Abstract
Membrane proteins diffuse within the membrane, form oligomers and supramolecular assemblies. Using high-speed atomic force microscopy, we present direct experimental measure of an in-membrane-plane interaction potential between membrane proteins. In purple membranes, ATP-synthase c-rings formed dimers that temporarily dissociated. C-ring dimers revealed subdiffusive motion, while dissociated monomers diffused freely. C-rings center-to-center distance probability distribution allowed the calculation and modeling of an in-membrane-plane energy landscape that presented repulsion at 80 Å, most stable dimer association at 103 Å (−3.5 kBT strength), and dissociation at 125 Å (−1 kBT strength). This first experimental data of nonlabeled membrane protein diffusion and the corresponding in-membrane-plane interaction energy landscape characterized membrane protein interaction with an attractive range of several kBT that reaches to a radius of ∼50 Å within the membrane plane.
With increasing knowledge about the structures of individual membrane proteins (1), the focus of investigations turns toward the study of integrated membrane structure and function (2). The amphiphilic nature of membrane proteins restricts their structure (3) and motion (4,5) to the two-dimensional membrane plane where they form oligomers, are densely packed (2) and form supramolecular assemblies (6). Such an ensemble is represented in the purple membrane (PM) by the light-driven proton pump bacteriorhodopsin (bR) (7) and the ATP-synthase using the protonmotive force (chemical potential energy due to the proton concentration difference across a membrane) generated by bR to form ATP (8) (see Section 1 in the Supporting Material).
Recently, atomic-force microscopy (AFM) has reached maturity and allowed acquisition at submolecular resolution of membrane proteins in native membranes. Unfortunately, the image acquisition frequency of ∼1 min or more did not allow real-time assessment of membrane protein movement in native membranes (6,9). Membrane protein dynamics has been monitored by single particle tracking experiments of labeled membrane proteins using optical microscopy (5), with the shortcoming that the protein itself and its molecular environment are not observed. High-speed atomic force microscopy (HS-AFM (10)) now allows us to the concomitant assessment of structure and dynamics of integrated nonlabeled membrane protein assemblies, and to calculate corresponding physical parameters (see Section 2 in the Supporting Material).
A transmembrane protein may locally modify the lipid environment, including the bilayer thickness, its flexibility, and its local curvature. Such perturbation bears an energetic cost and has a finite spatial extension. If two such proteins are in close vicinity, they will feel each other's presence through the overlap of their perturbation fields. Any type of protein-membrane coupling may lead to such membrane-mediated interactions, most notably hydrophobic mismatch and curvature-mediated interactions (for a review see (11)). The range of the interaction is set by the extent of the membrane perturbation around a protein, and depends on the type of perturbation involved. Variation of the bilayer thickness involves a (costly) stretching or compression of the lipid tails, which typically relax over a distance comparable to the bilayer thickness. Relaxation of the mean membrane curvature is dictated by the (weaker) membrane's mechanical tension, which relaxes over larger distances (10–100 nm). Although numerous theoretical and numerical studies have explored various aspects of membrane-mediated interactions between membrane proteins, direct experimental observation of such interaction have not been obtained until now. Based on dynamic molecule imaging by HS-AFM (10), we analyze the in-membrane-plane diffusion and interaction of two membrane proteins and derive the corresponding interaction potential landscape.
Here, we used HS-AFM to image the edge regions of native PM from Halobacterium (H.) salinarum (12,13) (see Section 2 in the Supporting Material). PMs (7) are mainly constituted of bR arrays, with lipid membrane border regions where bR and ATP-synthase c-rings (14) diffuse. It has been shown that the lipid membrane outside the bR lattice is in the fluid state (15). The predominant lipid in the H. salinarum membrane is phosphatidyl glycerophosphate, with C17 acyl-chain length, and the hydrophobic thickness is ∼28 Å (16,17). The hydrophobic thickness of the membrane bilayer is important as the thickness mismatch between proteins and the embedding lipid environment is the major source of membrane-mediated protein-protein interactions (18,19). At the array-lipid interface of PM, bR dynamics can be monitored, in agreement with a previous study (13). Detailed analysis of sequential images and the calculation of difference maps between sequential images allowed visualization of association and dissociation of individual bR trimers to the bR-lattice (see Section 2 and Movie S1 in the Supporting Material). Indeed, the bR lattice edge is in dynamic equilibrium with bR molecules attaching and detaching to and from the array. The ratio of attaching and detaching molecules is ∼1 during an observation period of 30 s. The molecules that undergo dynamic changes are only ∼2% of all molecules observed along the lattice edge at the applied imaging rate of one frame per second.
In the edge-region of PM, we observed ring-shaped molecules with 65 Å diameter. Based on their shape, size, and location, in addition to the bR-arrays we attributed the protein to membrane standing ATP-synthase c-rings (see Section 3 in the Supporting Material). This attribution was confirmed by mass-spectrometry: The c-ring subunit gene (atpk) of H. salinarum (CAP14578 (20)) codes for a 89-amino-acids-long hydrophobic protein. After cleavage of the 12 N-terminal amino acids upon membrane insertion (21), the calculated mass of the protein is 7516.84 Da, in very good agreement with the 7517 Da measured (see Section 3 in the Supporting Material).
The c-rings assembled into dimers with dynamic association distances ranging from tight (∼80 Å) to elongated (∼120 Å) (Fig. 1 A). More rarely, c-rings dissociated into monomers, but reformed after several hundreds of milliseconds (Fig. 1 B). Hence, overall, three situations could be distinguished using HS-AFM at imaging rate of 187 ms per frame (Movie S2):
-
1.
The close dimer where the two c-rings touched each other (Fig. 1 C, left).
-
2.
The elongated dimer where the two c-rings were not in physical contact (Fig. 1 C, middle). (Also note that intermediate situations were equally observed.)
-
3.
The monomer (Fig. 1 C, right).
Figure 1.
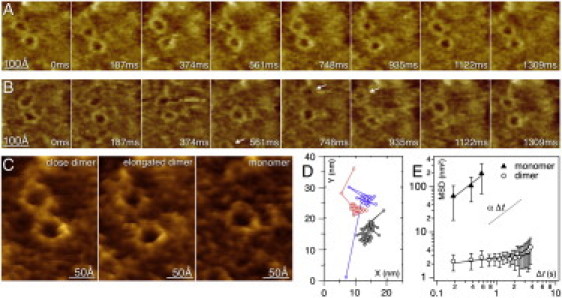
ATP-synthase c-ring motion monitored by HS-AFM (full false color scale: 35 Å). (A) Two c-rings in a dimer over a time span of 1309 ms (of a longer movie), swinging of the c-rings in the dimer is observed. (B) The c-ring dimer that dissociates and reforms. Intermediately one c-ring is monomeric for ∼400 ms (frames: 561 ms, 748 ms, 935 ms) and subsequently reforms a dimer that resembles the initial assembly. (C) Three examples of ATP-synthase c-ring assemblies. (Left to right) Close dimer (center-to-center distance 80 Å); elongated dimer (center-to-center distance 120 Å); and monomer. (D) Real-space representation of particle tracking. (Black dots and lines) The c-ring remaining in the frame. (Red and blue dots and lines) Trajectories of alternating c-rings. (E) Mean-square displacement for monomer (solid triangles) and dimer (open circles) c-rings fitted as described (see Section 5 in the Supporting Material). The obtained exponent values (±SD) were α = 1.0 ± 0.9 in the case of monomers, and α = 0.2 ± 0.1 for the dimer configuration. These results indicated diffusive and subdiffusive motion of the monomer and dimer, respectively.
The c-rings were characterized through particle tracking and the diffusion characteristics analyzed. The real-space positions of particle-tracking, where the c-rings were imaged as close dimer, elongated dimer, and monomer, illustrated the motion behavior of the molecules within a HS-AFM movie of 60 frames (Fig. 1 D) Most often dimers were found with inter-ring distances of ∼10 nm (frames 1–23 and frames 29–60), with an intermediate period of ∼400 ms during which dimer dissociation occurred (frames 24–28); the monomer c-ring detection is limited by the imaging frame size and acquisition rate (see Section 5 in the Supporting Material). The notable preservation of the assembly interaction angle between the c-rings was facilitated by remaining stator elements (see Section 6 in the Supporting Material). The strong interaction between c-rings influences their motion (Movie S2). While c-rings within dimers rattle about their localization during a period of seconds, c-ring monomers diffused fast in the membrane plane.
The analysis of the mean-square displacement (MSD) as a function of the lag-time Δt showed two different diffusion regimes for monomers and dimers (see Section 5 in the Supporting Material). These regimes were characterized by fitting the generalized diffusion equation MSD ∼ Δtα to the data, where α is the anomalous diffusion exponent (Fig. 1 E). The obtained exponent values (± SD) were α(monomer) = 1.0 ± 0.9 for monomers, and α(dimer) = 0.2 ± 0.1 for the dimer. Subdiffusion (α < 1) is characteristic of confined proteins on living cells (5). Furthermore, size and oligomerization dependence of membrane diffusion has been reported experimentally and theoretically (4,22), though these studies diverge concerning the magnitude of the effect and the interpretation of the underlying mechanism. The corresponding free diffusion constants are 125 nm2/s and 1.5 nm2/s for the monomer and the dimer, respectively, the monomer diffusion compares to diffusion of lipids in gel phase (23), while the dimer is almost static.
Quantitative information about the energy of interaction between two c-rings can be obtained from the probability of observing two proteins at a given distance. At thermal equilibrium, the probability distribution function p(d) (Fig. 2 A) is related to the energy of interaction U(d) (Fig. 2 B) by the Boltzmann law
where kBT is the thermal energy (see Section 8 in the Supporting Material). We observe a short-range repulsion, coupled with a long-range attraction between proteins. The former contribution is well fitted by a generic soft-core (quadratic) potential (24) and probably originates from structural perturbation of the protein/lipid organization upon close contact. Considering the extent of the long-range attractive potential, it likely corresponds to membrane-mediated interactions such as those arising from hydrophobic mismatch between the proteins and the surrounding lipid bilayer (25) (see Sections 4 and 8 in the Supporting Material). This hypothesis is further supported by the measurement of elevated lipid plateaus between the c-rings (see Section 7 in the Supporting Material).
Figure 2.
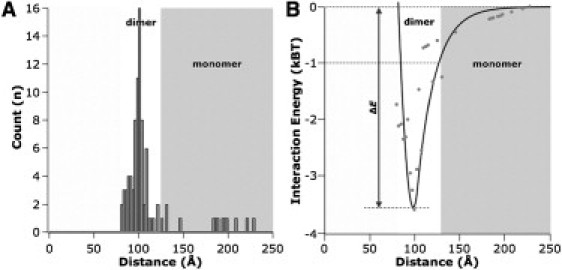
Interaction between ATP synthase c-rings. (A) Histogram (n = 91) of the center-to-center distance of c-rings. (B) Membrane-mediated two-protein interaction energy landscape.
Combining these two contributions results in a well-defined energy minimum of ∼−3.5 kBT at a center-to-center distance of 103 Å. The minimal distance we could observe was ∼80 Å, and the attractive interaction energy dropped below −1 kBT for distances larger than 125 Å, beyond which the attraction is insufficient to hold a dimer together against thermal fluctuations. Sporadically monomeric c-rings at larger distances were observed. However, because dissociated monomers moved fast and the size of the HS-AFM imaging frame was limited, we could only rarely observe them.
Due to the high image acquisition frequency of HS-AFM (∼187 ms per image), mobile ATP-synthase c-rings were analyzed in membranes of H. salinarum, undetectable by conventional AFM. The use of HS-AFM allowed the analysis of membrane diffusion of a nonlabeled protein, and enabled us to calculate the in-membrane-plane interaction energy between transmembrane molecules.
Acknowledgments
The authors thank Dr. Wolfgang Faigle for help with the mass spectrometry analysis.
This study was supported by the Agence Nationale de la Recherche and the City of Paris.
Supporting Material
References and Footnotes
- 1.White, S. 2009. Membrane proteins of known 3D structure. http://blanco.biomol.uci.edu/Membrane_Proteins_xtal.html.
- 2.Engelman D.M. Membranes are more mosaic than fluid. Nature. 2005;438:578–580. doi: 10.1038/nature04394. [DOI] [PubMed] [Google Scholar]
- 3.Bowie J.U. Solving the membrane protein folding problem. Nature. 2005;438:581–589. doi: 10.1038/nature04395. [DOI] [PubMed] [Google Scholar]
- 4.Gambin Y., Lopez-Esparza R., Urbach W. Lateral mobility of proteins in liquid membranes revisited. Proc. Natl. Acad. Sci. USA. 2006;103:2098–2102. doi: 10.1073/pnas.0511026103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kusumi A., Nakada C., Fujiwara T. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: high-speed single-molecule tracking of membrane molecules. Annu. Rev. Biophys. Biomol. Struct. 2005;34:351–378. doi: 10.1146/annurev.biophys.34.040204.144637. [DOI] [PubMed] [Google Scholar]
- 6.Scheuring S., Sturgis J.N. Chromatic adaptation of photosynthetic membranes. Science. 2005;309:484–487. doi: 10.1126/science.1110879. [DOI] [PubMed] [Google Scholar]
- 7.Oesterhelt D., Stoeckenius W. Functions of a new photoreceptor membrane. Proc. Nat. Amer. Soc. 1973;70:2853–2857. doi: 10.1073/pnas.70.10.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshida M., Sone N., Kagawa Y. ATP synthesis catalyzed by purified DCCD-sensitive ATPase incorporated into reconstituted purple membrane vesicles. Biochem. Biophys. Res. Commun. 1975;67:1295–1300. doi: 10.1016/0006-291x(75)90167-9. [DOI] [PubMed] [Google Scholar]
- 9.Buzhynskyy N., Hite R.K., Scheuring S. The supramolecular architecture of junctional microdomains in native lens membranes. EMBO Rep. 2007;8:51–55. doi: 10.1038/sj.embor.7400858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ando T., Kodera N., Toda A. A high-speed atomic force microscope for studying biological macromolecules. Proc. Nat. Amer. Soc. 2001;98:12468–12472. doi: 10.1073/pnas.211400898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phillips R., Ursell T., Sens P. Emerging roles for lipids in shaping membrane-protein function. Nature. 2009;459:379–385. doi: 10.1038/nature08147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casuso I., Kodera N., Scheuring S. Contact-mode high-resolution high-speed atomic force microscopy movies of the purple membrane. Biophys. J. 2009;97:1354–1361. doi: 10.1016/j.bpj.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamashita H., Voïtchovsky K., Ando T. Dynamics of bacteriorhodopsin 2D crystal observed by high-speed atomic force microscopy. J. Struct. Biol. 2009;167:153–158. doi: 10.1016/j.jsb.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 14.Ihara K., Abe T., Mukohata Y. Halobacterial A-ATP synthase in relation to V-ATPase. J. Exp. Biol. 1992;172:475–485. doi: 10.1242/jeb.172.1.475. [DOI] [PubMed] [Google Scholar]
- 15.Piknová B., Pérochon E., Tocanne J.F. Hydrophobic mismatch and long-range protein/lipid interactions in bacteriorhodopsin/phosphatidylcholine vesicles. Eur. J. Biochem. 1993;218:385–396. doi: 10.1111/j.1432-1033.1993.tb18388.x. [DOI] [PubMed] [Google Scholar]
- 16.Renner C., Kessler B., Oesterhelt D. Lipid composition of integral purple membrane by 1H and 31P NMR. J. Lipid Res. 2005;46:1755–1764. doi: 10.1194/jlr.M500138-JLR200. [DOI] [PubMed] [Google Scholar]
- 17.Dumas F., Lebrun M.C., Tocanne J.F. Is the protein/lipid hydrophobic matching principle relevant to membrane organization and functions? FEBS Lett. 1999;458:271–277. doi: 10.1016/s0014-5793(99)01148-5. [DOI] [PubMed] [Google Scholar]
- 18.Lewis B.A., Engelman D.M. Bacteriorhodopsin remains dispersed in fluid phospholipid bilayers over a wide range of bilayer thicknesses. J. Mol. Biol. 1983;166:203–210. doi: 10.1016/s0022-2836(83)80006-0. [DOI] [PubMed] [Google Scholar]
- 19.Botelho A.V., Huber T., Brown M.F. Curvature and hydrophobic forces drive oligomerization and modulate activity of rhodopsin in membranes. Biophys. J. 2006;91:4464–4477. doi: 10.1529/biophysj.106.082776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfeiffer F., Schuster S.C., Oesterhelt D. Evolution in the laboratory: the genome of Halobacterium salinarum strain R1 compared to that of strain NRC-1. Genomics. 2008;91:335–346. doi: 10.1016/j.ygeno.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 21.Ihara K., Watanabe S., Mukohata Y. Identification of proteolipid from an extremely halophilic archaeon Halobacterium salinarum as an N,N′-dicyclohexyl-carbodiimide binding subunit of ATP synthase. Arch. Biochem. Biophys. 1997;341:267–272. doi: 10.1006/abbi.1997.9972. [DOI] [PubMed] [Google Scholar]
- 22.Ohtsuki T., Okano K. Diffusion coefficients of interacting Brownian particles. J. Chem. Phys. 1982;77:1443–1450. [Google Scholar]
- 23.Tamm L.K., McConnell H.M. Supported phospholipid bilayers. Biophys. J. 1985;47:105–113. doi: 10.1016/S0006-3495(85)83882-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson K.L. Cambridge University Press; Cambridge, UK: 1985. Contact Mechanics. [Google Scholar]
- 25.Dan N., Pincus P., Safran S.A. Membrane-induced interactions between inclusions. Langmuir. 1993;9:2768–2771. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


