Abstract
The effect of Pb2+ ions on the Na+,K+-ATPase was investigated in detail by means of steady-state fluorescence spectroscopy. Experiments were performed by using the electrochromic styryl dye RH421. It is shown that Pb2+ ions can bind reversibly to the protein and do not affect the Na+ and K+ binding affinities in the E1 and P-E2 conformations of the enzyme. The pH titrations indicate that lead(II) favors binding of one H+ to the P-E2 conformation in the absence of K+. A model scheme is proposed that accounts for the experimental results obtained for backdoor phosphorylation of the enzyme in the presence of Pb2+ ions. Taken together, our results clearly indicate that Pb2+ bound to the enzyme stabilizes an E2-type conformation. In particular, under conditions that promote enzyme phosphorylation, Pb2+ ions are able to confine the Na+,K+-ATPase into a phosphorylated E2 state.
Introduction
The Na+,K+-ATPase was the first ion pump to be discovered (1), and it is a fundamental integral membrane enzyme for animal physiology. This enzyme, which belongs to the P-type ATPase family, actively transports sodium and potassium ions against their electrochemical gradients across the plasma membrane in most animal cells by utilizing the free enthalpy of ATP hydrolysis (2–4). The Na+ and K+ electrochemical gradients are required for basic cellular functions such as maintenance of membrane potential, regulation of cell volume, secondary transport of other solutes, and signal transduction. The molecular mechanism of ion transport in P-type ATPases is usually described by the so-called Albers-Post cycle (5,6). According to this model, the pump can exist in two main conformations: E1 and E2. The E1 conformation has a high affinity for sodium ions, can be phosphorylated by ATP, and presents ion binding sites to the cytoplasm. The E2 conformation has a high affinity for potassium ions, can be phosphorylated by inorganic phosphate (Pi), and presents ion binding sites to the extracellular aqueous phase. The recently published x-ray crystal structures of the Na+,K+-ATPase (7,8) support the concept that Na+ and K+ ions share common binding sites, with the exception of the third sodium ion, which binds to a distinct region.
Lead is a toxic heavy metal that poses a major public health problem around the world. The nervous system, kidneys, and blood are primary targets of the toxic effects of Pb2+. At the cellular and molecular level, it is reported that lead has the potential to induce oxidative stress and cause direct damage to the structure and function of biological membranes (9). It is known that several heavy metals, including lead, inhibit the activity of P-type ATPases. For instance, toxic heavy metal ions (Cd2+, Hg2+, Pb2+, Cu2+, Zn2+) have been shown to affect the activity of Ca2+-ATPase (10), Mg2+-ATPase, and Na+,K+-ATPase (11–14). In particular, the inhibitory effect of Pb2+ on the Na+,K+-ATPase has been investigated in different membrane preparations, i.e., microsomes from beef cerebellar cortex (11), human erythrocyte membranes (12), synaptic plasma membranes from rat brain (13), and Electrophorus electroplax microsomes (14).
Despite such experimental evidences, however, the molecular mechanism underlining Na+,K+-ATPase inhibition by Pb2+ is still unclear. Some information about the effect of Pb2+ on the phosphorylation and dephosphorylation of the Na+,K+-ATPase are provided in earlier studies. Siegel and Fogt (14) suggested that Pb2+ may act at a single independent binding site to produce or stabilize an enzyme conformation that can be phosphorylated but that cannot catalyze hydrolysis of enzyme phosphate. Swarts et al. (15) have shown that if Pb2+ is present during phosphorylation of Na+,K+-ATPase by ATP, the rate of phosphorylation increases and a phosphointermediate is formed which is insensitive toward K+ and ADP. In a previous article, we have investigated the inhibitory effect of Pb2+ on the transport cycle of the Na+,K+-ATPase by combining biochemical and electrical measurements (16). Our results led us to propose that the inhibitory effect of Pb2+ on the Na+,K+-ATPase may be due to an interference with hydrolytic cleavage of the phosphorylated intermediate P-E2, which occurs in the K+-related branch of the pump cycle.
The aim of this work is to unravel the effect of Pb2+ on the electrogenic partial reactions of the Na+,K+-ATPase enzymatic cycle. The experiments were performed by using a fluorescence technique that makes use of the electrochromic styryl dye RH421, whose spectral characteristics are affected by proximity to charge in the membrane (17,18). This method can monitor electrogenic steps within the pump cycle of the protein, and it has been applied in recent studies to identify and analyze the effect of various inhibitors on the reaction kinetics of the Na+,K+-ATPase (19,20). Our results reveal which conformational states of the enzyme are affected by Pb2+ and enable us to propose a molecular mechanism of the Na+,K+-ATPase inhibition by Pb2+.
Materials and Methods
Materials
Magnesium, sodium and potassium chloride, tris(hydroxymethyl)-aminomethane (TRIS), hydrochloric acid, and lead nitrate were obtained from Merck (Whitehouse Station, NJ) at analytical grade. Adenosine-5′-triphosphate disodium salt (ATP, >97%), TRIS phosphate (>99%), ouabain (≥99%), and digitoxigenin (≥99%) were purchased from Fluka (Buchs, Switzerland).
Phosphoenolpyruvate, pyruvate kinase/lactate dehydrogenase suspension, β-nicotinamide adenine dinucleotide (reduced disodium salt hydrate), and L-histidine were purchased from Sigma-Aldrich (St. Louis, MO) at highest quality available.
The electrochromic styryl dye RH421 was obtained from Molecular Probes (Eugene, OR).
Membrane protein preparation
Membrane fragments containing Na+,K+-ATPase were obtained by extraction from the outer medulla of rabbit kidneys using procedure C of Jørgensen (21). The specific ATPase activity was measured by the pyruvate kinase/lactate dehydrogenase suspension assay (22). Protein concentration was determined by the method of Lowry et al. (23) using bovine serum albumin as a standard. The preparation used in this work had a specific activity of 1700 μmol Pi/(h·mg protein) at 37°C. The total protein content of membrane fragments was 2.07 mg/mL.
Fluorescence measurements
The styryl dye RH421 is an amphiphilic molecule that inserts into the lipid domains of Na+,K+-ATPase membrane fragments and detects changes of electric field strength inside the membrane dielectric but not conformational transitions (18). Such changes in electric field strength may arise from ion binding or release and translocations in the course of the pump cycle of the protein. The fluorescence level refers to a particular charged state of the enzyme and the fluorescence change between states can be assumed to be proportional to the change of the electric field in the membrane (17,18). The styryl dye responds with a shift of the absorption spectra to longer (red) or shorter (blue) wavelengths corresponding to changes in the local electric potential inside the membrane to more negative or more positive values, respectively (18).
Fluorescence measurements were carried out with a FP-6500 spectrofluorometer (JASCO, Easton, MD). The excitation wavelength was set to 580 nm (slit-width 10 nm) and the emission wavelength was set to 650 nm (slit-width 20 nm). The thermostated cell holder was set at 20°C and was equipped with a magnetic stirrer. A high-pass optical filter (λcut = 590 ± 6 nm; Edmund Optics, Barrington, NJ) was put between the cuvette and the detector to cut off contributions to the emitted radiation due to higher harmonics.
If not otherwise stated, the standard buffer solution contained 25 mM histidine (pH 7.2), and 5 mM MgCl2. After an equilibration time of 10 min, the styryl dye RH421 was added to the buffer, followed by the aqueous suspension of protein containing membrane fragments after further 5 min. The final concentrations were typically 200 nM for the RH421 dye and ∼6 μg/mL for the protein suspension.
Data obtained from each fluorescence experiment were normalized according to the function ΔF/F0 = (F–F0)/F0 with respect to the initial fluorescence level before the first substrate addition, F0, so that different experiments can be compared easily.
When needed, the experimental data were fitted by the binding isotherm (Hill type function)
| (1) |
where (ΔF/F0)0 is the initial fluorescence level at 0 substrate concentration, (ΔF/F0)max is the maximal fluorescence change attained at saturating substrate concentrations, c is the substrate concentration, K0.5 is the half-saturation concentration, and n is a coefficient related to the cooperativity of the substrate binding process.
For pH titrations, data were fitted by the equation
where (ΔF/F0)∞ is the fluorescence level at high pH.
In the presence of ATP, free Pb2+ concentration was calculated by the MaxChelator software program (24).
Standard experiment
The typical trace of a standard experiment is reported in Fig. S1 in the Supporting Material (trace a). In standard buffer at pH 7.2 and in the absence of Na+ and K+, the Na+,K+-ATPase assumes a proton-bound conformation, HxE1 (1.5 < x < 1.8 (25)), that determines a fluorescence level taken as a reference zero. The subsequent addition of 50 mM NaCl shifts quantitatively the pump into the Na3E1 conformation. It is now well established that only the third sodium ion binds electrogenically to the pump (26), thus determining the fluorescence decrease (∼20% ΔF/F0) observed here (Fig. S1, trace a). Sodium occlusion, protein phosphorylation, conformational change from E1 to E2, and sodium release on the extracellular side are induced by subsequent addition of 500 μM ATP, which displaces the protein in the P-E2 conformation. The uncompensated release of three sodium ions is responsible for the observed 60% ΔF/F0 increase of the fluorescence intensity (Fig. S1, trace a). The final addition of 20 mM K+ promotes full turnover conditions, accompanied by a fluorescence decrease (Fig. S1, trace a) due to accumulation of positive charge inside the membrane. The final steady-state fluorescence level is due to the weighted contribution of the ion-bound conformations (20).
Usually, each experiment was repeated at least three times. Traces reported in the figures are representative of the experiment performed.
Backdoor phosphorylation
To perform backdoor phosphorylation, Na+,K+-ATPase-containing membrane fragments were equilibrated in the standard buffer (25 mM histidine, pH 7.2, and 5 mM MgCl2) with 200 nM RH421 until a constant fluorescence level was obtained. TRIS phosphate (TRIS-Pi) was then added up to 0.5 mM in appropriate aliquots from a 100-mM aqueous stock solution at pH 7.0 to titrate the pump into its phosphorylated state. The same experiment was repeated after equilibration of the pump with different free Pb2+ concentrations. The addition of inorganic phosphate and Pb2+ ions did not affect buffer pH.
Titration experiments
After equilibration of the protein in the standard buffer, titrations with Na+ in the E1 conformation were carried out by adding small aliquots of aqueous stock solutions of NaCl (5 M and diluted stocks). Analogous titrations were performed with Na+ and K+ ions, after confining the pump in the P-E2 conformation by addition of 0.5 mM TRIS-Pi at pH 7.0 (backdoor phosphorylation). All titrations were carried out in the absence and presence of 10-μM free Pb2+.
To perform pH titrations, the protein was equilibrated in a buffer composed by 25 mM His, pH 7.4, and 5 mM MgCl2 (for titrations in the E1 conformation) or 25 mM His-TRIS, pH 8.1, and 5 mM MgCl2 (for titrations in the P-E2 conformation produced by 50 mM NaCl plus 500 μM ATP). After equilibration, small aliquots of 0.25, 0.5, or 1 M HCl were added to the cuvette, depending on the experiment carried out. The same titration was performed in a second cuvette in which pH was measured by a glass microelectrode (model No. 1093B; Hanna Instruments, Woonsocket, RI). The final pH into the two cuvettes was found to differ by no more than 0.08 pH units.
Results
The effect of Pb2+ ions on the Na+,K+-ATPase was investigated in detail by means of steady-state fluorescence spectroscopy. Previous work has shown that Pb2+ inhibits the steady-state hydrolytic activity of the sodium pump in the 0–5 μM range (16). It was proposed that lead ions may interfere with the dephosphorylation step of the enzymatic cycle. The experiments reported in this article were carried out to unravel the mechanism of inhibition at the molecular level by determining the conformational states of the enzyme that are mostly affected by Pb2+ ions.
Control experiments
In order to follow protein-related fluorescence emission by the RH421 dye in the presence of Pb2+ ions, it is first necessary to check whether lead(II) interferes with the fluorescence emission mechanism of the probe. In particular, Pb2+ may interact nonspecifically with the membrane surface, being a divalent ion (27). In this manner, it may produce variations of the local field strength that may be sensed by the electrochromic dye. Therefore, we performed control experiments to exclude possible Pb-induced artifacts.
In our control experiments we used 10 μM free Pb2+, which is able to block completely the ATPase activity (KI = 0.5 μM (16),). We tested the effect of 10 μM free Pb2+ on membrane fragments containing a ouabain-inactivated Na+,K+-ATPase. For this purpose, the protein was incubated in standard buffer in the absence of Na+ and K+ ions and in the presence of Pi (from TRIS-Pi, pH 7.0), so that the protein is confined in the P-E2 conformation (28) (Fig. S1, trace b). Under these conditions, ouabain is able to bind to the protein inhibiting the steady-state hydrolytic activity (29). The addition of 100 μM ouabain determines a reduction of the steady-state fluorescence level as compared to that of the P-E2 level (30). The following addition of 10 μM Pb2+ leaves the fluorescence level unchanged (Fig. S1, trace b). An analogous fluorescence pattern was obtained by adding 100-μM digitoxigenin instead of ouabain (not shown). Moreover, the same concentration of lead(II) ions does not influence the fluorescence emission of the styryl dye partitioned in pure lipid vesicles (not shown). Based on these results, we can exclude an interference of Pb2+ with the RH421 dye up to 10 μM Pb2+.
Effect of Pb2+ on ion-bound conformations
We then added 10 μM free Pb2+ to the ion-bound conformations of the Na+,K+-ATPase, i.e., Na3E1, E2(K2), and HxE1 (Fig. 1, traces A–D). It is possible to confine the Na-pump in the Na3E1 or E2(K2) conformations by adding saturating concentrations of Na+ or K+ ions to the standard buffer (18,31). On the other hand, at pH 7.2 and in the absence of Na+ and K+, protons act as a congeneric ion species and produce a HxE1 conformation, with 1.5 < x < 1.8 (25).
Figure 1.
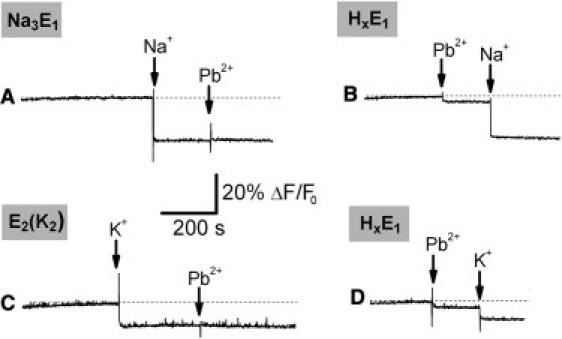
Addition of 10 μM Pb2+ to the ion-bound conformations Na3E1 (trace A), E2(K2) (C), and HxE1 (traces B and D). (Shading) Preferentially adopted conformation when Pb2+ is added. Where indicated (arrows), sodium and potassium ions are added at final concentrations of 50 mM and 20 mM, respectively. The initial fluorescence level, relative to HxE1 conformation, is due to RH421 dye partitioned into protein-containing membrane fragments. (Dashed line) Zero fluorescence level taken as reference.
The addition of 50 mM NaCl stabilizes the Na3E1 conformation of the pump (18,31), and it is accompanied by a fluorescence decrease that is associated to the electrogenic binding of the third sodium ion (26) (Fig. 1, trace A). On the other hand, the addition of 20 mM KCl produced an E2(K2) conformation through the reaction pathway HxE1 + 2K+ ⇆ K2E1 ⇆ E2(K2) (18,31). It has been shown that binding of potassium ions on the extracellular side is completely electroneutral at acidic pH, and it exhibits a small electrogenic contribution at higher pH values (25). In particular, at pH 7.2, the E1 conformation is partially protonated (25) and K+ binding determines a partial charge displacement. With our preparation, the addition of 20 mM KCl produced a 10% ΔF/F0 decrease (Fig. 1, trace C).
The fluorescence levels characteristic of these ion-bound states remained unchanged upon the addition of lead(II) (Fig. 1, traces A and C). This indicates that under these conditions Pb2+ ions do not bind to the protein in an electrogenic fashion, and they do not promote release of the bound ions.
On the other hand, it is interesting to observe that when Pb2+ ions are added to the membrane fragments suspension before Na+ or K+ ions, a small (∼4% ΔF/F0) fluorescence drop is observed (Fig. 1, traces B and D). In both experiments, Pb2+ interacts with the HxE1 conformation of the pump. The subsequent addition of saturating Na+ (Fig. 1, trace B) or K+ ions (Fig. 1, trace D) restores the fluorescence levels characteristic for the ion-bound conformations (in Fig. 1, compare trace A with B, and trace C with D). This suggests that the presence of 10 μM free Pb2+ do not prevent Na+ and K+ binding to the pump. Moreover, by titrating the HxE1 and the P-E2 conformations with Na+ or K+ ions, almost identical K0.5 and n values were obtained in the absence and presence of 10 μM free Pb2+ (Table S1 in the Supporting Material). Therefore, the presence of lead(II) does not affect the affinity of the protein for sodium and potassium ions. It is worth noting that charge measurements also demonstrated that Na+ binding to E1 and release from P-E2 are not affected by the presence of Pb2+ ions up to 20 μM free Pb2+ (16).
In summary, these experiments suggest that: 1), Pb2+ ions allow binding of Na+ and K+ to the protein with unaltered affinity; and 2), they can interact with the HxE1 conformation of the Na-pump.
Standard experiments
With this in mind, we carried out additions of 10 μM free Pb2+ in correspondence to the various fluorescence levels attained during a standard experiment (Fig. 2).
Figure 2.
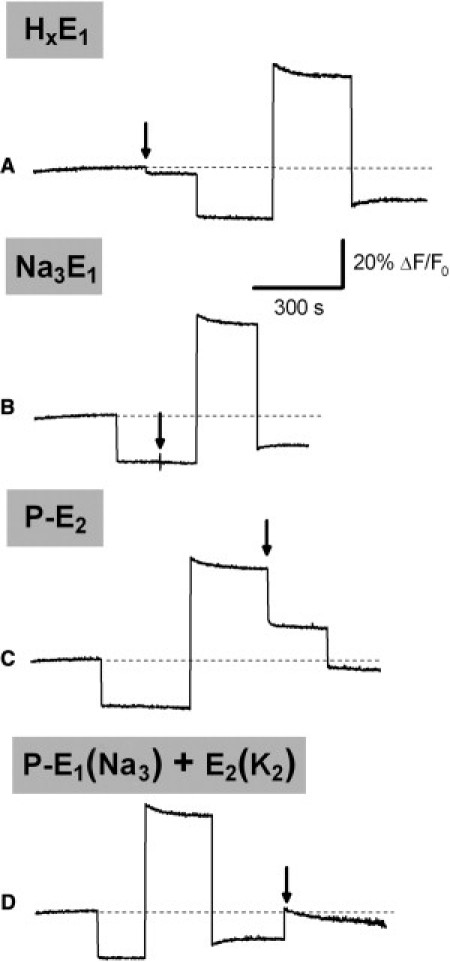
Representative series of standard experiments showing the traces obtained by adding 10 μM Pb2+ in correspondence of the various steady-state fluorescence levels. (Shaded) Preferentially adopted conformation when Pb2+ is added. (Arrows) Lead(II) ions are added where indicated. (Dashed line) Zero fluorescence level taken as reference. The standard experiments were carried out under the experimental conditions reported in Materials and Methods.
Pb2+ addition to HxE1 and Na3E1 conformations (Fig. 2, traces A and B) produces the fluorescence changes observed by adding lead(II) to the ion-bound conformations (Fig. 1, traces B and A, respectively). The following part of the standard experiment does not differ from the pattern observed in the absence of Pb2+ ions (Fig. S1, trace a). This can be explained by considering the very high stability constant of the Pb-ATP complex (log K(PbATP) = 7.02 (10)). In fact, the addition of 0.5 mM ATP removes all free Pb2+ ions from the solution and the standard experiment can proceed as in the absence of Pb2+. A similar behavior was obtained by chelating Pb2+ with EDTA before addition of Na+ (not shown). This result indicates the reversibility of lead(II) binding to the Na-pump.
Pb2+ addition to the P-E2 conformation determines a significant fluorescence drop (∼25% ΔF/F0, Fig. 2, trace C) that can be ascribed to positive charge accumulation inside the membrane dielectric. In this experiment, a total Pb(NO3)2 concentration of 310 μM was added to the standard buffer in order to obtain a 10 μM free Pb2+ concentration in the presence of 0.5 mM ATP (see Materials and Methods). The final addition of K+ ions determines a further drop of the fluorescence signal. This suggests that K+ ions can bind to the pump in the presence of lead(II) ions, in agreement with the results of the K+ titrations in the P-E2 conformation (Table S1).
A concentration of 10 μM free Pb2+ was also added in correspondence to the final step of the standard experiment (Fig. 2, trace D). After the addition of K+ ions, the pump is in turnover condition, and the final steady-state level is determined by the weighted contribution of the ion-occluded conformations, P-E1(Na3) and E2(K2) (20) (Fig. S1, trace a). It has been proposed that the Na3E1 conformation (32,33) and not the P-E1(Na3) (34) contributes to the final fluorescence level. Both sodium-bound conformations are expected to contribute equally to the steady-state fluorescence level, due to the electroneutrality of the phosphorylation process (31). The addition of lead(II) determines an increase of the steady-state fluorescence level (Fig. 2, trace D). It is interesting to note that the final level is the same as that obtained in trace C.
In summary, the standard experiments indicate that:
-
1.
Pb2+ ions bind reversibly to the Na-pump.
-
2.
They also bind to an E2-type conformation.
-
3.
The presence of lead(II) does not prevent K+ binding to the protein in the presence of Na+ ions and ATP.
pH titrations
We then performed pH titrations of E1 and P-E2 conformations in the absence and presence of 10 μM free Pb2+. The experiments begin at high pH values and the subsequent HCl additions titrate the unoccupied binding sites, shifting to the right the reaction sequences HxE1 ⇆ H2E1 ⇆ E2(H2) and P-E2 ⇆ P-E2(H2), respectively.
Results for the E1 conformation are reported in Fig. 3 A. The effect of Pb2+ ions is moderate, producing only a slight (5%) reduction of ΔF/F0 at the initial pH of 7.4. The subsequent HCl addtions titrated the residual empty binding sites. Similar experiments were performed in the P-E2 state produced by 50 mM NaCl plus 500 μM ATP. In this case, the addition of Pb2+ ions causes a major drop of the fluorescence signal at pH 8.1 (Fig. 3 B), similarly to that observed during the standard experiment (Fig. 2, trace C). In particular, it is interesting to observe that in the presence of Pb2+ the fluorescence drop over the pH range from 8 to 5 is approximately half that in the absence of Pb2+.
Figure 3.
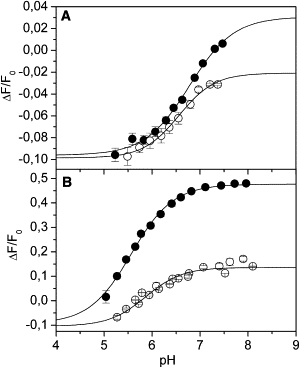
pH titrations performed in the E1 (A) and P-E2 (B) conformations, in the absence (solid circles) and presence (open circles) of 10 μM free Pb2+. Experimental data were fitted by Eq. 2. (A) pK = 6.8 ± 0.1, n = 0.9 ± 0.2 (no Pb2+); pK = 6.56 ± 0.08, n = 1.2 ± 0.3 (with Pb2+). (B) pK = 5.57 ± 0.02, n = 1.00 ± 0.04 (no Pb2+); pK = 5.87 ± 0.09, n = 1.1 ± 0.2 (with Pb2+).
All experimental data can be fitted by the binding isotherm of Eq. 2. In the case of E1 conformation, the pK values in the absence and presence of Pb2+ ions are 6.8 ± 0.1 and 6.56 ± 0.08, respectively. On the other hand, in P-E2 state, the pK value raises from 5.57 ± 0.02 (no Pb2+) to 5.87 ± 0.09 (10 μM free Pb2+).
It is worth noting that the fluorescence level at low pH is the same for all titrations independently of the presence of lead(II) ions.
Moreover, by comparing the fluorescence levels of the E2(H2) or P-E2(H2) (−0.1), HxE1 (0.0) and P-E2 (0.48) states in the absence of Pb2+, the number of protons, x, can be estimated to be 1.7 at pH 7.2.
Backdoor phosphorylation
Finally, we investigated the effect of Pb2+ on the phosphorylated form of the enzyme. It is known that the Na+,K+-ATPase can be phosphorylated by inorganic phosphate (Pi) in the presence of Mg2+ ions but in the absence of Na+ and K+ ions (28). The reaction is known as backdoor phosphorylation and it can be split into two reaction steps. In the first step, the enzyme undergoes a conformational transition, H2E1 ⇆ E2(H2), and in the second step phosphorylation occurs, E2(H2) + Pi ⇆ P-E2H2 ⇆ P-E2 (28). In the absence of Na+ and K+, H+ acts as a congeneric ion species because the formation of the occluded state with empty binding sites, E2(), is energetically unfavorable. The electrogenicity of the backdoor phosphorylation is due to the release of both H+ in the P-E2 conformation, P-E2H2 ⇆ P-E2, because the affinity for protons in the P-E2 conformation is significantly lower than in the E1 conformation (25).
Backdoor phosphorylation experiments were carried out as described in Materials and Methods. The experimental data relative to Pi titrations in the presence of 0, 0.1, 1, 2.5, 5, and 10 μM free Pb2+ are reported in Fig. 4 (A–F, respectively).
Figure 4.
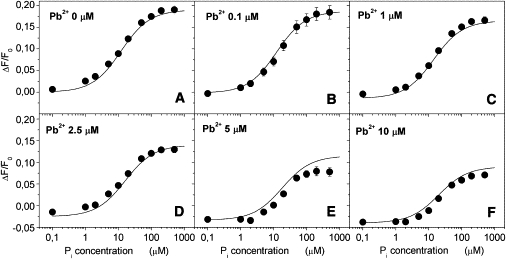
Backdoor phosphorylation titrations carried out in the presence of different Pb2+ concentrations. Experimental data are obtained by averaging two-to-three independent titrations for each concentration of Pb2+ ions. (Error bars) Standard deviation (where not visible, they are hidden by the data points). (Solid lines) Simulated curves obtained by using the six-intermediates model of Fig. 6 (upper panel) as described in the text.
Each titration curve was fitted by a Hill type function (Eq. 1), and the characteristic parameters K0.5 and n were determined (Table 1). In the absence of Pb2+ the half-saturation constant for Pi was found to be 12.1 ± 0.8 μM, which is in reasonable agreement with the values of 23 μM (28), 29 μM (35), and 32 μM (36) reported in the literature for the kidney enzyme. The K0.5 value rises to 21 ± 2 μM when Pb2+ is increased to 10 μM (Table 1). This behavior indicates an apparent decrease in affinity of the sodium pump for Pi. As a consequence, a possible competition between Pi and Pb2+ binding to the protein cannot be excluded. Furthermore, the Hill coefficient also increases from 0.9 ± 0.1 in the absence of Pb2+ to 1.6 ± 0.2 in the presence of 10 μM free Pb2+ (Table 1).
Table 1.
Experimental (K0.5 and n) and simulated (K0.5) parameters are reported for each concentration of lead(II) ions
| Free Pb2+ concentration (μM) | Experimental |
Simulated |
|
|---|---|---|---|
| K0.5 (μM) | n | K0.5 (μM) | |
| 0 | 12.1 ± 0.8 | 0.9 ± 0.1 | 12.1 |
| 0.1 | 15.3 ± 0.7 | 1.05 ± 0.04 | 12.5 |
| 1 | 15.4 ± 0.5 | 1.11 ± 0.04 | 14.9 |
| 2.5 | 13.8 ± 0.9 | 1.12 ± 0.07 | 16.9 |
| 5 | 18 ± 2 | 1.7 ± 0.2 | 18.5 |
| 10 | 21 ± 2 | 1.6 ± 0.2 | 19.8 |
Note that a value for the cooperativity coefficient, n, cannot be obtained from the simulation (see text).
The initial fluorescence level of the titration curves is affected by lead(II). This is not unexpected, because a fluorescence decrease caused by Pb2+ addition to the HxE1 conformation is also evident from experiments carried out on ion-bound conformations (Fig. 1, traces B and D) and from pH titrations in the E1 conformation in the presence of Pb2+ (Fig. 3 A). Such fluorescence decrease must be determined by proton binding to the protein. The dependence of the initial fluorescence levels on Pb-concentration is reported in Fig. 5 A. The experimental data can be fitted by a binding isotherm (Eq. 1) that describes the equilibrium Pb2+ + HxE1 + yH+ ⇆ Hx+yE1Pb. The fitting provides a maximal fluorescence drop of ∼4% ΔF/F0, K0.5 = 3.1 ± 0.9 μM and n = 2 ± 1 (Fig. 5 A).
Figure 5.
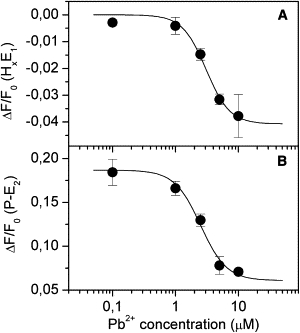
Dependence of the steady-state fluorescence levels of states HxE1 (A) and P-E2 (B) on Pb2+ concentration. (Solid lines) Binding isotherms describing the equilibria Pb2+ + HxE1 + yH+ ⇆ Hx+yE1Pb (A) and Pb2+ + P-E2 + zH+ ⇆ P-E2Pb (Hz) (B). Best fitting with Eq. 1 gives the following values: (ΔF/F0)0 = 0, (ΔF/F0)max = −0.04 ± 0.01, K0.5 = 3.1 ± 0.9 μM, and n = 2 ± 1 (A); (ΔF/F0)0 = 0.187 ± 0.006, (ΔF/F0)max = −0.13 ± 0.02, K0.5 = 2.5 ± 0.4 μM, and n = 2.1 ± 0.7 (B).
The final fluorescence level is also affected by the presence of Pb2+ ions as shown in Fig. 5 B. The decrease of the final fluorescence level with increasing Pb2+ concentration has to be attributed by H+ binding to the phosphorylated enzyme, as suggested by pH titrations in the P-E2 conformation (Fig. 3 B). The best curve that fits the experimental data is the binding isotherm relative to the equilibrium Pb2+ + P-E2 + zH+ ⇆ P-E2Pb (Hz). In this case, the maximal variation of ΔF/F0 is ∼12%. The K0.5 (2.5 ± 0.4 μM) and n (2.1 ± 0.7) values are almost the same as those determined for Pb2+ binding to HxE1. The K0.5 value denotes an affinity for Pb2+ very similar to that in the E1 state. Considering the high uncertainty related to n, it does not make sense to propose a cooperative binding of more than one Pb2+ ion to the pump in both equilibria.
Discussion
Fluorescence measurements by means of styryl dyes are useful to characterize electrogenic partial reactions of the Na+,K+-ATPase (33,37–41) and the pump interaction with drugs (19,20). We recently found that Pb2+ ions inhibit the enzyme activity of the sodium pump almost quantitatively at concentration of 10 μM and above (KI = 0.5 μM (16)). We now use steady-state fluorescence measurements to characterize in detail the effect of Pb2+ ions on the Na-pump and to pinpoint the reaction step(s) of the enzymatic cycle at which the heavy-metal ions provoke their inhibitory action.
We first determined the effect of Pb2+ on the ion binding process. Our results indicate that Na+ and K+ ions can bind to the pump also in the presence of Pb2+ ions (Fig. 1, traces B and D). Although charge measurements previously indicated that sodium binding and release are not affected by lead(II) ions (16), here we demonstrated for the first time, to our knowledge, that Pb2+ ions do not modify the binding affinity of the pump for sodium and potassium ions (Table S1). We can therefore conclude that Pb2+ binding to the protein does not block the access pathway to ion-binding sites.
The experiments performed by adding Pb2+ to the ion-bound conformations also indicate that lead(II) appears to bind to the HxE1 conformation (Fig. 1, traces B and D). Under the adopted experimental conditions (i.e., absence of Na+ and K+ ions), we observed a slight reduction of the fluorescence signal (∼5%), which is confirmed by pH titrations in the E1 state (Fig. 3 A) and the experimental data of Fig. 5 A. This reduction has to be ascribed to a corresponding further 5% protonation of the pump, producing an Hx+yE1Pb state, with y ≈ 0.1. It is worth mentioning that pH titrations in the E1 state indicate a Pb2+-induced moderate decrease in proton affinity.
On the other hand, a significant fluorescence decrease (∼25%) is observed after addition of Pb2+ to the P-E2 conformation (Fig. 2, trace C). Because Pb2+ was added when K+ ions were still absent, the species responsible for the decrease in fluorescence can be Na+, H+, or Pb2+ ions themselves. Pb2+ ions do not affect the fluorescence level attained by the ouabain-induced P-E2 state (Fig. S1, trace b), therefore it is unlike that they can be responsible for the fluorescence drop observed in this case. On the other hand, it seems unreasonable to propose a Pb-induced re-binding of Na+ ions to the pump, because it is known that the P-E2 conformation has a low affinity for sodium (2). Therefore, the fluorescence level obtained after the addition of Pb2+ to the P-E2 conformation has to be attributed to a protonated P-E2 state. We can indicate this conformation as P-E2Pb(Hz). From pH titrations of the P-E2 state (Fig. 3 B), z can be estimated to be ∼1 at pH 7.2. Interestingly, these experiments also reveal that the presence of 10 μM Pb2+ increases the pK value and, therefore, the affinity of the pump for H+.
Under turnover conditions and in the absence of Pb2+, the pump is present partially in an E1 state with three Na+ bound. This causes a lower steady-state fluorescence level (Fig. S1, trace a). In the presence of lead(II), the final level is significantly higher (Fig. 2, trace D), indicating that Pb2+-induced sodium-bound states (Na3E1Pb or P-E1Pb(Na3)) are not present. This reveals that the enzyme is almost completely blocked in an E2-type conformation in which two K+ are bound (and no turnover occurs). According to traces A and C in Fig. 2, the fluorescence level of states [HxE1 + Pb2+] and [P-E2Pb(H) + K+] are practically the same. This can be explained by two monovalent cations (H+ or K+) bound to the Pb2+-modified pump.
Simulations
To obtain further insight into the mechanism of interaction of Pb2+ ions with the Na-pump, we adopted a model scheme of six intermediates for the chemical reactions involved in the backdoor phosphorylation in the presence of Pb2+ ions (Fig. 6, upper panel). Given the equilibrium constants for each step of the scheme, Ki, it is possible to solve the resulting linear equations system for the equilibrium concentration of each intermediate at different concentrations of Pi and of free Pb2+. The fluorescence data can be reproduced by considering the specific fluorescence level, fi, of each intermediate (see the Appendix in the Supporting Material).
Figure 6.
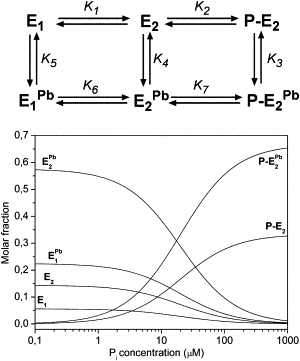
(Upper panel) Proposed model scheme for the reactions involved in backdoor phosphorylation in the presence of Pb2+. For each step, the relative equilibrium constant, Ki, is indicated. For simplicity, protons are omitted in the scheme. (Lower panel) Distribution of the concentrations for all intermediates against Pi concentration as obtained from the simulation. The concentration of Pb2+ is 10 μM. For each curve, the corresponding intermediate is indicated.
In the proposed model scheme, steps 1 and 2 correspond to the backdoor phosphorylation. Steps 3, 4, and 5 refer to binding of Pb2+ to the P-E2, E2, and E1 conformations respectively, as discussed above. Steps 6 and 7 are introduced assuming that Pb2+ does not prevent the E1 to E2 conformational transition nor enzyme phosphorylation.
A satisfying reproduction of the experimental data is obtained by assigning to Ki and to fi the values reported in Table 2 (Fig. 4, solid lines). The values for the equilibrium constants K1 and K2 were taken from Apell et al. (28). In particular, the value of K2 has been adjusted to obtain the best fit of the experimental data.
Table 2.
Values of Ki and fi used in the simulation according to the six-intermediates model scheme
| i | Ki | Source | Specific fluorescence levels |
Source | |
|---|---|---|---|---|---|
| fi | Intermediate | ||||
| 1 | 0.39 | Apell et al. (28) | 0 | E1 | By definition |
| 2 | 8.7 μM | Adapted from Apell et al. (28) | 0 | E2 | By definition |
| 3 | 5.0 μM | Simulation | +0.19 | P-E2 | Fig. 5B |
| 4 | 2.5 μM | Fig. 5A | −0.05 | E1Pb | Fig. 5A |
| 5 | 2.5 μM | Fig. 5A | −0.05 | E2Pb | Fig. 5A |
| 6 | 0.39 | See Appendix in the Supporting Material | +0.04 | P-E2Pb | Fig. 5B |
| 7 | 17.4 μM | See Appendix in the Supporting Material | |||
See Fig. 6 for the six-intermediates model scheme.
The equilibrium constants K4 and K5 are identical (2.5 μM), to indicate the same affinity of Pb2+ ions for the E1 and the E2 conformations. This value is in agreement with that determined experimentally, 3.1 ± 0.9 μM (Fig. 5 A). The difference between the two values is negligible, considering the experimental uncertainty.
The value of the K3 equilibrium is twice that of K4 and K5 to obtain the best simulation curves. In fact, by setting K3 = K4 = K5 = 2.5 μM, no variation of the K0.5 for Pi is observed with increasing Pb2+ concentration, whereas a value of K3 < K4 = K5 = 2.5 μM determines an apparent increase of affinity for Pi (not shown), contrary to that observed experimentally.
Finally, the values of K6 and K7 are not independent parameters, and are calculated according to the principle of detailed balance (see (42) and Appendix in the Supporting Material).
The specific fluorescence levels of the P-E2, E1Pb, E2Pb, and P-E2Pb intermediates, that is f3, f4, f5, and f6, respectively, are related to the degree of protonation of these states and were taken from Fig. 5 (Table 2).
The simulated curves so obtained are in very good agreement with the experimental data (Fig. 4). In particular, the initial and final fluorescence levels nicely agree with the experimental values, as well as the half-saturation constants, K0.5 (Table 1). Moreover, by assigning a nonzero value to the specific fluorescence levels of the intermediates E1Pb and E2Pb, we can reproduce the Pb-induced fluorescence drop observed after addition of Pb2+ ions to the pump in the absence of any other reagent, i.e., Na+, K+, Pi, or ATP (Fig. 5 A). However, the reaction scheme of Fig. 6 (upper panel) cannot simulate the increase of the Hill coefficient experimentally observed. This increase of n is probably an apparent effect.
Finally, it can be useful to analyze the concentration distribution for all the intermediates to determine the most stable conformations at equilibrium. As shown in Fig. 6 (lower panel), in the presence of 10 μM Pb2+ the preferentially adopted conformation is the P-E2Pb form (∼65%) at saturating Pi. On the other hand, the E2Pb form is the main species at low Pi.
Mechanism of interaction
In conclusion, with the experiments reported here we analyzed the effects of Pb2+ on the electrogenic partial reactions of the Na+,K+-ATPase enzymatic cycle. Taken together, our fluorescence measurements allow us to propose a possible molecular mechanism for the interaction of Pb2+ ions with the Na+,K+-ATPase.
One important finding of this study is that Pb2+ ions do not affect the Na+ and K+ binding affinities in the E1 and P-E2 conformations of the enzyme. Another interesting observation is that Pb2+ binds with almost the same affinity to the E1 and P-E2 conformations. In the absence of Na+ and K+ and in the presence of Pb2+, an E1 state is formed with approximately two H+ (Hx+yE1Pb, x + y = 1.8), whereas in the presence of Na+, ATP, and K+, Pb2+ ions stabilize a P-E2 state with two K+ (P-E2Pb(K2)). It is also worth noting that Pb2+ promotes at pH>7 binding of one H+ to the P-E2 conformation in the absence of K+ (P-E2Pb(H)).
In summary, our experimental data clearly indicate that Pb2+ bound to the enzyme stabilizes E2-type conformations, as also confirmed by the results of the simulation. In particular, under conditions that promote enzyme phosphorylation, Pb2+ ions are able to confine the Na+,K+-ATPase into a phosphorylated E2 state. This conclusion supports our hypothesis about the possible interference of lead(II) ions with the dephosphorylation step (16). Moreover, it can explain the experimental results previously reported (15), where a Pb2+-induced decrease of the dephosphorylation rate constant was observed.
Finally, in addition to the significance of the Pb2+ effect with regard to the catalytic mechanism, we consider that stabilization of the (phosphorylated) E2 conformation of the enzyme by Pb2+ ions may be a very useful tool in structural and crystallization studies of cation transport ATPases. In fact, it is well known that, very often, ATPase crystal structures of the highest resolution are obtained by incorporation of one or even two inhibitors, as recently reported in the case of sarcoplasmic reticulum Ca2+-ATPase (43).
Acknowledgments
The authors thank Professor H.-J. Apell for providing them with membrane fragments containing the Na+,K+-ATPase.
The Ministero dell'Istruzione, dell'Università e della Ricerca (PRIN 2008), and the Ente Cassa di Risparmio di Firenze are gratefully acknowledged for financial support.
Supporting Material
References
- 1.Skou J.C. The influence of some cations on an adenosine triphosphatase from peripheral nerves. Biochim. Biophys. Acta. 1957;23:394–401. doi: 10.1016/0006-3002(57)90343-8. [DOI] [PubMed] [Google Scholar]
- 2.Jørgensen P.L., Hakansson K.O., Karlish S.J.D. Structure and mechanism of Na,K-ATPase: functional sites and their interactions. Annu. Rev. Physiol. 2003;65:817–849. doi: 10.1146/annurev.physiol.65.092101.142558. [DOI] [PubMed] [Google Scholar]
- 3.Kaplan J.H. Biochemistry of Na,K-ATPase. Annu. Rev. Biochem. 2002;71:511–535. doi: 10.1146/annurev.biochem.71.102201.141218. [DOI] [PubMed] [Google Scholar]
- 4.Cornelius F. The sodium pump. In: Lee A.G., editor. Biomembranes. JAI Press; Greenwich, CT: 1996. pp. 133–184. [Google Scholar]
- 5.Albers R.W. Biochemical aspects of active transport. Annu. Rev. Biochem. 1967;36:727–756. doi: 10.1146/annurev.bi.36.070167.003455. [DOI] [PubMed] [Google Scholar]
- 6.Post R.L., Hegyvary C., Kume S. Activation by adenosine triphosphate in the phosphorylation kinetics of sodium and potassium ion transport adenosine triphosphatase. J. Biol. Chem. 1972;247:6530–6540. [PubMed] [Google Scholar]
- 7.Morth J.P., Pedersen B.P., Nissen P. Crystal structure of the sodium-potassium pump. Nature. 2007;450:1043–1049. doi: 10.1038/nature06419. [DOI] [PubMed] [Google Scholar]
- 8.Shinoda T., Ogawa H., Toyoshima C. Crystal structure of the sodium-potassium pump at 2.4 Å resolution. Nature. 2009;459:446–450. doi: 10.1038/nature07939. [DOI] [PubMed] [Google Scholar]
- 9.Gurer H., Ercal N. Can antioxidants be beneficial in the treatment of lead poisoning? Free Radic. Biol. Med. 2000;29:927–945. doi: 10.1016/s0891-5849(00)00413-5. [DOI] [PubMed] [Google Scholar]
- 10.Hechtenberg S., Beyersmann D. Inhibition of sarcoplasmic reticulum Ca2+-ATPase activity by cadmium, lead and mercury. Enzyme. 1991;45:109–115. doi: 10.1159/000468875. [DOI] [PubMed] [Google Scholar]
- 11.Hexum T.D. Studies on the reaction catalyzed by transport (Na, K) adenosine triphosphatase. I. Effects of divalent metals. Biochem. Pharmacol. 1974;23:3441–3447. doi: 10.1016/0006-2952(74)90347-5. [DOI] [PubMed] [Google Scholar]
- 12.Caspers M.L., Siegel G.J. Inhibition by lead of human erythrocyte (Na+ + K+)-adenosine triphosphatase associated with binding of 210Pb to membrane fragments. Biochim. Biophys. Acta. 1980;600:27–35. doi: 10.1016/0005-2736(80)90408-3. [DOI] [PubMed] [Google Scholar]
- 13.Vasic V., Kojic D., Stojic D. Time-dependent inhibition of Na+/K+-ATPase induced by single and simultaneous exposure to lead and cadmium. Russ. J. Phys. Chem. A. 2007;81:1402–1406. [Google Scholar]
- 14.Siegel G.J., Fogt S.M. Inhibition by lead ion of electrophorus electroplax (Na+ + K+)-adenosine triphosphatase and K+-p-nitrophenylphosphatase. J. Biol. Chem. 1977;252:5201–5205. [PubMed] [Google Scholar]
- 15.Swarts H.G., Zwartjes H.A., de Pont J.J. Pb2+ and imidazole-activated phosphorylation by ATP of (Na+ + K+)-ATPase. Biochim. Biophys. Acta. 1987;903:525–532. doi: 10.1016/0005-2736(87)90060-5. [DOI] [PubMed] [Google Scholar]
- 16.Gramigni E., Tadini-Buoninsegni F., Moncelli M.R. Inhibitory effect of Pb2+ on the transport cycle of the Na+,K+-ATPase. Chem. Res. Toxicol. 2009;22:1699–1704. doi: 10.1021/tx9001786. [DOI] [PubMed] [Google Scholar]
- 17.Grinvald A., Hildesheim R., Anglister L. Improved fluorescent probes for the measurement of rapid changes in membrane potential. Biophys. J. 1982;39:301–308. doi: 10.1016/S0006-3495(82)84520-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bühler R., Stürmer W., Läuger P. Charge translocation by the Na,K-pump: I. Kinetics of local field changes studied by time-resolved fluorescence measurements. J. Membr. Biol. 1991;121:141–161. doi: 10.1007/BF01870529. [DOI] [PubMed] [Google Scholar]
- 19.Harmel N., Apell H.-J. Palytoxin-induced effects on partial reactions of the Na,K-ATPase. J. Gen. Physiol. 2006;128:103–118. doi: 10.1085/jgp.200609505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartolommei G., Devaux N., Apell H.J. Effect of clotrimazole on the pump cycle of the Na,K-ATPase. Biophys. J. 2008;95:1813–1825. doi: 10.1529/biophysj.108.133546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jørgensen P.L. Isolation of (Na+ plus K+)-ATPase. Methods Enzymol. 1974;32(Part B):277–290. [PubMed] [Google Scholar]
- 22.Schwartz A.K., Nagano M., Allen J.C. The sodium- and potassium-activated adenosinetriphosphatase system. Methods Pharmacol. 1971;1:368–371. [Google Scholar]
- 23.Lowry O.H., Rosebrough N.J., Randall R.J. Protein measurement with the folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 24.Patton C., Thompson S., Epel D. Some precautions in using chelators to buffer metals in biological solutions. Cell Calcium. 2004;35:427–431. doi: 10.1016/j.ceca.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 25.Apell H.-J., Diller A. Do H+ ions obscure electrogenic Na+ and K+ binding in the E1 state of the Na,K-ATPase? FEBS Lett. 2002;532:198–202. doi: 10.1016/s0014-5793(02)03675-x. [DOI] [PubMed] [Google Scholar]
- 26.Domaszewicz W., Apell H.-J. Binding of the third Na+ ion to the cytoplasmic side of the Na,K-ATPase is electrogenic. FEBS Lett. 1999;458:241–246. doi: 10.1016/s0014-5793(99)01162-x. [DOI] [PubMed] [Google Scholar]
- 27.Garcia-Celma J.J., Hatahet L., Fendler K. Specific anion and cation binding to lipid membranes investigated on a solid supported membrane. Langmuir. 2007;23:10074–10080. doi: 10.1021/la701188f. [DOI] [PubMed] [Google Scholar]
- 28.Apell H.-J., Roudna M., Trentham D.R. Kinetics of the phosphorylation of Na,K-ATPase by inorganic phosphate detected by a fluorescence method. Biochemistry. 1996;35:10922–10930. doi: 10.1021/bi960238t. [DOI] [PubMed] [Google Scholar]
- 29.Glynn I.M. The Na+,K+-transporting adenosine triphosphatase. In: Martonosi A.N., editor. Membrane Transport. Plenum Press; New York: 1985. pp. 35–114. [Google Scholar]
- 30.Stürmer W., Apell H.-J. Fluorescence study on cardiac glycoside binding to the Na,K-pump. Ouabain binding is associated with movement of electrical charge. FEBS Lett. 1992;300:1–4. doi: 10.1016/0014-5793(92)80151-6. [DOI] [PubMed] [Google Scholar]
- 31.Stürmer W., Bühler R., Läuger P. Charge translocation by the Na,K-pump: II. Ion binding and release at the extracellular face. J. Membr. Biol. 1991;121:163–176. doi: 10.1007/BF01870530. [DOI] [PubMed] [Google Scholar]
- 32.Kane D.J., Grell E., Clarke R.J. Dephosphorylation kinetics of pig kidney Na+,K+-ATPase. Biochemistry. 1998;37:4581–4591. doi: 10.1021/bi972813e. [DOI] [PubMed] [Google Scholar]
- 33.Kane D.J., Fendler K., Clarke R.J. Stopped-flow kinetic investigations of conformational changes of pig kidney Na+,K+-ATPase. Biochemistry. 1997;36:13406–13420. doi: 10.1021/bi970598w. [DOI] [PubMed] [Google Scholar]
- 34.Heyse S., Wuddel I., Stürmer W. Partial reactions of the Na,K-ATPase: determination of rate constants. J. Gen. Physiol. 1994;104:197–240. doi: 10.1085/jgp.104.2.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fedosova N.U., Cornelius F., Klodos I. E2P phosphoforms of Na,K-ATPase. I. Comparison of phosphointermediates formed from ATP and Pi by their reactivity toward hydroxylamine and vanadate. Biochemistry. 1998;37:13634–13642. doi: 10.1021/bi980703h. [DOI] [PubMed] [Google Scholar]
- 36.Campos M., Beaugé L. Na+-ATPase activity of Na+,K+-ATPase. Reactivity of the E2 form during Na+-ATPase turnover. J. Biol. Chem. 1994;269:18028–18036. [PubMed] [Google Scholar]
- 37.Apell H.-J. Structure-function relationship in P-type ATPases—a biophysical approach. Rev. Physiol. Biochem. Pharmacol. 2003;150:1–35. doi: 10.1007/s10254-003-0018-9. [DOI] [PubMed] [Google Scholar]
- 38.Clarke R.J., Kane D.J. Two gears of pumping by the sodium pump. Biophys. J. 2007;93:4187–4196. doi: 10.1529/biophysj.107.111591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khalid M., Fouassier G., Clarke R.J. Interaction of ATP with the phosphoenzyme of the Na+,K+-ATPase. Biochemistry. 2010;49:1248–1258. doi: 10.1021/bi9019548. [DOI] [PubMed] [Google Scholar]
- 40.Fedosova N.U., Cornelius F., Klodos I. Fluorescent styryl dyes as probes for Na,K-ATPase reaction mechanism: significance of the charge of the hydrophilic moiety of RH dyes. Biochemistry. 1995;34:16806–16814. doi: 10.1021/bi00051a031. [DOI] [PubMed] [Google Scholar]
- 41.Pratap P.R., Robinson J.D. Rapid kinetic analyses of the Na+/K+-ATPase distinguish among different criteria for conformational change. Biochim. Biophys. Acta. 1993;1151:89–98. doi: 10.1016/0005-2736(93)90075-b. [DOI] [PubMed] [Google Scholar]
- 42.Hill T.L. Academic Press; New York: 1977. Free Energy Transduction in Biology. [Google Scholar]
- 43.Takahashi M., Kondou Y., Toyoshima C. Interdomain communication in calcium pump as revealed in the crystal structures with transmembrane inhibitors. Proc. Natl. Acad. Sci. USA. 2007;104:5800–5805. doi: 10.1073/pnas.0700979104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


