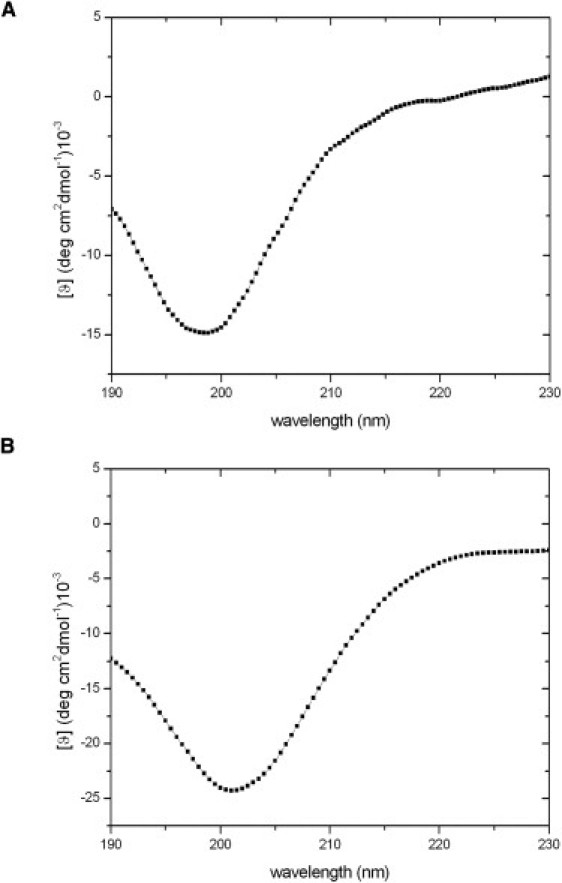
Far-UV CD spectra of (A) NT4-Nter and (B) NGF-Nter.
MD simulations
The MD simulations were conducted on the N-terminal fragments (residues 1–16) of NGF and NT4 (Table 1). To avoid any bias in the MD analysis, the simulations were performed by assuming an extended conformation for both peptides. To emulate the state of the peptides in the corresponding proteins, their N- and C-termini were kept charged and uncharged, respectively. Because the overall charge of NT4-Nter was +1, a Cl− ion was added to neutralize the simulated system. The AMBER03 force field (29) and the Tip3p water model implemented in the program GROMACS (30) were used in the simulations. For both systems, to relax bond geometries, the potential energy of the system (peptides and water) was minimized by using the steepest-descent method until convergence was reached. The solvent was then relaxed by 50 ps of MD at 300 K, restraining protein atomic positions with a harmonic potential. The system temperature was brought to 300 K in a stepwise manner: 30 ps MD runs were carried out at 50, 100, 150, 200, 250, and 300 K. We checked to ensure that in each step of the heating of the different systems, the potential energy reached a stable value. The timescale of the individual simulations is reported in Table 2. The simulations were run with periodic boundary conditions. Bond lengths were constrained by the LINCS algorithm. The electrostatic interactions were calculated using the particle mesh Ewald algorithm with a cutoff of 0.9 nm. The cutoff radius for the Lennard-Jones interactions was set to 0.9 nm. A dielectric constant of 1, and a time step of 2 fs were used. We used the NVT ensemble in all simulations. The temperature was maintained constant using the Berendsen thermostat with a time constant of 0.1 ps. Trajectory structures were analyzed by using GROMACS (30) and VMD (31) routines.
Table 2.
Summary of the simulations performed in this study
| Model | No. atom peptide/ No. atom water/Cl− | Box dimension (Å3) | Simulation time (ns) | T(K) |
|---|---|---|---|---|
| NT4-Nterm | 227/23991/1 | 62 × 62 × 62 | 200 | 300 |
| NGF-Nter | 240/23952 | 62 × 62 × 62 | 200 | 300 |
| QK-peptide | 280/32103/2 | 69 × 69 × 69 | 200 | 300 |
Results and Discussion
To characterize the intrinsic conformational preferences of the N-terminal regions of both NT4 and NGF, we adopted an integrated experimental (CD) and computational (MD) approach.
CD spectroscopy
To avoid any bias caused by chain termination, the length of the peptides corresponding to the N-terminus of the two NTs considered here was larger than the regions that become helicoidal upon binding to Trk receptors. As shown in Table 1, the first 16 residues of the two proteins were considered. The far-UV spectrum of NT4-Nter measured at room temperature shows a minimum at ∼200 nm (Fig. 1 A). A similar trend is exhibited by the spectrum of NGF-Nter (Fig. 1 B). Although, given the intrinsic limitations of the technique, these analyses do not preclude the possibility that structured forms of these peptides occur in solution as minor conformational states, these spectra clearly indicate that both peptides show a preference for random coil states.
MD simulations
To investigate the structural ensemble of these peptide at the atomic level and to verify the occurrence of low-populated structured states, we performed rather long MD simulations (200 ns) on both NT4-Nter and NGF-Nter by using the AMBER03 (29) force field, which has been proven to be well suited for the analysis of the peptide structured states (32), implemented in the program GROMACS (30). We also checked that a test simulation carried out on the α-helical QK-peptide (sequence Ac-KLTWQELYQLKYKGI-NH2) (33) using the potential, parameters, and equilibration protocol employed in NT4-Nter and NGF-Nter MD analyses was able to reproduce the expected conformational behavior of QK-peptide (see Fig. S1 in the Supporting Material).
NT4 N-terminal peptide
To minimize the bias caused by specific structural features of the starting model of the peptide used in the MD simulations, it was built in an extended conformation (Fig. 2 A). The initial stages of the simulation were characterized by a collapse of the extended conformer. These transitions led to the formation of transient turns and bends within the peptide. The analysis of the MD structures was conducted by excluding the first 50 ns of the trajectory, which could have been influenced by the starting state. As shown in Fig. 2, the NT4-Nter displays a remarkable dynamical behavior. The ensemble of the structures detected in the MD trajectory is highly variable. In line with the results of the CD analysis, most of the NT4-Nter structures do not exhibit regular secondary structure elements. Nevertheless, in a small but significant (∼20%) fraction of conformers, a 3–10 helix motif is detected in the central region of the peptide. As shown in Fig. 2, B–E, residues spanning from 8 to 10 form a short 3–10 helix that folds and unwinds many times in the simulation. A deeper inspection of the trajectory shows that the helical structure is more persistent in the 55–70 and 150–170 ns intervals (Fig. 2, D and E). Because this secondary structure motif relies on the formation of backbone hydrogen bonds, we analyzed the evolution of distances between potentially interacting atoms. We detected the formation of transient hydrogen bonds between 1), the O atom of Pro7 and the N atom of Arg10; and 2), the O atom of Ala8 and the N atom of Arg11 (Fig. 3 A). Although these atoms were quite distant in the starting extended NT4-Nter model (∼15 Å), they frequently came closer than 3.0 Å. It is worth noting that the interaction between O Pro7 and N Arg10 is more preserved than the one between O Ala8 and N Arg11. Of note, these hydrogen bonds are also detected in the structure of NT4 in its complex with TrkB (20). Although the NT4-Nter region contains charged groups, its helical state is not stabilized by specific electrostatic interactions. The structural similarity of the 3–10 conformers identified in the MD ensemble with the structure of the NT4 N-terminus in the complex with TrkB is of particular interest. As shown in Fig. 4, a representative example of the 3–10 conformers detected in the simulation perfectly fits the structure of the protein in the complex. This finding suggests that although it is highly flexible, the conformation needed for the binding to the receptor is present in the repertoire of structures intrinsically accessible to NT4-Nter.
Figure 2.
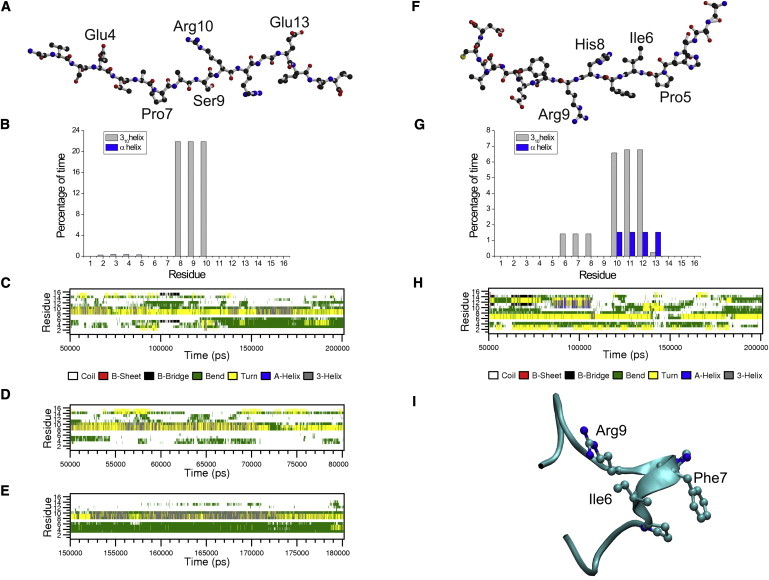
MD of NT4 N-terminal peptide (A–E) and NGF N-terminal peptide (F–I): (A) starting model, (B) percentage of time that each residue spent in helical conformation on the simulation timescale (50–200 ns), (C) secondary structure formation along the trajectory, (D and E) snapshots of the secondary structure formation of specific regions of the trajectory, (F) starting model, (G) percentage of time that each residue spent in helical conformation on the simulation timescale (50–200 ns), (H) secondary structure formation along the trajectory, and (I) a representative helical structure selected from the trajectory.
Figure 3.
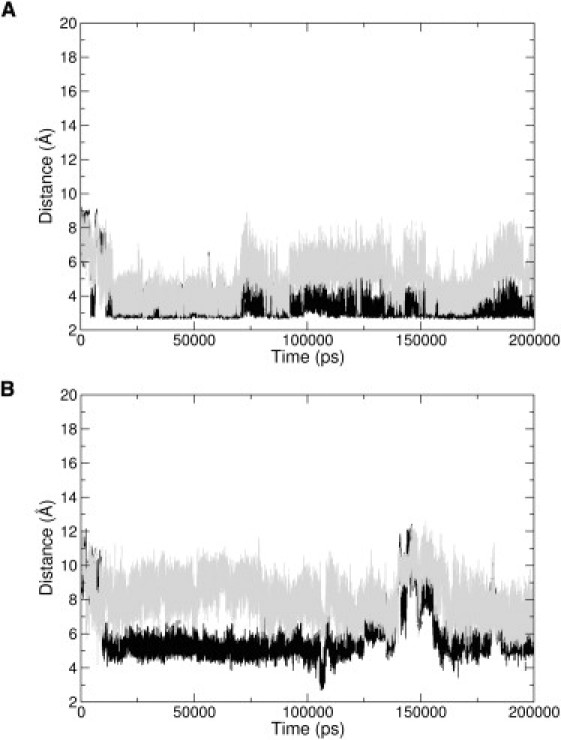
Evolution of the distances between atoms involved in the hydrogen bonds that stabilize (A) the 3–10 helix in the complex NT4-TrkB: O Pro7-N Arg10 (black) and O Ala8-N Arg11 (gray), and (B) the α-helix in the complex NGF-TrkA: O Pro5-N Arg9 (black) and O Ile6-N Gly10 (gray).
Figure 4.
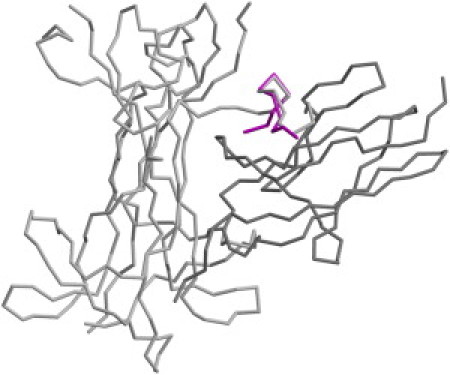
Representative example of a 3–10 helical model of NT4-Nter derived from the simulation superimposed on the N-terminal region of the NT4 (light gray) in the NT4-TrkB complex. The TrkB receptor is colored in dark gray. The model of the complex was generated from the coordinates of PDB entry 1HCF.
MD simulation on the NGF N-terminal peptide
The overall procedure adopted for investigating the intrinsic conformational preferences of NT4-Nter was extended to NGF-Nter, the N-terminal region of NGF (see Table 1 for the sequence). As found for NT4-Nter, the 200 ns MD simulation on NGF-Nter clearly shows that this peptide is intrinsically highly flexible (Fig. 2, F–I). In line with the CD experiments, most of the NGF-Nter conformations do not present a regular secondary structure. A helical structure is detected for a limited fraction of conformers present in the ensemble. In contrast to what is observed for NT4-Nter, the helical structure is detected for a NGF-Nter region (residues 10–12) that is buried in the native protein. Only an almost negligible portion (∼1%) of NGF-Nter simulation structures presents a helical structure in the region that the NGF N-terminus uses for TrkA binding (residues 6–8). The analysis of the distances between NGF-Nter atoms that are hydrogen-bonded in the NGF-TrkA complex shows that these atoms infrequently come close in the simulation (Fig. 3 B). This is particularly evident for the hydrogen-bonding partners O Ile6-N Gly10, whose distance is rarely less than 6 Å in the simulation. In conclusion, the MD simulation carried out on NGF-Nter is in line with the CD results. It also highlights significant differences in the conformational behavior of this peptide compared to that of NT4-Nter.
Implications for the molecular recognition of NTs by receptors
Because previous structural studies have revealed that the N-terminal regions of NT4 and NGF are not involved in specific interactions with the rest of the protein (20,24), the findings reported above, generated from an analysis of the isolated peptides, are also significant in the framework of the entire proteins. The observation that both peptides display a variety of different states is in line with crystallographic data. Indeed, these previous studies showed that they assume different conformations in the unliganded NTs, or when the NTs are in complex either with Trk or with p75 receptors. This conformational versatility is an important structural feature that allows these proteins to interact with diversified receptors. The data presented here indicate that for NT4-Nter, and to a lesser extent for NGF-Nter, the conformation of the peptide that is prone to Trk binding is already present among the energetically accessible states. Considered in a more general context, these findings show that Trk recognition by NTs is accomplished by the general mechanism known as population shift (34,35). In other words, rather than remodeling the structure of the N-terminal region of the NTs (induced fit), the receptor selects states that are already populated.
The MD simulations reported here also suggest that the N-termini of the two NTs have a distinct conformational behavior. This is not surprising, as the sequences of the N-termini of these NTs do not display any significant similarity (Table 1) despite an overall 47% sequence identity for the rest of the proteins. The lower tendency of NGF-Nter to intrinsically assume the helical state that is essential for binding to TrkA may be related to the presence of an Ile residue in its central region (position 6). Indeed, it is well established that Ile residues have a poor helical propensity. The crystallographic structure of the NGF-TrkA complex shows that this residue makes extensive hydrophobic contacts with the receptor. Evidently, these interactions compensate for the lower helical preorganization of the peptide due to the presence of this residue. Of note, in 48 NGF sequences isolated from different organisms reported in the Swiss-Prot database (http://www.expasy.ch/sprot), this residue is either conserved or replaced with a valine. Since valine has a similar low propensity for helical structures and a similar hydrophobic side chain, the considerations reported above hold for all known NGF variants.
It is also important to note that a secondary-structure prediction analysis, using the PSIPRED Protein Structure Prediction Server (http://www.psipred.net/psiform.html), of the peptide fragments examined here indicates a general agreement with the MD simulations. Indeed, this analysis suggests that NT4-Nter has a limited intrinsic tendency to form helical structure in region 8–12 (confidence score: 3 out of 9), whereas the highly helical QK-peptide shows a confidence score of 8 out of 9 in its N-terminal region, in line with the MD results. On the other hand, according to the prediction, the peptide NGF-Nter does not display any significant tendency to form helical structure. The preference of NT N-termini for unstructured states also provides insights into the ineffectiveness of peptides designed on the sequence of NGF N-terminus as NTs mimetics. In fact, it has been shown that when the sequence of the NGF N-terminus is conjugated to sequences of loops of the protein, negligible effects in their agonist activity are observed (7). The limited population of the helical state, which is required for binding, straightforwardly explains these observations. This consideration also prompts feasible strategies for the design of effective NT agonist/antagonists. Indeed, variants of this peptide with an increased helical propensity likely better mimic the NT functions. Several previous studies on other systems conducted using this strategy provide indirect support for the proposed approach (33,36,37).
Conclusions
The combined experimental and theoretical approach here reported provides a detailed picture of the NT N-terminal regions, which are considered hot spots in the molecular recognition between NT and their Trk receptors. Previous MD analyses focused on the evolution of the determinants of NGF-Nter recognition in its helical state by TrkA (25,26). Here, we complement those studies by providing information on the intrinsic conformational properties of NT N-terminal regions. Our MD simulations, which are compatible with the experimental CD analysis, give insights into the structural features of low-populated species. Not only do our findings provide a structural explanation for the observed activity of synthetic peptides containing these NT regions, they also suggest strategies for the development of effective peptide-based, biologically active molecules. In particular, the design of peptide mimetics of these regions, containing amino acids endowed with enhanced preferences for these structural motifs, is likely to yield high-affinity compounds for these receptors. Depending on their monomeric or dimeric state, these peptides may act as NT antagonists or agonists. Moreover, these peptides may be used to create conjugates with bioactive oligonucleotides for the selective targeting of cells overexpressing Trk receptors (38).
Acknowledgments
We thank Luca De Luca for technical assistance, and CINECA for providing computational resources (project 932 cne0fm4f).
This study was supported by the Ministero dell'Istruzione, dell'Università e della Ricerca (PRIN 2007).
Footnotes
Antonella Paladino's present address is Laboratorio di Bioinformatica e Biologia Molecolare, Istituto di Scienze Alimentari, CNR, Avellino, Italy.
Contributor Information
Luciana Esposito, Email: luciana.esposito@unina.it.
Luigi Vitagliano, Email: luigi.vitagliano@unina.it.
Supporting Material
References
- 1.Levi-Montalcini R. The nerve growth factor 35 years later. Science. 1987;237:1154–1162. doi: 10.1126/science.3306916. [DOI] [PubMed] [Google Scholar]
- 2.Hallböök F. Evolution of the vertebrate neurotrophin and Trk receptor gene families. Curr. Opin. Neurobiol. 1999;9:616–621. doi: 10.1016/S0959-4388(99)00011-2. [DOI] [PubMed] [Google Scholar]
- 3.Lanave C., Colangelo A.M., Alberghina L. Molecular evolution of the neurotrophin family members and their Trk receptors. Gene. 2007;394:1–12. doi: 10.1016/j.gene.2007.01.021. [DOI] [PubMed] [Google Scholar]
- 4.Williams B.J., Eriksdotter-Jonhagen M., Granholm A.C. Nerve growth factor in treatment and pathogenesis of Alzheimer's disease. Prog. Neurobiol. 2006;80:114–128. doi: 10.1016/j.pneurobio.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 5.Covaceuszach S., Capsoni S., Cattaneo A. Development of a non invasive NGF-based therapy for Alzheimer's disease. Curr. Alzheimer Res. 2009;6:158–170. doi: 10.2174/156720509787602870. [DOI] [PubMed] [Google Scholar]
- 6.Apfel S.C. Nerve growth factor for the treatment of diabetic neuropathy: what went wrong, what went right, and what does the future hold? Int. Rev. Neurobiol. 2002;50:393–413. doi: 10.1016/s0074-7742(02)50083-0. [DOI] [PubMed] [Google Scholar]
- 7.Colangelo A.M., Bianco M.R., Martegani E. A new nerve growth factor-mimetic peptide active on neuropathic pain in rats. J. Neurosci. 2008;28:2698–2709. doi: 10.1523/JNEUROSCI.5201-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peleshok J., Saragovi H.U. Functional mimetics of neurotrophins and their receptors. Biochem. Soc. Trans. 2006;34:612–617. doi: 10.1042/BST0340612. [DOI] [PubMed] [Google Scholar]
- 9.Saragovi H.U., Hamel E., Di Polo A. A neurotrophic rationale for the therapy of neurodegenerative disorders. Curr. Alzheimer Res. 2009;6:419–423. doi: 10.2174/156720509789207912. [DOI] [PubMed] [Google Scholar]
- 10.O'Leary P.D., Hughes R.A. Design of potent peptide mimetics of brain-derived neurotrophic factor. J. Biol. Chem. 2003;278:25738–25744. doi: 10.1074/jbc.M303209200. [DOI] [PubMed] [Google Scholar]
- 11.Xie Y., Tisi M.A., Longo F.M. Nerve growth factor (NGF) loop 4 dimeric mimetics activate ERK and AKT and promote NGF-like neurotrophic effects. J. Biol. Chem. 2000;275:29868–29874. doi: 10.1074/jbc.M005071200. [DOI] [PubMed] [Google Scholar]
- 12.Massa S.M., Xie Y., Longo F.M. Alzheimer's therapeutics: neurotrophin small molecule mimetics. J. Mol. Neurosci. 2002;19:107–111. doi: 10.1007/s12031-002-0019-1. [DOI] [PubMed] [Google Scholar]
- 13.Beglova N., Maliartchouk S., Gehring K. Design and solution structure of functional peptide mimetics of nerve growth factor. J. Med. Chem. 2000;43:3530–3540. doi: 10.1021/jm990441x. [DOI] [PubMed] [Google Scholar]
- 14.Bruno M.A., Clarke P.B., Saragovi H.U. Long-lasting rescue of age-associated deficits in cognition and the CNS cholinergic phenotype by a partial agonist peptidomimetic ligand of TrkA. J. Neurosci. 2004;24:8009–8018. doi: 10.1523/JNEUROSCI.1508-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDonald N.Q., Lapatto R., Blundell T.L. New protein fold revealed by a 2.3-A resolution crystal structure of nerve growth factor. Nature. 1991;354:411–414. doi: 10.1038/354411a0. [DOI] [PubMed] [Google Scholar]
- 16.Blundell T.L., Burke D.F., Sibanda B.L. Protein-protein interactions in receptor activation and intracellular signalling. Biol. Chem. 2000;381:955–959. doi: 10.1515/BC.2000.117. [DOI] [PubMed] [Google Scholar]
- 17.Bax B., Blundell T.L., McDonald N.Q. Structure of mouse 7S NGF: a complex of nerve growth factor with four binding proteins. Structure. 1997;5:1275–1285. doi: 10.1016/s0969-2126(97)00280-3. [DOI] [PubMed] [Google Scholar]
- 18.Wehrman T., He X., Garcia K.C. Structural and mechanistic insights into nerve growth factor interactions with the TrkA and p75 receptors. Neuron. 2007;53:25–38. doi: 10.1016/j.neuron.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 19.He X.L., Garcia K.C. Structure of nerve growth factor complexed with the shared neurotrophin receptor p75. Science. 2004;304:870–875. doi: 10.1126/science.1095190. [DOI] [PubMed] [Google Scholar]
- 20.Banfield M.J., Naylor R.L., Brady R.L. Specificity in Trk receptor:neurotrophin interactions: the crystal structure of TrkB-d5 in complex with neurotrophin-4/5. Structure. 2001;9:1191–1199. doi: 10.1016/s0969-2126(01)00681-5. [DOI] [PubMed] [Google Scholar]
- 21.Gong Y., Cao P., Jiang T. Crystal structure of the neurotrophin-3 and p75NTR symmetrical complex. Nature. 2008;454:789–793. doi: 10.1038/nature07089. [DOI] [PubMed] [Google Scholar]
- 22.Holland D.R., Cousens L.S., Matthews B.W. Nerve growth factor in different crystal forms displays structural flexibility and reveals zinc binding sites. J. Mol. Biol. 1994;239:385–400. doi: 10.1006/jmbi.1994.1380. [DOI] [PubMed] [Google Scholar]
- 23.Wiesmann C., Ultsch M.H., de Vos A.M. Crystal structure of nerve growth factor in complex with the ligand-binding domain of the TrkA receptor. Nature. 1999;401:184–188. doi: 10.1038/43705. [DOI] [PubMed] [Google Scholar]
- 24.Wiesmann C., de Vos A.M. Nerve growth factor: structure and function. Cell. Mol. Life Sci. 2001;58:748–759. doi: 10.1007/PL00000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Settanni G., Cattaneo A., Carloni P. Molecular dynamics simulations of the NGF-TrkA domain 5 complex and comparison with biological data. Biophys. J. 2003;84:2282–2292. doi: 10.1016/S0006-3495(03)75034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berrera M., Cattaneo A., Carloni P. Molecular simulation of the binding of nerve growth factor peptide mimics to the receptor tyrosine kinase A. Biophys. J. 2006;91:2063–2071. doi: 10.1529/biophysj.106.083519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saragovi H.U., Zaccaro M.C. Small molecule peptidomimetic ligands of neurotrophin receptors, identifying binding sites, activation sites and regulatory sites. Curr. Pharm. Des. 2002;8:2201–2216. doi: 10.2174/1381612023393215. [DOI] [PubMed] [Google Scholar]
- 28.Granata V., Graziano G., Zagari A. Chemical denaturation of the elongation factor 1α isolated from the hyperthermophilic archaeon Sulfolobus solfataricus. Biochemistry. 2006;45:719–726. doi: 10.1021/bi050479d. [DOI] [PubMed] [Google Scholar]
- 29.Duan Y., Wu C., Kollman P. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 30.Van Der Spoel D., Lindahl E., Berendsen H.J. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 31.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 32.Best R.B., Buchete N.-V., Hummer G. Are current molecular dynamics force fields too helical? Biophys. J. 2008;95:L07–L09. doi: 10.1529/biophysj.108.132696. (Erratum in Biophys. J. 2008. 95:4494) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diana D., Ziaco B., D'Andrea L.D. Structural determinants of the unusual helix stability of a de novo engineered vascular endothelial growth factor (VEGF) mimicking peptide. Chemistry. 2008;14:4164–4166. doi: 10.1002/chem.200800180. [DOI] [PubMed] [Google Scholar]
- 34.Boehr D.D., Nussinov R., Wright P.E. The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 2009;5:789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gunasekaran K., Ma B., Nussinov R. Is allostery an intrinsic property of all dynamic proteins? Proteins. 2004;57:433–443. doi: 10.1002/prot.20232. [DOI] [PubMed] [Google Scholar]
- 36.D'Andrea L.D., Iaccarino G., Pedone C. Targeting angiogenesis: structural characterization and biological properties of a de novo engineered VEGF mimicking peptide. Proc. Natl. Acad. Sci. USA. 2005;102:14215–14220. doi: 10.1073/pnas.0505047102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walensky L.D., Kung A.L., Korsmeyer S.J. Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science. 2004;305:1466–1470. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma N., Wu S.S., Wang S. Nerve growth factor receptor-mediated gene transfer. Mol. Ther. 2004;9:270–281. doi: 10.1016/j.ymthe.2003.11.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


