Abstract
The mechanism of oligomerization and its role in the regulation of activity in large GTPases are not clearly understood. Human guanylate binding proteins (hGBP-1 and 2) belonging to large GTPases have the unique feature of hydrolyzing GTP to a mixture of GDP and GMP with unequal ratios. Using a series of truncated and mutant proteins of hGBP-1, we identified a hydrophobic helix in the connecting region between the two domains that plays a critical role in dimerization and regulation of the GTPase activity. The fluorescence with 1-8-anilinonaphthalene sulfonate and circular dichroism measurements together suggest that in the absence of the substrate analog, the helix is masked inside the protein but becomes exposed through a substrate-induced conformational switch, and thus mediates dimerization. This is further supported by the intrinsic fluorescence experiment, where Leu298 of this helix is replaced by a tryptophan. Remarkably, the enzyme exhibits differential GTPase activities depending on dimerization; a monomer produces only GDP, but a dimer gives both GDP and GMP with stimulation of the activity. An absolute dependence of GMP formation with dimerization demonstrates a cross talk between the monomers during the second hydrolysis. Similar to hGBP-1, hGBP-2 showed dimerization-related GTPase activity for GMP formation, indicating that this family of proteins follows a broadly similar mechanism for GTP hydrolysis.
Introduction
Human guanylate binding proteins (hGBPs) belong to a class of large GTPases that include Dynamin, Mx, etc. Despite limited sequence homology, these proteins share similar biochemical features distinct from the canonical small GTPases (1). These include higher molecular mass (67–100 kDa), lower affinity for the substrate GTP (micromolar), high intrinsic GTPase activity (kcat ∼2–100 min−1), and the ability to dimerize/oligomerize in the presence of substrate. Unlike small GTPases, these proteins do not require external GTPase-activating protein (GAP); instead, they self-assemble upon the substrate binding and stimulate the activity. The guanylate binding proteins in vertebrates comprise a conserved family of GTPases with five mouse and seven human homologs (2). Among these, hGBP-1 and hGBP-2 are mostly induced in all tissues upon induction with interferon-γ (3,4). hGBP-1 has been shown to block the replication of RNA viruses such as the vesicular stomatitis virus (VSV) and the encephalomyocarditis virus (EMCV) (5). It also regulates endothelial cell proliferation by inhibiting matrix metalloproteinase-1 (6). The higher level of expression of this gene in multidrug-resistant human ovarian cancer implicated that it may have a role in drug resistance (7). This protein has been shown to associate with the Golgi apparatus in the GTP-bound state (8). Similarly, mouse GBP-2 inhibits VSV and EMCV (9). Recently, hGBP-2 has been shown to be highly expressed in proliferative squamous cells, which suggests that it may represent a marker of esophageal squamous cell carcinomas (10).
The most striking feature for both hGBP-1 and hGBP-2 is their ability to hydrolyze GTP to a mixture of GDP and GMP with unequal ratios (11,12). With hGBP-1, GMP is the major product of the reaction, whereas GDP predominates when the hydrolysis is catalyzed by hGBP-2. The structure and biochemical function of hGBP-1 has been investigated extensively, whereas for hGBP-2 it is studied at a preliminary level. The crystal structure of hGBP-1 is reported with and without the substrate analog, GppNHp, and the overall structure is similar (13,14). Notably, the structure of the full-length protein has been solved only as a monomer in the presence of the analog, although it dimerizes with the analog. Pyrophosphate was not obtained as a reaction product of hGBP-1 catalyzed reaction and the protein cannot hydrolyze the β-γ resistant GTP-analog GppNHp, suggesting that GMP formation occurs through successive cleavages of GTP and the second phosphate cleavage cannot occur without the first, respectively (11). The protein is 592-residues-long and the structure can be classified into two domains—a globular domain (residues 1–278) and a purely helical domain (residues 311–592). These two domains are joined by a small connecting region (residues 279–310), which consists of a two stranded β-sheet and an α-helix. hGBP-1 dimerizes in the presence of GppNHp, and dimerization is essential for stimulation of the GTPase activity (13,15,16). Studies with the mutant as well as truncated proteins showed that the 103DxEKGD108 motif and the connecting region together play important roles in GMP formation (17). The single mutant D108A hydrolyzes GTP to both GDP and GMP with a ratio of GMP/GDP ∼0.9:1 (for the wild-type, the ratio is ∼6:1), whereas the double-mutant D103L.D108L completely abolishes GMP formation (hydrolyzes GTP to only GDP). The truncated hGBP-1278 and hGBP-1289 produce only GDP, and no GMP was observed. On the other hand, hGBP-1307 hydrolyzes GTP to both GDP and GMP with a ratio of GMP/GDP ∼0.6:1.
The intermolecular interactions are clearly important in self-assembly and regulation of the GTPase activity in large GTPases. Although these proteins oligomerize/dimerize upon binding with the substrate, the mechanism of self-association and its role in the regulation of GTPase activity are not clearly understood because of the absence of structure of the full-length protein in the oligomeric/dimeric state. Using hGBP-1, an extensively studied GTP-binding protein among large GTPases that display unique second activity, we addressed the following key issues:
-
1.
Which region of the multidomain protein is involved in the dimerization and how does the protein dimerize?
-
2.
Is there a cross talk between the two monomers that regulates the GTPase activity?
Additionally, hGBP-2, which shares almost 75% sequence identity with hGBP-1, deviates markedly in the ratio of product formation and has been used to understand whether the dimerization-related GTPase activity is similar in this family of protein. With a series of truncated globular and helical proteins of hGBP-1, we identify a hydrophobic helix in the connecting region that plays a critical role in the dimerization. Our data suggest that, in the absence of the substrate analog, the helix is buried inside the protein but upon binding with the analog becomes exposed and mediates dimerization. The monomer produces only GDP but the dimer gives both GDP and GMP, demonstrating a cross talk between the two monomers during GMP formation. To our knowledge, this is the first GTPase that has differential activities depending on the dimerization of the protein. This study illustrates how the large GTPases self-regulate their dimerization-associated stimulation of GTP hydrolysis.
Materials and Methods
See the Supporting Material.
Results
GMP formation is regulated with dimerization of the protein
To understand whether GMP formation is regulated with dimerization in hGBP-1, we carried out analytical gel filtration assays with the wild-type, mutant, and truncated proteins (where the globular domain, i.e., nucleotide binding domain, was kept intact) as shown in Fig. 1 in the absence and presence of the substrate analog GppNHp. The wild-type protein eluted with a single peak (Fig. 2) in the presence of the analog that corresponds to a dimer (Table 1). It is interesting to note from Fig. 2 that D108A, a catalytically important mutant, exists as a mixture of monomer and dimer with the analog (Table 1). In contrast, D103L.D108L, defective in the second hydrolysis, elutes with a single peak that corresponds to a monomer, indicating that the double mutant did not dimerize in the presence of the analog (Fig. 2). The truncated hGBP-1278 and hGBP-1289, impaired in the second hydrolysis, do not dimerize in the presence of the analog (Table 1). Interestingly, hGBP-1307 (which is 18-residues-longer than the hGBP-1289, consisting of a helix) eluted with a single peak that corresponds to a dimer with the analog (Fig. 2). All these data clearly demonstrate that, in the presence of analog GppNHp, the α-helix of the connecting region plays a critical role in dimerization. Because hGBP-1307 has been shown to produce GMP with lower catalytic efficiency than that of the full-length (17), taken together it suggests that GMP formation is regulated with dimerization of the proteins. To examine whether hGBP-2, a family of hGBP-1, follows dimerization-linked GMP formation, we carried out similar gel filtration assays. Like hGBP-1, hGBP-2 dimerizes with the analog (Fig. 2), further confirming that GMP formation occurs through dimerization of the protein and the mechanism of the second hydrolysis appears to be conserved in this family of proteins.
Figure 1.
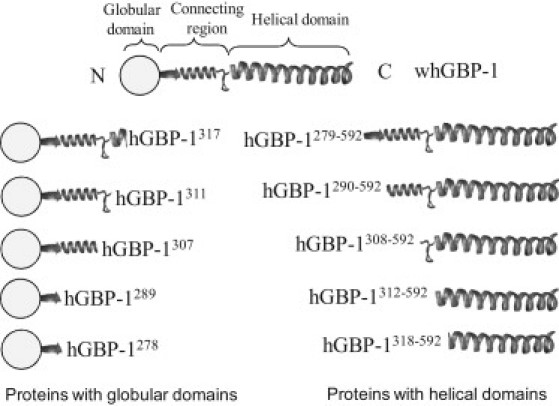
Schematic representation showing the structural elements and domains in the full-length and truncated hGBP-1 proteins.
Figure 2.
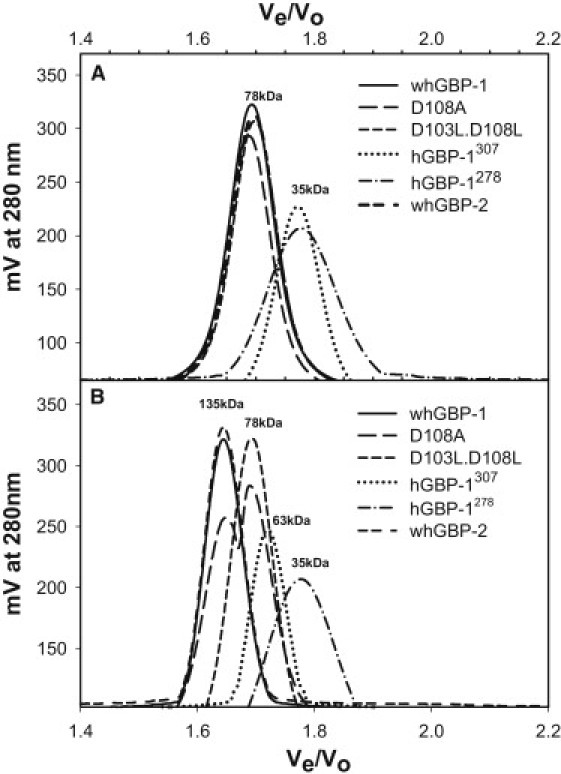
Analytical gel filtration assay of various proteins was carried out without (A) and with (B) GTP-analog GppNHp. For experiment with the analog, 200 μM GppNHp was incubated with protein with a buffer containing 50 mM Tris, 100 mM KCl, and 5 mM MgCl2, pH 8.0. The mixture was kept for at least 1 h before injecting into the column. The mobile phase contained the buffer and 200 μM GppNHp. In either case, the protein concentration was kept at 20 μM.
Table 1.
Molecular masses and dimerization status of the wild-type, mutant, and truncated proteins in the presence and absence of GTP-analog GppNHp based on the analytical size-exclusion chromatography
| Protein | Molecular mass with GppNHp (kDa) | Molecular mass without GppNHp (kDa) | Dimer/monomer with GppNHp | Ability to produce GMP |
|---|---|---|---|---|
| whGBP-1 | 135 ± 4.5 | 78 ± 2 | Dimer | Yes |
| D108A | 135 ± 4 | 78 ± 2.5 | Dimer, Monomer | Yes |
| D103L.D108L | 79 ± 3 | 78 ± 2 | Monomer | No |
| hGBP-1317 | 62.5 ± 3 | 34.6 ± 2.5 | Dimer | Yes |
| hGBP-1311 | 62.5 ± 4 | 34.6 ± 3 | Dimer | Yes |
| hGBP-1307 | 62.5 ± 3 | 34.6 ± 2.5 | Dimer | Yes |
| hGBP-1289 | 34.6 ± 3 | 34.6 ± 2.5 | Monomer | No |
| hGBP-1278 | 34.6 ± 3 | 34.6 ± 3 | Monomer | No |
| hGBP-1279-592 | ND | 143 ± 3, 46 ± 2 | Dimer, Monomer | — |
| hGBP-1290-592 | ND | 143 ± 3, 46 ± 2.5 | Dimer, Monomer | — |
| hGBP-1308-592 | ND | 46 ± 2 | Monomer | — |
| hGBP-1312-592 | ND | 46 ± 2 | Monomer | — |
| hGBP-1318-592 | ND | 46 ± 2 | Monomer | — |
Experiments were carried out in triplicate and the results were found to be consistent. ND, not determined.
α-Helix of the connecting region has a pivotal role in dimerization
To identify which region of the multidomain protein hGBP-1 is involved in the dimerization, we made a series of truncated proteins as shown in Fig. 1, where the nucleotide-binding domain has been removed. The analytical gel filtration assay of these proteins was carried out. The presence of the analog was not required because the nucleotide-binding domain has been deleted. Remarkably, hGBP-1290-592 eluted with two peaks (Fig. 3); the higher retention volume corresponds to a molecular mass of 46 kDa, consistent with a monomer (calculated molecular mass of a monomer ∼38 kDa), whereas the smaller one with molecular mass of ∼143 kDa may correspond to a dimer. We also obtained a similar result with hGBP-1279-592. Interestingly, hGBP-1308-592, hGBP-1312-592, and hGBP-1318-592, which do not contain the α-6 helix, eluted with a single peak (Fig. 3) that corresponds to a monomer. The estimated molecular mass of a dimer in hGBP-1290-592 and hGBP-1279-592 is higher than the calculated (∼76 kDa). To confirm whether it is a dimer, we carried out chemical cross-linking experiments of these truncated proteins. This was employed by using the zero-length cross-linker 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride in the presence of n-hydroxy succinimide. We also carried out similar experiments on the other truncated proteins as shown in Fig. 4. The reaction products were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. As observed in Fig. 4, in the presence of the cross-linking reagents hGBP-1290-592 and hGBP-1279-592 exist as a dimer. On the other hand, even after a long incubation period (3 h), hGBP-1308-592 and hGBP-1318-592 exist primarily as a monomer (Fig. 4). A very faint band of dimer was observed in these truncated proteins. This could be due to some contaminations and/or nonspecific interactions. All these data confirm that the truncated proteins containing at least α-6 helix (hGBP-1289-592 and hGBP-1279-592) are able to dimerize without the globular domain. Thus, the α-6 helix of the connecting region plays a critical role in the dimerization.
Figure 3.

Analytical gel filtration assay of the truncated hGBP-1 proteins with a buffer containing 50 mM Tris, 100 mM KCl, and 5 mM MgCl2, pH 8.0. The mobile phase did not contain analog because the nucleotide binding domain has been removed. The concentration of the protein was 20 μM.
Figure 4.
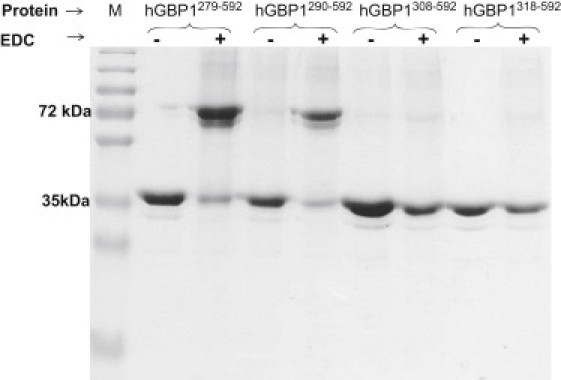
Chemical cross-linking of the truncated hGBP-1 proteins. This was done in the presence of 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride and n-hydroxy succinimide as described in the Materials and Methods in the Supporting Material and the products were run on sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The experiments were done both in the absence and presence of the cross-linker. The experiments were carried out in duplicate and the results were highly reproducible.
α-Helix of the connecting region gets exposed upon nucleotide binding and mediates dimerization
As already known, hGBP-1 is a monomer without the substrate analog but dimerizes upon binding with the analog. Our analytical gel filtration and cross-linking assays suggest that the α-6 helix may be buried inside the protein but becomes exposed upon nucleotide binding and thus the protein dimerizes. Because the α-6 helix has a large number of hydrophobic residues (see the Discussion), its exposure may increase the hydrophobicity of the protein. To determine whether the binding of the nucleotide to the wild-type hGBP-1 increases the total hydrophobicity of the protein, fluorescence measurements were carried out using a hydrophobic dye 1-8-anilinonaphthalene sulfonate (ANS). This was done in the absence and presence of the analog GppNHp. The increase in the fluorescence of ANS is a measure of the total surface hydrophobicity of the proteins (18,19). Fig. 5 A shows an increase in fluorescence with increasing concentrations of ANS in the wild-type protein with and without the substrate analog. However, the increase in fluorescence with the nucleotide-bound protein is higher than the unbound, indicating that the nucleotide-bound protein (dimer) exhibits higher surface hydrophobicity than that of the unbound (monomer). Similar experiments were carried out with the double-mutant to reveal whether the hydrophobicity increases in the presence of the analog. As observed in Fig. 5 B, the fluorescence of ANS is almost the same in both the nucleotide-bound and free forms of the double-mutant, indicating that the exposed hydrophobicity did not alter upon binding with the nucleotide. The circular dichroism measurement on the wild-type hGBP-1 showed that the protein undergoes a conformational change upon binding with the analog (17). We also carried out a similar experiment with the double-mutant in the presence of the analog, but it did not show any change in the molar ellipticity (see Fig. S1 in the Supporting Material). The structure of the full-length hGBP-1 showed that the α-6 helix of the connecting region is buried inside the protein (Fig. 6 A). All these data suggest that the binding of the nucleotide induces a conformational change in the wild-type protein, which increases the total hydrophobicity upon exposure of the α-6 helix and thus mediates dimerization (see the Discussion).
Figure 5.
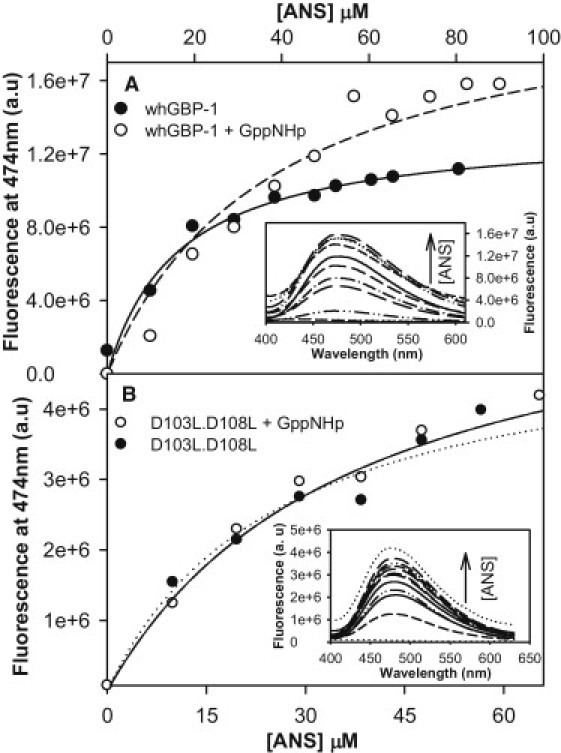
ANS binding studies. (A) The fluorescence intensity at 474 nm was plotted with increasing concentrations of ANS in the presence of 1 μM wild-type hGBP-1 with and without 200 μM GppNHp. (Inset) Representative case for the titration of whGBP-1 plus analog with ANS concentration. (B) A similar experiment was carried out with a double-mutant D103L.D108L. The concentration of the double-mutant and analog was 1 and 200 μM, respectively. (Inset) Titration of the double-mutant plus analog with increasing concentrations of ANS. Each spectrum was recorded as the average of three fluorescence emission wavelength scans. The experiments were carried out in triplicate and the results were consistent.
Figure 6.
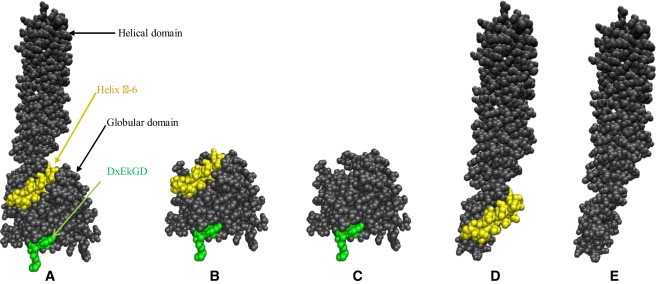
Space-filled structures of the full-length as well as truncated hGBP-1 proteins without the nucleotide were shown using a PDB code-1dg3. These are generated using VMD software. The figure shows the gradual removal of the different domains. This shows that the connecting region is partly buried but becomes exposed upon removal of the different domains as shown here: (A) full-length whGBP-1, (B) hGBP-1307, (C) hGBP-1278, (D) hGBP-1279-592, and (E) hGBP-1318-592.
To further consolidate this idea and to directly investigate whether the helix is indeed involved in dimerization, we decided to introduce a tryptophan residue in the place of Leu298 of the α-6 helix, which is facing outside so that, after binding with the analog, it can be in the dimeric interface. We therefore carried out intrinsic tryptophan fluorescence of the wild-type and L298W mutant proteins in the absence and presence of the analog GppNHp. The wild-type protein has four tryptophan residues. The wild-type protein showed similar fluorescence in the absence and presence of the analog—indicating that the environment of tryptophans in the wild-type protein did not alter upon dimerization, which is consistent with the earlier report (15). As expected, L298W showed higher fluorescence than the wild-type without the analog. Because we introduced a tryptophan in the α-6 helix and this helix becomes exposed upon nucleotide binding and is involved in dimerization, the fluorescence of the L298W should alter. In contrast to the wild-type, L298W showed a significant decrease in fluorescence upon nucleotide binding (Fig. 7). This may be explained by the energy transfer from the tryptophan residue (L298W) of one monomer to the same residue in the other monomer (i.e., homotransfer), when these are in close proximity. The decrease in the intensity may also be contributed by the energy transfer from Tyr300 (in the α-6 helix) to L298W (i.e., heterotransfer) between the two intermonomers. We also carried out similar experiments by taking an equimolar mixture of the wild-type and L298W in the presence of the analog in support of homotransfer. In this case, homo- (WT.WT, L298W.L298W) as well as heterodimers (WT.L298W) are expected to be formed. Interestingly, the fluorescence of the mixture is more than L298W with the analog, but is less than L298W without the analog (Fig. 7). The increase in the fluorescence clearly indicates the absence of homotransfer in 33% of heterodimer. It is to be noted that L298W showed a GMP/GDP ratio similar that of the wild-type when the assay was carried out with radiolabeled [α-32P] GTP (Fig. S2). The homotransfer has been reported in many proteins where tryptophan residues were close within the R0 (the distance corresponding to 50% energy transfer) of ∼6–12 Å (20,21). All of these results clearly suggest that L298W residues of the two monomers are in close proximity in the presence of the analog and thus the helix is involved in dimerization.
Figure 7.
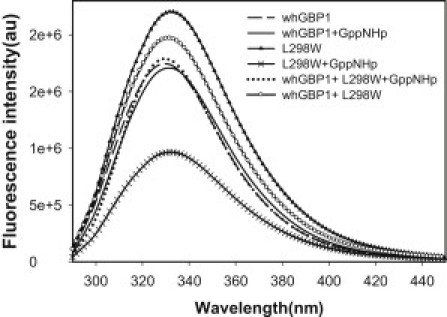
Intrinsic tryptophan fluorescence of the wild-type and L298W hGBP-1 proteins in the absence and presence of the substrate analog GppNHp. A similar experiment was carried out with equimolar mixture of the wild-type and L298W. The concentration of total protein was kept at 0.5 μM. One-hundred micromole of the analog was used for the assay. The samples were excited at 280 nm and the fluorescence emission spectra were recorded between 290 and 450 nm. A blank containing the buffer as described in the Materials and Methods was subtracted from each spectrum. Each spectrum was recorded as the average of three fluorescence emission wavelength scans. The experiments were carried out in triplicate and the results were consistent.
A cross talk of monomers is essential for GMP formation
Our studies so far suggest that dimerization is essential for GMP formation as well as for the stimulation of GTPase activity in both hGBP-1 and hGBP-2. This suggests that there may be a cross talk between the two monomers, which allows the second hydrolysis. This could be tested by preventing dimerization of the protein. This was done by immobilizing the proteins in two different ways in the absence of the substrate analog to eliminate the method-dependent specificities. Both methods are well documented for proteins and are known not to alter the binding sites of enzymes and antibodies (22–25). This ensures that the immobilized protein will remain in the monomer in the presence of the substrate. These proteins were used for the activity assay with the radiolabeled [α-32P] GTP. As observed in Fig. 8, in contrast to the unimmobilized wild-type hGBP-1, the immobilized proteins produced only GDP, and GMP formation was abolished. We also carried out similar assays with the mutant proteins D108A and D103L.D108L. The single mutant showed result similar to the wild-type but as expected, the double-mutant did not produce GMP (Fig. 8). A positive control was carried out, i.e., the protein was allowed to dimerize first with the analog and then immobilized, and the radioactive assay was done (Fig. S3). We failed to observe GMP (see the Discussion). We also carried out similar experiments on the truncated proteins hGBP-1317 and hGBP-1307, and these showed results similar to the wild-type (Fig. 8). All these validate our immobilization assays and confirm that the monomer can hydrolyze GTP to only GDP. Similar studies were carried out with hGBP-2 to examine whether the interactions between the monomers are also essential for GMP formation. Interestingly, hGBP-2 showed results similar to the hGBP-1, confirming that the monomer can hydrolyze GTP to only GDP (Fig. 8). The absence of GMP formation by preventing dimerization in the immobilized proteins clearly demonstrates a cross talk between the two monomers that allows the second hydrolysis. The data on hGBP-2 suggests that this family of proteins primarily follows a common mechanism of dimerization-associated GTP hydrolysis to GMP.
Figure 8.
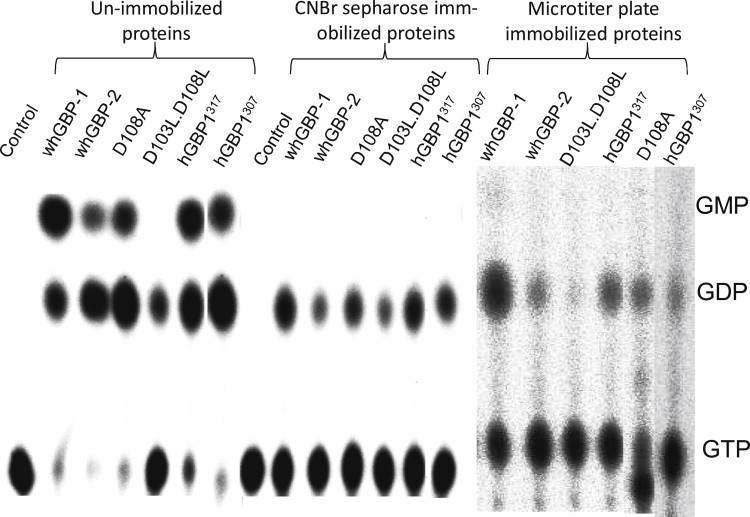
GTPase assays of the unimmobilized and immobilized proteins. The assay was carried out by mixing the proteins with a trace amount of [α-32P] GTP plus 50 μM of unlabeled GTP at 37°C for 1 h. The reaction was terminated with 250 mM EDTA (final concentration). The reaction mixture was separated as described earlier. These experiments were carried out in triplicate and the results were reproducible.
Discussion
Protein-protein interaction is a central theme in GTPase biology. Ras and other small GTPases play many crucial roles because they interact with a number of regulatory proteins. Although large GTPases differ sharply from the small ones, the basic interactions are retained in the G-domain. Among large GTPases, Dynamin and MxA have been studied extensively because they play important roles in vesicle formation and antiviral activity against large numbers of RNA and DNA viruses, respectively. Although these proteins have been shown to undergo the substrate-induced oligomerization and activation without the external GAP (26–28), which region of the multidomain proteins is involved in the self-assembly is not clearly understood because of the absence of crystal structure for the full-length protein in the oligomeric state. Using limited proteolysis, it has been shown that an α-helical domain of 13 kDa between the pleckstrin homology and the proline- and arginine-rich domains in Dynamin is important for self-assembly and stimulation of the GTPase activity (26). Similarly, for Mx, by using yeast two hybrid assays, the C-terminal domain has been shown to be important for oligomerization (28). Among large GTPases, hGBP-1 and 2 have the unique feature of substrate-induced dimerization with additional product (GMP) formation. Although the biological role of GMP formation by hGBP-1 is not yet known, it is possible that the dimerization-associated GMP formation may be related with the antiviral activity against VSV and EMCV.
By using analytical gel filtration assays on a series of truncated globular as well as helical proteins of hGBP-1, we have identified that a helix of 18 residues in the connecting region, rather than the whole helical domain, plays a critical role in dimerization. This is further confirmed by the cross-linking experiments where the helical domain containing the α-6 helix can dimerize without the globular domain. We also previously showed that the presence of this helix with the globular domain is able to provide GMP formation (17). The crystal structure of the full-length protein dimer is not yet known. However, the structure of a truncated protein hGBP-1317 dimer in complex with GMP.AlF4 has been reported (29). Analysis of the structure reveals that the connecting region has not been found to interact in dimerization, but the conserved 103DxEKGD108 motif is positioned at the dimeric interface. It is possible that the orientation of the dimer in the truncated protein may be slightly altered from the full-length. Nevertheless, our data collectively provide an important role for the helix in the dimerization and regulation of the GTPase activity. hGBP-2 exhibits ∼75% of sequence identity with the hGBP-1 and shows substrate-induced dimerization similar to hGBP-1. Because dimerization has also been found to be essential for GMP formation in hGBP-2, the connecting region of this protein may play a similar role in self-assembly. Although both hGBP-1 and hGBP-2 exhibited a similar mechanism of GTP hydrolysis, the marked difference in the ratio of GMP/GDP formation in these two enzymes could be due to differences in the dynamics of dimer dissociation after the first hydrolysis.
Because the α-6 helix of the connecting region in hGBP-1 plays a critical role in dimerization, a key question is, despite having this in the full-length hGBP-1, why is the protein a monomer without the substrate analog?
Our earlier studies with CD measurement showed that upon binding with the substrate analog, the wild-type protein undergoes a conformational change (17). This is essential for dimerization of the protein, which is supported by the CD measurement of the double-mutant D103L.D108L, where it did not undergo conformational change with the analog and hence did not dimerize. Analysis of the structure in the full-length protein shows that the α-6 helix of the connecting region is partly buried inside the protein (Fig. 6 A) and that the helix has a large number of hydrophobic residues such as iso-leucine, leucine, valine, etc. The dimerization of the protein upon exposure of this helix is supported by the ANS-binding experiment, where the total hydrophobicity increased upon binding with the analog. This is also validated by the double-mutant D103L.D108L where the surface hydrophobicity is virtually unaltered in the presence of the analog. The role of this helix in dimerization is further supported by an increase in the fluorescence of tryptophan with a mixture of the wild-type and L298W (1:1) in the presence of the analog compared to the L298W alone and analog. Therefore, our data strongly suggest that the α-6 helix of the connecting region becomes exposed through a conformational switch (from a closed to open conformation, see Scheme 1) and is involved in dimerization. This correlated well with the structures of the full-length as well as several truncated variants of hGBP-1 using VMD software (http://www.ks.uiuc.edu/), where the α-6 helix of the connecting region in the full-length protein is partly buried (Fig. 6, A–E). As observed in Fig. 6 (compare panels A and D), the connecting region of the truncated hGBP-1279-592 and hGBP-1289-592 is exposed. This elucidates why hGBP-1279-592 and hGBP-1289-592 dimerize without the globular domain, and the truncated globular proteins that lack the connecting region (hGBP-1278 and hGBP-1289) fail to dimerize. Similarly, the absence of this helix in hGBP-1308-592, hGBP-1312-592, and hGBP1318-592 rules out the possibility of dimerization. This provides insight into why the full-length hGBP-1 exists as a monomer despite having the connecting region.
Scheme 1.
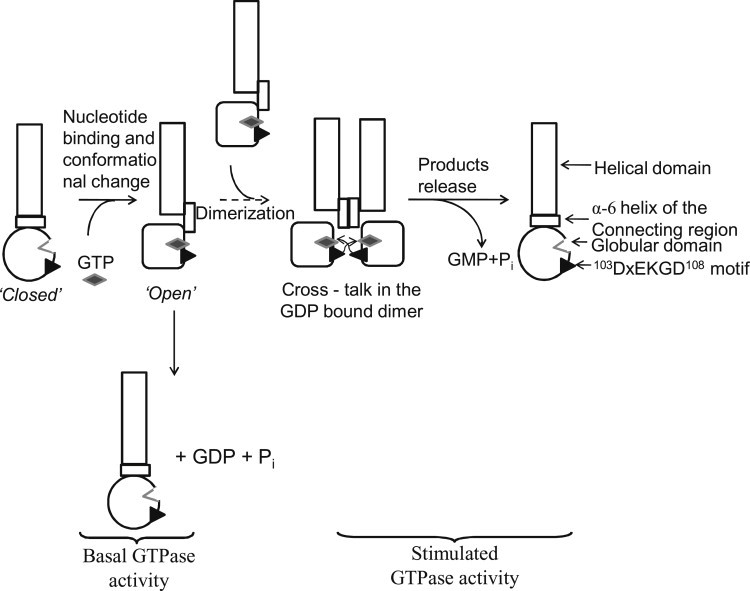
Schematic representation of the GTP hydrolysis by hGBP-1. The helix in the connecting region gets exposed upon binding with GTP from a closed to open conformation and mediates dimerization. For the clarity, the first catalysis has been omitted in the stimulated GTPase activity. Reciprocal trans-activation of whGBP-1 in the GDP-bound dimer after the first catalysis has been shown. The 103DxEKGD108 motif of one monomer could activate the other monomer for the second catalysis in trans.
With a series of truncated and mutant hGBP-1 proteins, our data strongly suggest that GMP formation is directly associated with the dimerization of the protein. This correlated well with the specific activities of the products formation and their ratios (Fig. S4). The immobilization assays confirm that monomer fails to hydrolyze GTP to GMP. The absence of GMP formation in the immobilized dimeric protein is most likely due to the protein going back to monomer after removal of the analog. Nevertheless, our other assays strongly indicate that dimer produces both GDP and GMP with stimulation of the activity. This obviously demonstrates a cross talk between the two monomers during the second catalysis. This is more evident in the truncated hGBP-1317. Unlike the full-length hGBP-1, the truncated hGBP-1317 can utilize GDP as another substrate and hydrolyze to GMP (29).
As observed in Fig. 8, GDP, which is produced after the first catalysis by the immobilized hGBP-1317, cannot be utilized further by the same protein for the second catalysis. The absence of GMP in the immobilized hGBP-1317 clearly indicates that the cross talk is essential for the second catalysis. Thus, the immobilization assay and the gel filtration analysis together show that the monomer and dimer of both hGBP-1 and hGBP-2 exhibit different GTPase activities. Monomer can hydrolyze GTP to only GDP (first hydrolysis), whereas dimerization is absolutely essential for the second hydrolysis (GDP to GMP) and regulates this activity. Although, the second catalysis is associated with the cross talk, the mechanism is still unknown. In the case of small GTPases, external GAPs bind and stimulate the GTPase activity by providing catalytic residues or stabilizing transition states (30,31). Although oligomerization-induced stimulation of GTPase activity is a hallmark in large GTPases, why the hGBP-1 family exceptionally exhibits additional product GMP formation (i.e., second hydrolysis) is still not known.
Based on our data and available literature, we hypothesize that the cross talk could happen in two possible ways.
First, as observed in the truncated dimeric protein (hGBP-1317) in complex with GMP.AlF4, after the first hydrolysis there is a change in the nucleotide conformation at the ribose sugar that keeps the guanine base intact but moves the β-phosphate in the position of the γ-phosphate to utilize the same catalytic machinery for the second hydrolysis. In this way, it prevents the dissociation of bound GDP after the first phosphate cleavage so that it can carry out the second cleavage. The conformational change may occur only when the protein is a dimer.
Second, there could be a direct influence on the second catalysis of one monomer by the catalytic residues of another monomer. As described earlier, hGBP-1 family has a unique 103DxEKGD108 motif that is not present in other GTP-binding proteins. The conservation of the motif in this family clearly suggests that it may have a significant role in GTPase activity. Our mutational studies earlier showed that this motif plays a very critical role in GMP formation. As discussed earlier, the motif in the truncated hGBP-1317 is positioned at the dimer interface and could be potentially important for the catalysis.
As described in Scheme 1, it is possible that the 103DxEKGD108 motif of one monomer could provide a catalytic residue to the other monomer for the second catalysis in trans and thus they reciprocally stimulate the GTPase activity of each other without the need for external regulatory factors. These features perhaps made this family of protein catalytically exceptional with the formation of a second product GMP compared to other GTPases. Further investigations are necessary to distinguish between these two possibilities.
Conclusions
Our data with hGBP-1 and hGBP-2 provide a unique example that shows differential GTPase activities depending on the dimerization of the protein (Scheme 1); a monomer can hydrolyze GTP to only GDP, whereas a dimer hydrolyzes GTP to both GDP and GMP with stimulation of the activity through a cross talk between the two monomers. The monomer may be associated with the basal GTPase activity, where the protein is expressed at low level without IFN-γ induction. However, upon induction, the proteins dimerize as the intracellular concentration of GTP is ∼100 μM. Our study provides insight as to how the large GTPases self-regulate their activities through the substrate-induced conformational switch that leads to dimerization and in turn, stimulation of the activity in response to the inflammatory cytokines. The presence of the second product GMP in both hGBP-1 and hGBP-2 suggests that this family of protein has a catalytic machinery that is unique compared to other GTPases. Although the underlying mechanism for GMP formation remains to be established, this study presents new insights as to the regulation of GTPase activity in the cell. The ability of this protein to dimerize and produce GMP may be related with the antiviral activity against VSV and EMCV.
Acknowledgments
We acknowledge the use of the advanced instrumentation facility provided by Jawaharlal Nehru University for carrying out CD measurements. We thank Prof. G. Krishnamoorthy, Tata Institute of Fundamental Research, Mumbai, for his comments on the manuscript.
This work was supported by the National Institute of Immunology and Department of Biotechnology, India. N.A. thanks the Council of Scientific and Industrial Research, India, for a senior research fellowship.
Supporting Material
References
- 1.Praefcke G.J., McMahon H.T. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat. Rev. Mol. Cell Biol. 2004;5:133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- 2.Olszewski M.A., Gray J., Vestal D.J. In silico genomic analysis of the human and murine guanylate-binding protein (GBP) gene clusters. J. Interferon Cytokine Res. 2006;26:328–352. doi: 10.1089/jir.2006.26.328. [DOI] [PubMed] [Google Scholar]
- 3.Cheng Y.S., Colonno R.J., Yin F.H. Interferon induction of fibroblast proteins with guanylate binding activity. J. Biol. Chem. 1983;258:7746–7750. [PubMed] [Google Scholar]
- 4.Boehm U., Guethlein L., Howard J.C. Two families of GTPases dominate the complex cellular response to IFN-γ. J. Immunol. 1998;161:6715–6723. [PubMed] [Google Scholar]
- 5.Anderson S.L., Carton J.M., Rubin B.Y. Interferon-induced guanylate binding protein-1 (GBP-1) mediates an antiviral effect against vesicular stomatitis virus and encephalomyocarditis virus. Virology. 1999;256:8–14. doi: 10.1006/viro.1999.9614. [DOI] [PubMed] [Google Scholar]
- 6.Guenzi E., Töpolt K., Stürzl M. The guanylate binding protein-1 GTPase controls the invasive and angiogenic capability of endothelial cells through inhibition of MMP-1 expression. EMBO J. 2003;22:3772–3782. doi: 10.1093/emboj/cdg382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duan Z., Foster R., Seiden M.V. GBP1 over-expression is associated with a paclitaxel resistance phenotype. Cancer Chemother. Pharmacol. 2006;571:25–33. doi: 10.1007/s00280-005-0026-3. [DOI] [PubMed] [Google Scholar]
- 8.Modiano N., Lu Y.E., Cresswell P. Golgi targeting of human guanylate-binding protein-1 requires nucleotide binding, isoprenylation, and an IFN-γ-inducible cofactor. Proc. Natl. Acad. Sci. USA. 2005;102:8680–8685. doi: 10.1073/pnas.0503227102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carter C.C., Gorbacheva V.Y., Vestal D.J. Inhibition of VSV and EMCV replication by the interferon-induced GTPase, mGBP-2: differential requirement for wild-type GTP binding domain. Arch. Virol. 2005;150:1213–1220. doi: 10.1007/s00705-004-0489-2. [DOI] [PubMed] [Google Scholar]
- 10.Guimarães D.P., Oliveira I.M., Hainaut P. Interferon-inducible guanylate binding protein (GBP)-2: a novel p53-regulated tumor marker in esophageal squamous cell carcinomas. Int. J. Cancer. 2009;124:272–279. doi: 10.1002/ijc.23944. [DOI] [PubMed] [Google Scholar]
- 11.Schwemmle M., Staeheli P. The interferon-induced 67-kDa guanylate-binding protein (hGBP1) is a GTPase that converts GTP to GMP. J. Biol. Chem. 1994;269:11299–11305. [PubMed] [Google Scholar]
- 12.Neun R., Richter M.F., Schwemmle M. GTPase properties of the interferon-induced human guanylate-binding protein 2. FEBS Lett. 1996;390:69–72. doi: 10.1016/0014-5793(96)00628-x. [DOI] [PubMed] [Google Scholar]
- 13.Prakash B., Praefcke G.J., Herrmann C. Structure of human guanylate-binding protein 1 representing a unique class of GTP-binding proteins. Nature. 2000;403:567–571. doi: 10.1038/35000617. [DOI] [PubMed] [Google Scholar]
- 14.Prakash B., Renault L., Wittinghofer A. Triphosphate structure of guanylate-binding protein 1 and implications for nucleotide binding and GTPase mechanism. EMBO J. 2000;19:4555–4564. doi: 10.1093/emboj/19.17.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kunzelmann S., Praefcke G.J., Herrmann C. Nucleotide binding and self-stimulated GTPase activity of human guanylate-binding protein 1 (hGBP1) Methods Enzymol. 2005;404:512–527. doi: 10.1016/S0076-6879(05)04045-0. [DOI] [PubMed] [Google Scholar]
- 16.Praefcke G.J., Kloep S., Herrmann C. Identification of residues in the human guanylate-binding protein 1 critical for nucleotide binding and cooperative GTP hydrolysis. J. Mol. Biol. 2004;344:257–269. doi: 10.1016/j.jmb.2004.09.026. [DOI] [PubMed] [Google Scholar]
- 17.Abdullah N., Srinivasan B., Sau A.K. Role of individual domains and identification of internal gap in human guanylate binding protein-1. J. Mol. Biol. 2009;386:690–703. doi: 10.1016/j.jmb.2008.12.060. [DOI] [PubMed] [Google Scholar]
- 18.Cardamone M., Puri N.K. Spectrofluorimetric assessment of the surface hydrophobicity of proteins. Biochem. J. 1992;282:589–593. doi: 10.1042/bj2820589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matulis D., Lovrien R. 1-Anilino-8-naphthalene sulfonate anion-protein binding depends primarily on ion pair formation. Biophys. J. 1998;74:422–429. doi: 10.1016/S0006-3495(98)77799-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moens P.D.J., Helms M.K., Jameson D.M. Detection of tryptophan to tryptophan energy transfer in proteins. Protein J. 2004;23:79–83. doi: 10.1023/b:jopc.0000016261.97474.2e. [DOI] [PubMed] [Google Scholar]
- 21.de Oliveira A.H.C., Giglio J.R., Ward R.J. The effect of resonance energy homotransfer on the intrinsic tryptophan fluorescence emission of the bothropstoxin-I dimer. Biochem. Biophys. Res. Commun. 2001;284:1011–1015. doi: 10.1006/bbrc.2001.5073. [DOI] [PubMed] [Google Scholar]
- 22.Parikh I., March S., Cuatercasas P. Topics in the methodology of substitution reactions with agarose. Methods Enzymol. 1974;34:77–102. doi: 10.1016/s0076-6879(74)34009-8. [DOI] [PubMed] [Google Scholar]
- 23.Hermanson G.T., Mallia A.K., Smith P.K. Academic Press; London, UK: 1992. Immobilized Affinity Ligand Techniques. [Google Scholar]
- 24.Hermanson I. Preparing antibody resins. In: Dean P.G., Johnson W.S., Middle F.A., editors. Affinity Chromatography: A Practical Approach. IRL/Oxford University Press; Oxford, UK: 1985. pp. 31–34. [Google Scholar]
- 25.Engvall E. Enzyme immunoassay ELISA and EMIT. Methods Enzymol. 1980;70:419–439. doi: 10.1016/s0076-6879(80)70067-8. [DOI] [PubMed] [Google Scholar]
- 26.Muhlberg A.B., Warnock D.E., Schmid S.L. Domain structure and intramolecular regulation of dynamin GTPase. EMBO J. 1997;16:6676–6683. doi: 10.1093/emboj/16.22.6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramachandran R., Surka M., Schmid S.L. The dynamin middle domain is critical for tetramerization and higher-order self-assembly. EMBO J. 2007;26:559–566. doi: 10.1038/sj.emboj.7601491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Di Paolo C., Hefti H.P., Pavlovic J. Intramolecular backfolding of the carboxyl-terminal end of MxA protein is a prerequisite for its oligomerization. J. Biol. Chem. 1999;274:32071–32078. doi: 10.1074/jbc.274.45.32071. [DOI] [PubMed] [Google Scholar]
- 29.Ghosh A., Praefcke G.J., Herrmann C. How guanylate-binding proteins achieve assembly-stimulated processive cleavage of GTP to GMP. Nature. 2006;440:101–104. doi: 10.1038/nature04510. [DOI] [PubMed] [Google Scholar]
- 30.Scheffzek K., Ahmadian M.R., Wittinghofer A. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277:333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- 31.Rittinger K., Walker P.A., Gamblin S.J. Structure at 1.65 A of RhoA and its GTPase-activating protein in complex with a transition-state analogue. Nature. 1997;389:758–762. doi: 10.1038/39651. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


