Abstract
The replicative helicase for Escherichia coli is DnaB, a hexameric, ring-shaped motor protein that encircles and translocates along ssDNA, unwinding dsDNA in advance of its motion. The microscopic mechanisms of DnaB are unknown; further, prior work has found that DnaB's activity is modified by other replication proteins, indicating some mechanistic flexibility. To investigate these issues, we quantified translocation and unwinding by single DnaB molecules in three tethered DNA geometries held under tension. Our data support the following conclusions: 1), Unwinding by DnaB is enhanced by force-induced destabilization of dsDNA. 2), The magnitude of this stimulation varies with the geometry of the tension applied to the DNA substrate, possibly due to interactions between the helicase and the occluded ssDNA strand. 3), DnaB unwinding and (to a lesser extent) translocation are interrupted by pauses, which are also dependent on force and DNA geometry. 4), DnaB moves slower when a large tension is applied to the helicase-bound strand, indicating that it must perform mechanical work to compact the strand against the applied force. Our results have implications for the molecular mechanisms of translocation and unwinding by DnaB and for the means of modulating DnaB activity.
Introduction
The replicative helicase for Escherichia coli, DnaB, is a ring-shaped hexameric motor protein that catalyzes the separation of double-stranded DNA (dsDNA) into its component single-stranded DNAs (ssDNAs) during replication. DnaB binds to and encircles ssDNA, and uses chemical energy from ATP hydrolysis to translocate in the 5′–3′ direction. Upon encountering an ss/dsDNA fork, DnaB continues translocating along the encircled strand, displacing the complementary strand and thus unwinding the dsDNA (1). In vivo, DnaB localizes to the lagging strand of the bacterial replisome and unwinds the mother dsDNA, providing the ssDNA templates for daughter dsDNA synthesis by Pol III DNA polymerases.
Previous measurements of DnaB activity indicate that the motor has a range of possible forward rates. In ideal conditions in vivo, the replisome progresses at ∼1000 bp/s (2). In vitro, DnaB's unwinding rate in isolation has been estimated in one work to be 35 bp/s (3), and 291 bp/s in another (4). DnaB is accelerated to 400 bp/s when coupled by τ to Pol III (3), and slowed to 1–10 bp/s when acting alongside translesion polymerases (5).
The microscopic basis for DnaB's translocation and unwinding activities, and thus the mechanisms underlying variability, are not well understood. Translocation requires coordination between the enzyme's ATPase and mechanical activities. The best clues for how this might work are found in crystal structures of other hexameric helicases, which suggest a mechanism wherein all six monomers are active both mechanically and as ATPases, with ATP hydrolysis proceeding sequentially around the ring (6–9). In particular, the structures of the papillomavirus E1 and E. coli Rho helicases complexed with ssNAs show that each monomer binds a single nucleotide, causing the bound ssNA to adopt a compact spiral geometry (6,8).
As with all helicases, DnaB's translocation activity must be intimately connected to its unwinding activity, because a motor that solely translocates along ssDNA will be blocked upon reaching a junction with dsDNA. Unwinding requires opening of the proximal basepairs, which can be actuated in a passive, helicase-independent manner through rectification of thermal fluctuations. Alternatively, the helicase can actively destabilize the basepairs (10,11). This active/passive scheme has been quantified (12,13), permitting more precise classification of helicases through model-derived estimates of the free energy of destabilization of nearby basepairs. In this manner, other ring-shaped helicases have been found to be either moderately active (gp4 from T7 phage (14)) or passive (gp41 from T4 phage (15)), while active models have been determined for non-ring-shaped helicases such as NS3 from hepatitis C virus (16), and the bacterial helicases PcrA (17), Rep (18), and UvrD (19,20).
In this single-molecule study, we quantify unwinding and translocation by individual DnaB hexamers moving along tethered DNA substrates held under externally applied tension with magnetic tweezers. We use three different substrate geometries, permitting force-based modulation both of basepair stability and of interactions between DnaB and DNA. This novel multifaceted approach permits us to uncover previously unknown aspects of DnaB activity. In particular, we find that DnaB has a significant pausing activity, and that DnaB's pausing and unwinding activities vary with force-induced basepair destabilization. Further, our data suggest that DnaB's activeness is not solely an intrinsic function of the helicase, but varies with the geometry of the substrate on which it travels. Finally, we show that the unwinding rate is significantly slowed when tension is applied to the ssDNA bound by DnaB, indicating that this strand is highly compact relative to protein-free ssDNA. This furthers the hypothesis that a compact, helical geometry of bound ssDNA is a common feature of hexameric helicases (6). We present a model of DnaB activity that synthesizes all aspects of our results, and relate that model to other hexameric helicases and to the regulation of DnaB activity by other replication proteins.
Materials and Methods
Proteins and DNA
Proteins were expressed and purified as described: DnaB (21) and DnaC (22). The stem of the hairpin was prepared by polymerase chain reaction amplification of a segment of lambda-phage genome and digestion with BtgI and BamHI, leaving a 349-bp construct with sticky ends. The forked region for the hairpin assay was prepared by annealing two oligonucleotides (one 5′ biotin labeled and one unlabeled) each at 1 μM in annealing buffer (10 mM Tris, 1 mM EDTA, 50 mM NaCl) at 37°C overnight. The hairpin stem was then ligated (with T4 DNA ligase) to the annealed fork and an oligonucleotide that forms a hairpin with a 4-nt single-stranded loop. The 3′ end of the resulting construct was then labeled using terminal transferase and digoxigenin-labeled nucleotides. The double-stranded region of the final 389-bp product has 51% GC content. For the fork assay, a 5.3-kb double-stranded region with 57% GC content was prepared by polymerase chain reaction from the lambda-phage genome with one digoxigenin-labeled primer and digested with BamHI to provide a sticky end. The fork region was prepared by annealing two oligonucleotides, with either a 3′ (occluded-strand assay) or 5′ (encircled-strand assay) biotin label, and ligation to the double-stranded region. Assembly diagrams and sequences of fork oligonucleotides are given in Fig. S1 of the Supporting Material.
Single-molecule assays
DNA tethers were made by incubating the digoxigenin-labeled DNA substrate with antidigoxigenin for 30 min at room temperature, and then incubating at 10 pM in a Sigmacote-treated (Sigma-Aldrich, St. Louis, MO) glass flow cell for 30 min, allowing the antidigoxigenin to nonspecifically bind to the glass surface. The surface was then passivated with 0.1% Triton X-100 and 0.1% F127 Pluronic. Streptavidin-coated paramagnetic beads were then added: 1 μm Dynal (Invitrogen, Carlsbad, CA) for the hairpin and either 1 μm MagSense (MagSense Life Sciences, West Lafayette, IN) or 2.8 μm Dynal (Invitrogen) for the fork. Enzymes were added in helicase buffer (23): 20 mM Tris, 10 mM magnesium acetate, 100 μM EDTA, 20% glycerol, 40 μg/mL bovine serum albumin, 5 mM dithreitol, 0.01% Triton X-100, and 0.01% F127 Pluronic; with 250 nM DnaB hexamer, 680 nM DnaC, and 5 mM ATP. Data were collected using real-time tracking at 60 Hz in three dimensions with multiplexed magnetic tweezers as described previously (24,25). For the fork assay, force was determined for each tether by analyzing the power spectra of bead positions, as described previously (26). For the hairpin assay, force was determined using a calibration of the magnet position/force relation, which is possible because 1 μm Dynal magnetic beads are monodisperse in size and magnetic content. Calibration was achieved using power spectrum methods and 16.5-μm-long dsDNA tethers. All experiments were conducted at 22°C.
Velocity measurement
To extract velocities from measured trajectories, traces were first smoothed with a third-order Savitzky-Golay filter and differentiated. The corresponding velocities were then median-filtered. To apply the appropriate smoothing for the noise level, the window size of these filters was chosen to be inversely proportional to the spring constant of the tether in the vertical direction (which varies with applied force, tether length, and tether stiffness (27)). The window size varied from two to nine frames for the hairpin assay, and from eight to 18 frames for the fork assay. Pauses were found by thresholding the velocity. Threshold values for each assay were determined by fitting histograms of the largest data sets (20 pN for the fork and 11.3 pN for the hairpin) to the sum of two Gaussians (one peak for pausing, one for nonzero velocity), and determining the threshold that gives a value that agrees with the fit. Threshold values were 25 bp/s for unwinding on both the fork and the hairpin, and 50 bp/s for hairpin rezipping.
Bootstrap estimates of compaction parameters
The six data points representing the ratio uenc/uocc seen later in Fig. 5 A were bootstrap-resampled 5000 times (28), and each sample was fit to the compaction model of translocation (Eq. 4). The means of the bootstrap best-fit values of A and xc were the reported values for each parameter, and the empirical standard deviations of the bootstrap best-fit values were the estimates of their standard errors.
Figure 5.
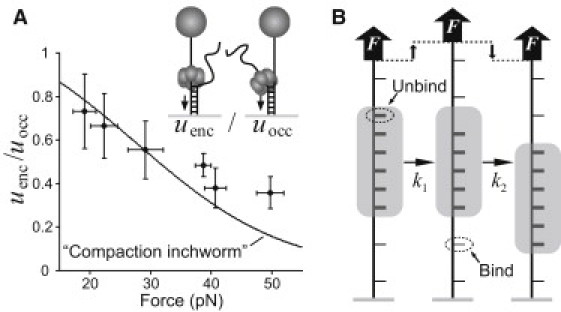
ssDNA compaction during DnaB translocation. (A) The ratio uenc/uocc versus force, calculated using the measured values of uenc in Fig. 3 B and the interpolation from the measurements of uocc in Fig. 3B. The line indicates a bootstrap estimate of the best fit of Eq. 4 to the ratio uenc/uocc. (B) Compaction-inchworm model of helicase translocation: in one enzymatic cycle, the helicase modulates the tether extension by releasing 1 nt from the rear and binding 1 nt at the front. Under applied tension (as in the encircled-strand assay, but not the occluded-strand assay), these extension changes involve mechanical work that alter the helicase translocation rate (Eq. 4).
Results
DNA hairpin unwinding and rezipping assay
We measured both the unwinding and translocation rates of single hexamers of DnaB by using a tethered hairpin assay (15,16,29). A 389-bp DNA hairpin with a 4-nt loop was tethered by single-stranded handles between the bottom of a glass flow cell and a magnetic bead (Fig. 1 A). We applied force to the hairpin and measured its extension using magnetic tweezers (25): force was applied by positioning a pair of permanent magnets above the sample while a microscope-based image analysis was used to track the position of multiple tethered beads in three dimensions in real-time (24). The mechanical unfolding force of the hairpin was 17 ± 1 pN (mean ± SD); below this critical force, with no enzyme present, the hairpin remained in the stably folded configuration. When the helicase DnaB, the helicase loader DnaC, and ATP were added to the flow cell, we observed transient changes in bead height. The most common event was a steady increase in bead height up to the entire length of the hairpin followed by a faster decrease back to the original position (Fig. 1 B). We interpreted each event as the loading of DnaB onto the ssDNA handle, followed by DnaB-catalyzed unwinding of the hairpin stem, then rezipping in the wake of the helicase as it translocates along the second half of the hairpin. Although most events were of full length, occasionally the hairpin abruptly returned to the completely folded state midevent, corresponding to unbinding of the helicase and reannealing of the hairpin. All hairpin data presented in this report are from 66 full-length events, observed at a range of forces between 5 and 15.5 pN.
Figure 1.
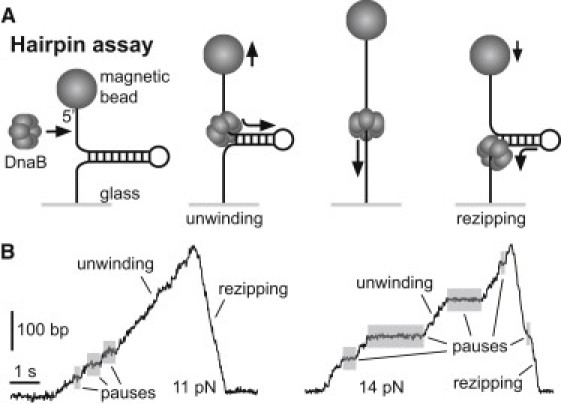
Overview of hairpin assay. (A) Cartoon of hairpin geometry. DnaB loads onto a 60-nt ssDNA handle and unwinds the 389-bp dsDNA stem, causing the bead to move up. The hairpin then rezips in the wake of the translocating helicase, causing the bead to move down. (B) Two example traces of hairpin events. At a given force, events exhibit varying rates of activity, along with pauses (highlighted) that interrupt unwinding and (more rarely) rezipping.
The rising (unwinding) and falling (rezipping) edges of each event were characterized by constant rates interrupted by pauses that endured up to several seconds. Pauses were observed more frequently during unwinding than rezipping, and more frequently at low force. Throughout this work, DnaB's velocity is reported after removal of pauses in the trajectories (see Materials and Methods). The mean pause-removed unwinding rate increased with force (see later in Fig. 3 A), which we attribute to the decreased free energy cost of breaking basepairs with increased applied force, as discussed below. By contrast, the mean rezipping rates were constant with force, and were greater than the unwinding rates at all forces (see later in Fig. 3 A).
Figure 3.
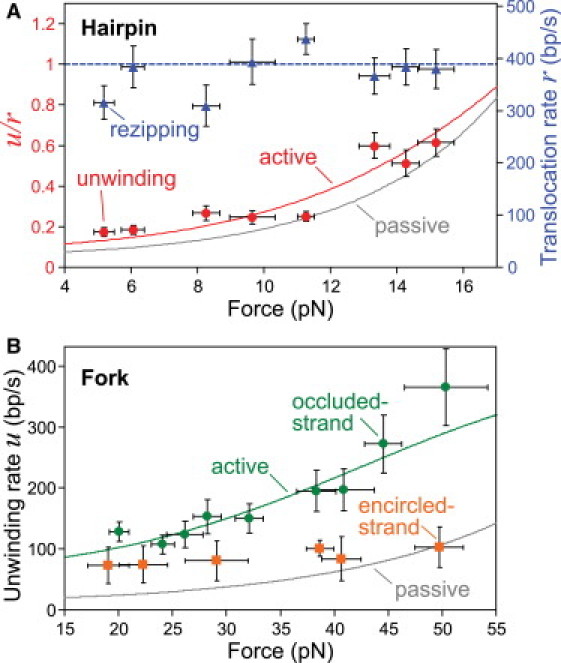
DnaB unwinding and translocation rates and fits to active/passive theory. (A) Hairpin unwinding and rezipping rates (mean ± SE) measured at various forces (± SD). The mean rezipping rate, which is equivalent to the translocation rate of DnaB on ssDNA, is force-independent, with mean r = 390 ± 15 nt/s. The unwinding data are well fit by a weakly active model with a step size n = 1 bp and a destabilization energy U = 0.5 kBT (Fig. S5 and Table S3). (B) Unwinding rate as measured in the fork assays (mean ± SE) at various forces (± SD). The occluded-strand data are well fit by a moderately active model with a step size n = 1 bp and a destabilization energy U = 1.6 kBT (Fig. S5 and Table S3). Analysis of the unwinding rates from the encircled-strand assay requires further consideration (see text and Fig. 5).
During rezipping, DnaB moves along ssDNA unhindered by dsDNA in front of it, implying that the rezipping rate is equivalent to the translocation rate of DnaB along ssDNA. This would not be true if the rehybridization of the hairpin behind the hexamer affects the rezipping rate. If reannealing exerted a significant driving force on the translocating helicase, the effect would be reduced at high applied force due to the decreased stability of the basepairs. However, the mean pause-removed rezipping rate does not decrease with applied force (see later in Fig. 3 A). In addition, as was similarly shown for T4 gp41 helicase (15), an analysis of events at forces near the mechanical unfolding force indicates that the presence of the closing junction has no effect on helicase translocation (Fig. S2 and Table S1). Therefore, we conclude that the mean measured rezipping rate is equal to the translocation rate of DnaB on ssDNA: 390 ± 15 nt/s (mean ± SE).
The events described here occurred infrequently (on average, approximately once every 5 min while the DnaB was active). This is consistent with rare DnaB binding, and events caused by single helicases. Alternatively, event rarity could be explained if binding occurs frequently, but events are only seen in the rare case that multiple hexamers are loaded simultaneously. To determine whether individual or multiple hexamers unwind DNA under the conditions in this study, we performed an experiment using a hairpin with a 20-nt ssDNA loading region that could accommodate only a single hexamer (30). DnaB was loaded onto the hairpin with AMP-PNP (31), and excess protein was washed out. ATP was then added to start the single preloaded helicase (4). Upon ATP addition, we immediately observed a single full length event, quantitatively similar to others described here (Fig. S3 A). This indicates that single DnaB hexamers are fully capable of achieving the unwinding and translocation rates measured in this study, and they are characterized by processivities of at least 389 bp (the hairpin length). This contrasts with a previous bulk study that measured a very low processivity of ∼9 bp (4).
Fork unwinding assays
To further investigate the unwinding mechanism of DnaB, we performed measurements of the unwinding rate on a different DNA substrate in a different force regime. In these fork assays (19), we tethered a 5.3-kb dsDNA between a glass surface and a magnetic bead. One end of the dsDNA was forked, with a 60-nt single-stranded region serving as a helicase loading area (Fig. 2, A and B). Because DnaB moves in the 5′–3′ direction, unwinding activity can only be caused by DnaB that has loaded onto the fork's 5′ tail. Therefore, by choosing whether the 5′ or 3′ tail is attached to the bead, we control whether the encircled or occluded strand is held under tension (Fig. 2, A and B). We will refer to these tethered DNA geometries by the strand to which force is applied (i.e., either the occluded- or encircled-strand assay, or collectively as the fork assays).
Figure 2.
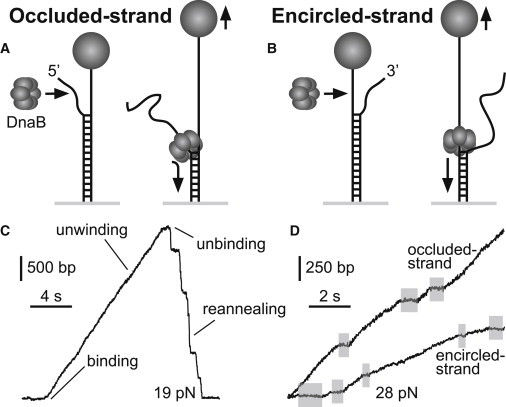
Overview of fork assays. A 5.3-kb dsDNA terminates in a fork with a 60-nt ssDNA loading region. DnaB loads onto the 5′ end and unwinds the dsDNA, causing the bead to move up. Force is applied either to (A) the strand occluded by the DnaB ring (occluded-strand assay), or (B) the strand encircled by DnaB (encircled-strand assay). (C) A typical fork event, showing DNA unwinding and helicase unbinding, followed by recovery of the initial bead position after reannealing of the unwound strands. Pauses during recovery are the result of secondary structure formation on the unconstrained strand that temporarily blocks reannealing. (D) Unwinding trajectories from events in each of the fork geometries. At a given force, events exhibit varying unwinding rates and occasional pauses (highlighted). Unwinding rates are consistently faster in the occluded-strand assay.
When DnaB, DnaC, and ATP were added and a force of at least ∼15 pN was applied, we observed events consisting of increases in bead height interrupted occasionally by pauses, followed by an abrupt decrease back to the original position (Fig. 2, C and D). The increase in bead height corresponds to helicase unwinding because, with forces >∼15 pN, the end-to-end extension of ssDNA is significantly greater than that of dsDNA (Fig. S4). The abrupt decrease after unwinding then corresponds to unbinding of DnaB followed by reannealing of dsDNA, which is occasionally blocked by hairpin formation on the free tail. Similar to the hairpin activity, events in the fork assays were infrequent (every ∼5 min). Along with other observations (Fig. S3 B), this supports the single-helicase nature of the observed events.
Helicase unwinding was measured in the fork assays for applied forces between 20 and 50 pN. In the occluded-strand assay, we observed 92 events with a mean processivity of ∼1 kb (Table S2). The mean unwinding rate increased markedly with force due to decreased basepair stability (Fig. 3 B), and was consistently less than the ssDNA translocation rate as measured in the hairpin assay. In the encircled-strand assay, we observed 33 events with similar processivity (Table S2). At low forces, the occluded- and encircled-strand unwinding rates are similar, but at higher force, the encircled-strand rate is significantly slower (Fig. 3 B). We attributed this slowed unwinding to compaction of bound ssDNA within DnaB's central channel, as discussed below.
Force dependence of DnaB unwinding rate does not support a purely passive unwinding mechanism
Because DnaB must overcome the binding energy of the basepairs during unwinding, it is slowed from its translocation rate. The unwinding rate should therefore depend on the basepair binding free energy ΔGbp. In turn, because ΔGbp decreases with force F, we expect the unwinding rate to increase with force, as was observed in both the hairpin and occluded-strand assays (Fig. 3). This dependence can be quantified by modeling the interaction between the helicase and the ss/dsDNA junction using the Betterton-Jülicher (12,13) model of active/passive helicases. This model posits that the helicase/junction system acts as a Brownian ratchet: fast fraying of the proximal basepairs is rectified by the relatively slow forward motion of the helicase. In the absence of significant backstepping, this predicts that a helicase's dsDNA unwinding rate u(F) is given by u = rP (12,13), where r(F) is the helicase's translocation rate along ssDNA, and P(F) is the probability that the n proximal basepairs are open, where n is the helicase step size.
Quantifying unwinding requires an assumption of the form of P(F). The simplest assumption is that the helicase is passive—i.e., that the helicase has no effect on P(F), and the basepairs are opened only by thermal fluctuations. P(F) is then given by the Boltzmann factor of the basepairing free energy, so the unwinding rate, upassive, is
| (1) |
ΔGbp (F) is the force-dependent free energy cost of opening 1 bp. This can be estimated by integrating the change in the equilibrium tether extension versus force upon unwinding of 1 bp (32):
| (2) |
where ΔG0bp is the binding free energy of a single basepair at zero force, which is ∼2.8 kBT per basepair for dsDNA with 50% GC content at room temperature (1.2 kBT and 3.4 kBT for AT and GC basepairs, respectively (14,33)). Δx(F) is the extension added to the tethered DNA with each unwound basepair (32). In the hairpin assay, Δx(F) = 2xss(F), while in the fork assays,
where xss(F) and xds(F) are the extensions per nucleotide of ssDNA and dsDNA, respectively, at force F (Fig. S4).
The following considerations support the use of Eq. 2 to model force-dependent dsDNA destabilization for both the hairpin and fork geometries. First, Eq. 2 correctly predicts the force at which mechanical unfolding occurs for each geometry: for the hairpin, ΔGbp (F) = 0 at 17.8 pN, consistent with the measured unfolding force of 17 ± 1 pN; for the fork, ΔGbp (F) = 0 at 72 pN, also in good agreement with prior measurements (32,34,35). Further, Eq. 2 has been used to analyze helicase activity in the hairpin geometry (14,15), and to predict that the DNA overstretching transition in the fork geometry was in fact due to denaturation (32); this prediction was recently confirmed (36). Therefore, we can accurately compare unwinding rates in each assay at a given value of ΔGbp (F) using Eq. 2, despite the different force regimes of the assays; this is discussed in the next section.
Equations 1 and 2 provide a prediction for the force-dependence of the ratio u/r, given the step size n, in each tethered DNA geometry. To compare this with the data from the hairpin assay, we extracted the pause-removed unwinding and translocation rates, u and r, from each event (see Materials and Methods), calculated the average ratio u/r for each event, then the average ratio for all events at a particular force. For the fork assays, we extracted the pause-removed u from each event and calculated the average u for all events at each force. In Fig. 3, we plot the mean values of these data alongside that predicted by the passive model. The passive prediction, for any n, is low compared to the measured values; thus, we conclude that DnaB does not unwind dsDNA with a purely passive mechanism. For the encircled-strand data (Fig. 3 B), further theoretical considerations are necessary due to the high forces applied to the DnaB-bound strand (discussed below).
DnaB unwinding is weakly or moderately active, depending on tethered DNA geometry
The results of the previous section imply that DnaB interacts with the junction to destabilize basepairing, i.e., that DnaB unwinding is at least partially active. Further, the data suggest that the activeness of the helicase varies with substrate geometry: Fig. 3 shows that DnaB unwinding is faster, relative to upassive, in the occluded-strand assay than in the hairpin assay. To see this more clearly, we note that the parameter that directly affects the unwinding rate is ΔGbp (F), rather than F. Unwinding rates are thus more meaningfully compared at constant ΔGbp (F): faced with a basepair of equivalent equilibrium stability, DnaB unwinds DNA at least twofold faster in the occluded-strand geometry than the hairpin geometry (Fig. 4 A).
Figure 4.
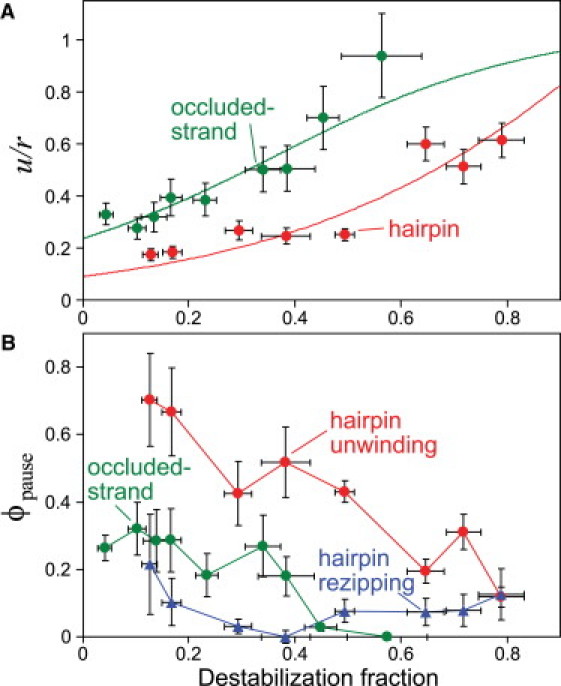
DnaB's unwinding and pausing activities vary with basepair stability and substrate geometry. (A) Unwinding rates for each assay (same data shown in Fig. 3, plotted as a fraction of the mean translocation rate from hairpin rezipping; ± SE) versus destabilization fraction, 1 − (ΔGbp(F)/ΔG0bp) (± SD). The destabilization fraction is 0 with no force-induced destabilization, and 1 for complete destabilization. The corresponding curves are the same fits to the active/passive helicase theory shown in Fig. 3. (B) ϕpause, the fraction of time spent pausing by DnaB while bound to the DNA substrate (mean ± SE).
This geometry dependence relies only on the model of basepair stability, Eq. 2, and not any aspect of the Betterton-Jülicher helicase theory. That said, we can quantify the difference in activeness by applying the Betterton-Jülicher model for an active helicase (12,13). Along with ΔGbp (F), the active model relies on four free parameters: the step size n and three parameters that model the details of the helicase-junction repulsive interaction. The quality of our data does not allow statistically reliable conclusions to be drawn from a fit to a model with four free parameters. However, our data are best fit by models with a 1-bp step size (Table S3), in agreement with results of a prior study of DnaB (4). Therefore, we can reduce the complexity somewhat by setting n = 1. Of the three remaining parameters, we find that only one, the strength U of the helicase/junction repulsion, can be robustly estimated from the data; thus we focus on that here (for details of the theoretical model, see Fig. S5; for a discussion of the best fit values of all parameters, see Table S3). The effect of a repulsion of amount U is to reduce the binding free energy of the proximal basepair by an amount U. This destabilizes the basepair, making it more likely to be open than due to thermal fluctuation alone and increasing the unwinding rate over that predicted by the passive model.
In both the hairpin and occluded-strand assays, the best-fit active model provides a good description of the dependence of unwinding rate on force (Fig. 3, A and B). The resulting values of destabilization are U/kBT = 0.5 ± 0.1 in the hairpin assay and U/kBT = 1.6 ± 0.1 in the occluded-strand assay. Thus, quantitative analysis of the measured unwinding rates provides further support for the conclusion that DnaB is more active in the fork geometry than the hairpin geometry.
Pausing observed during DnaB unwinding is dependent on force and substrate geometry
In both the hairpin and fork assays, DnaB unwinding events were often interrupted by pauses. Pausing cannot be caused by unbinding, because unbinding would allow the dsDNA to reanneal and the bead height to recover its initial value. We interpret these pauses as an off-pathway state from which DnaB is able to resume unwinding.
To analyze the tendency of DnaB to pause, we first identified pauses by applying a velocity threshold to all events (see Materials and Methods), identifying all event portions of the trajectories with velocities below the threshold as pause states. We then found the fraction of time spent by DnaB in the pause state, ϕpause, in each geometry as a function of applied force (Fig. 4 B). Ideally, we would estimate the distributions of individual pause times; however, we were unable to confidently do so, as variations in the signal/noise with force and between the two assays create systematic errors in resolving individual pauses.
In both the occluded-strand assay and the hairpin assay, pausing during unwinding decreased sharply with applied force (Fig. 4 B). Pausing also occurred during ssDNA translocation (hairpin rezipping), although far less frequently than during unwinding, with ϕpause ≈ 10% at all forces. Both the strong force dependence of ϕpause during unwinding and the less frequent pausing during free translocation suggest that the presence of proximal basepairs and their stability ΔGbp (F) contribute to DnaB pausing. At comparable values of ΔGbp (F), pausing was more substantial in the hairpin assay than in the fork assay (Fig. 4 B), suggesting that the tethered DNA geometry also plays a role in pausing. Generally, DnaB's pausing activity is inversely related to its velocity: conditions favoring faster unwinding display less pausing. This is discussed in more detail below.
Slowed unwinding with force applied to encircled strand suggests compaction of ssDNA within the central channel
In addition to providing an alternative measurement of the unwinding rate, the fork assays allow us to investigate the mechanism of DnaB translocation along ssDNA. In both the occluded- and encircled-strand assays, tension is applied to the dsDNA in an identical manner. Therefore, the destabilizing effect of the applied force on the basepairs is identical in both geometries, which means Penc(F) = Pocc(F). It follows that in equilibrium, the ratio of unwinding rates is equal to the ratio of translocation rates, uenc/uocc = renc/rocc. We further assume that r is only affected by force when applied to the strand encircled by the helicase. This means that rocc is independent of force, and is equal to the zero-force rate rocc = r0. Thus, the force dependence of uenc/uocc is related only to the force dependence of renc:
| (3) |
In Fig. 5 A, we plot the prediction for renc/r0 versus F by calculating uenc/uocc: we divide the discrete binned values of uenc by values interpolated from the active model fit of the measurements of uocc (interpolation is needed because difference in the sampling of unwinding rates disallows using bins of equal force in both assays).
The force dependence of renc can be explained if the ssDNA bound by the helicase adopts a relatively compact configuration within the central channel. Fluorescence titration experiments have shown that DnaB binds ssDNA with a footprint of ∼20 nt (30), whereas the depth of the hexameric ring is ∼7.5 nm (37). Therefore, the extension of each bound nucleotide can be no more than ∼0.38 nm, whereas the contour length per base of ssDNA is ∼0.58 nm (32). At the high forces used in the fork assays, the end-to-end extension of ssDNA is close to its contour length. Therefore, in the encircled-strand geometry, DnaB must perform mechanical work to compact the ssDNA against the applied force, slowing the helicase (Fig. 5 B).
Other studies have successfully modeled the effect of mechanical load on motor protein velocity by describing the enzymatic cycle as a combination of force-dependent mechanical transitions and force-independent biochemical transitions (38). In this case, the rate-limiting mechanical transition is compaction of n bound nucleotides (Fig. 5 B). The applied load slows the mechanical transitions by an amount proportional to the Boltzmann factor of the work done per enzymatic cycle (39), giving a translocation rate
| (4) |
where A is a positive, dimensionless parameter that indicates the degree to which either the biochemical transitions (A << 1) or the mechanical transitions (A > 1) are rate-limiting at zero force, and Ec(F) is the work to compact a single nucleotide. If the mechanical transition is limited by an activation energy with distance δ to the transition state, then Ec = Fδ. The simplest interpretation for δ is δ = xss(F) – xc, where xss(F) is the force-extension relation of ssDNA, and xc is the compacted length of a single nucleotide.
To fit Eq. 4 to the data, we fix n = 1 (4) as previously and produce bootstrap estimates of the best-fit values of A and xc (see Materials and Methods and Fig. 5 A). The data fit well to our theoretical model of compaction, with xc = 0.30 ± 0.09 nm and A = 0.24 ± 0.17. This value of xc is consistent with the maximum value of 0.38 nm based on aforementioned structural considerations. This validates the hypothesis that compaction of ssDNA within the central channel is responsible for the slowing of the unwinding rate.
Our model of hairpin unwinding does not account for the compaction effect on r. Ignoring the compaction effect in this geometry is directly justified by the lack of variation in the measured translocation (rezipping) rate with force (Fig. 3 A). Further, this is entirely consistent with the results from the fork assays: for the low forces used in the hairpin assay, xss(F) is never significantly greater than xc, so the work of compaction is negligible. Quantitatively, using the best-fit compaction parameters, Eq. 4 predicts that the translocation rate is > 90% of maximum across the entire range of forces used in the hairpin assay. The effect of compaction is thus only significant at the high forces available in the fork assay, and can be safely ignored in the hairpin assay.
Discussion
Pausing
A strength of single-molecule measurements is the ability to directly track individual protein trajectories. Here, this allowed us to find that DnaB unwinding is frequently interrupted by pauses, which could not be seen in previous bulk measurements (3,4). Each pause was usually terminated by continued unwinding, indicating that DnaB has an inactive DNA-bound state that can be both entered from, and exited to, an actively translocating state. The existence of pauses helps to resolve the disparity in unwinding rates seen in those prior bulk measurements: a rate of 35 bp/s was measured using a 3.7-kb DNA strand (3), while 291 bp/s was measured using much shorter strands (15–30 bp) (4). It is likely that the increased chance of significant pausing when unwinding a long strand explains some of the difference in unwinding rates reported in the two studies. These bulk assays are best compared to our occluded-strand assay, because that geometry limits the constraints on the orientation of the ssDNA tails (see Discussion below). Extrapolating the best-fit active model (Fig. 3 B) to zero force gives a pause-removed unwinding rate of ∼80 bp/s. Accounting for the maximum measured pause fraction (ϕpause ≈ 35%; Fig. 4 B) gives a net unwinding rate (with pauses) of ∼50 bp/s, in good agreement with the long-strand bulk assay (3).
Geometry dependence of DnaB unwinding is consistent with stimulation by the occluded strand
Our results show that DnaB is a moderately processive helicase whose unwinding rate increases with applied force due to decreased basepair stability, indicating that the separation of strands is a considerable barrier to dsDNA unwinding. Previous single-molecule experiments on other hexameric helicases have come to qualitatively similar conclusions, but with quantitatively different results. A study of gp41 helicase from bacteriophage T4 using a hairpin assay revealed a passive unwinding mechanism with a 1-bp step size (15), while measurements of gp4 helicase from bacteriophage T7 showed a moderately active unwinding mechanism (U ≈ 1.2 kBT) and a 2-bp step size (14). By repeating our measurements of the DnaB unwinding rate using multiple tethered DNA geometries, we have shown for the first time to our knowledge that the geometry of applied force can affect the activity of a helicase. In the hairpin assay, DnaB behaves as a nearly passive helicase (U ≈ 0.5 kBT), similar to T4 gp41. However, in the occluded-strand geometry, we find DnaB to be moderately active (U ≈ 1.6 kBT), similar to T7 gp4. That DnaB activity varies with substrate geometry suggests that the previous studies of T7 gp4 and T4 gp41, which were both done in the hairpin geometry, may not have fully realized these enzymes' potential for activeness. Similar issues complicate the interpretation of hairpin-based measurements of other helicases (16,29).
We suggest that the difference in activity between the two assays is likely due to the geometry of force application: in the hairpin geometry, the occluded and encircled strands are pulled away from each other, whereas in the fork geometry, only one strand is constrained by force while the other is free, so DnaB can assume an optimal configuration with respect to the occluded strand. Thus, if there is a stimulating interaction between the occluded strand and the DnaB ring exterior, it would be disallowed in the hairpin geometry but permitted in the fork geometry. Indeed, a prior study suggested that interactions with the 3′ tail can promote DnaB activity (40). Mechanistically, a stimulation by the occluded strand could come from a binding interaction with the ring exterior that leads to a helicase-derived mechanical force that destabilizes the basepairs, as suggested for the eukaryotic hexameric helicase MCM (41). Alternatively, the occluded strand interaction could be due to negative charges on the ring exterior that would repel the occluded strand to aid strand separation, as has been suggested for T7 gp4 (42).
We cannot rule out the possibility that other geometric effects contribute to the observed difference in activity. For example, the helicase could somehow interact differently with basepairs subject to shear forces (as in the fork assays) compared to those subject to normal forces (as in the hairpin assay). However, Eq. 2 provides a well-supported estimate for the equilibrium basepair stabilities in both geometries, and equilibrium differences are most relevant to the commonly accepted active/passive theory; thus we have correctly accounted for the most salient geometric difference.
In addition to the slower unwinding rates, DnaB also pauses more frequently during hairpin unwinding than in the occluded-strand assay. In contrast, the hairpin rezipping trajectories display little pausing despite also being in the hairpin geometry. Thus, the hairpin geometry alone does not intrinsically cause the motor to pause. Instead, the infrequency of pauses during rezipping suggests that pausing is a result of the presence of dsDNA in advance of the motor. Because the proximity of dsDNA during unwinding is dictated by the magnitude of the repulsive helicase/junction interaction, we speculate that pausing is a consequence of the geometry dependence of the helicase's active nature. This suggests the following model: the fork geometry provides a means to repel the ss/dsDNA junction, leading to the increased activeness in the occluded-strand assay compared to the hairpin assay. This also insures that the junction is farther away, resulting in less frequent pausing in the occluded-strand assay. This idea is consistent with the relation of pausing frequency to applied force in both assays (Fig. 4 B): higher forces destabilize the basepairs, leading to larger junction-DnaB distances, and lower propensities to pause.
Multiple means to modulate DnaB activity
Other replisome components need to control the helicase unwinding rate, because helicase unwinding must proceed in concert with both leading- and lagging-strand synthesis. Indeed, prior studies indicate that interactions with other proteins can significantly alter DnaB activity (3,5), although they provide no suggestion as to the means of this control. While we only study DnaB in isolation, our results provide what we believe to be two novel suggestions of pathways to modulation of DnaB activity.
First, we demonstrate an ability to alter the unwinding rate through control of DNA substrate geometry, possibly due to modulation of access to the occluded strand. In this light, it is notable that some of the proteins previously found to affect DnaB activity are polymerases that are localized to occluded strand (3,5). Thus, polymerases are ideally situated to control DnaB activity by altering the geometry of the fork, or by directly promoting DnaB contact with the occluded strand. This is consistent with the notion that DNA polymerase exerts a driving force on the helicase, which has been shown for T7 phage (43).
Second, other replisome proteins might modulate access of DnaB to the inactive/pause state, either through a direct allosteric interaction, or again, through an occluded strand-mediated interaction. We stress that this discussion is speculative; there are other microscopic mechanisms (such as effector-induced alteration of the intrinsic translocation rate) that might underlie the variation in the literature values of the DnaB unwinding rate.
Compaction-inchworm model of translocation is likely a common feature of hexameric helicases
The fork assays permitted us to determine the effect of tension applied to the encircled ssDNA on helicase activity. The results (Fig. 3 B and Fig. 5 A) show that such a force significantly decreases the unwinding rate, which suggests a compaction-inchworm model of DnaB translocation along stretched ssDNA. In this picture (Fig. 5 B), a single DnaB enzymatic cycle consists of binding and compaction of one base at the leading edge of the helicase, and release and decompaction of one base at the trailing edge. The applied force in the fork assays would speed the enzymatic cycle by accelerating the release step, just as it slows the compaction step. However, because uenc < uocc at all forces, our results indicate that compaction is the rate-limiting step in the conditions used here.
While our measurements are consistent with this model of ssDNA compaction, we cannot rule out the importance of other geometrical considerations in the fork assays. For example, the putative occluded strand interaction discussed above could be modulated by applying tension to that strand. However, we would expect this effect to slow the unwinding in the occluded-strand assay compared to the encircled-strand assay, which is contrary to what was measured (Fig. 3 B). Thus, the effect of compaction work provides the best explanation for the velocity difference.
There is evidence from structural assays that other hexameric ring-shaped helicases also compact bound ssNA to an extension less than its contour length. A crystal structure of the papillomavirus E1 helicase complexed with ssDNA found the nucleotides to be bound in a helical pattern with an extension of ∼0.35 nm per base (6). A crystal structure of the E. coli transcription termination factor Rho complexed with ssRNA found a similar compact helical binding pattern (8). It has also been shown that ssDNA is bound by T7 gp4 with an extension decreased to ∼0.3 nm per base (44), and a helical binding scheme has been proposed for the Geobacillus kaustophilus helicase DnaC (45). In combination with our results, these observations support the hypothesis that ssDNA compaction is a characteristic common to the translocation mechanisms of hexameric helicases.
Conclusions
We have undertaken a comprehensive single-molecule study of the unwinding and translocation rates of the hexameric helicase DnaB. We have found that DnaB's activities (i.e., its unwinding rate, translocation rate, and a newly discovered pausing activity) are dependent on applied force and DNA substrate geometry. DnaB unwinds DNA more efficiently (unwinding rate is increased, and pausing decreased) when basepairs are destabilized by applied force. Unwinding efficiency also increases in the occluded-strand assay, consistent with prior indications that DnaB is stimulated by an interaction with the occluded strand (40). Our data are consistent with a picture where increased basepair destabilization (either induced by external force or by increased activeness of the helicase) increases unwinding rate and decreases pausing. We have successfully modeled unwinding using the active model of helicase activity (12,13), and used the model to quantify the variation with substrate geometry of helicase-induced basepair destabilization. Finally, we find that high forces applied to the helicase-bound strand decrease the unwinding rate, consistent with the existence of a rate-limiting ssDNA compaction step, which indicates DnaB might share the spiral-staircase translocation mechanism that has been found for other hexameric helicases. Generally, our results show that even in isolation, DnaB's activity can be altered in a variety of ways, perhaps shedding light on the origin of the high degree of variability reported in the literature. Going forward, we expect our results will provide a firm basis to quantify the effects of protein-protein interactions on the activity of DnaB and the replisome.
Acknowledgments
This work was supported by the National Science Foundation under grant No. PHY-0748564.
Supporting Material
References
- 1.LeBowitz J.H., McMacken R. The Escherichia coli DnaB replication protein is a DNA helicase. J. Biol. Chem. 1986;261:4738–4748. [PubMed] [Google Scholar]
- 2.Chandler M., Bird R.E., Caro L. The replication time of the Escherichia coli K12 chromosome as a function of cell doubling time. J. Mol. Biol. 1975;94:127–132. doi: 10.1016/0022-2836(75)90410-6. [DOI] [PubMed] [Google Scholar]
- 3.Kim S.S., Dallmann H.G., Marians K.J. Coupling of a replicative polymerase and helicase: a τ-DnaB interaction mediates rapid replication fork movement. Cell. 1996;84:643–650. doi: 10.1016/s0092-8674(00)81039-9. [DOI] [PubMed] [Google Scholar]
- 4.Galletto R., Jezewska M.J., Bujalowski W. Unzipping mechanism of the double-stranded DNA unwinding by a hexameric helicase: quantitative analysis of the rate of the dsDNA unwinding, processivity and kinetic step-size of the Escherichia coli DnaB helicase using rapid quench-flow method. J. Mol. Biol. 2004;343:83–99. doi: 10.1016/j.jmb.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 5.Indiani C., Langston L.D., O'Donnell M. Translesion DNA polymerases remodel the replisome and alter the speed of the replicative helicase. Proc. Natl. Acad. Sci. USA. 2009;106:6031–6038. doi: 10.1073/pnas.0901403106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enemark E.J., Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- 7.Liao J.C., Jeong Y.J., Oster G. Mechanochemistry of T7 DNA helicase. J. Mol. Biol. 2005;350:452–475. doi: 10.1016/j.jmb.2005.04.051. [DOI] [PubMed] [Google Scholar]
- 8.Thomsen N.D., Berger J.M. Running in reverse: the structural basis for translocation polarity in hexameric helicases. Cell. 2009;139:523–534. doi: 10.1016/j.cell.2009.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singleton M.R., Sawaya M.R., Wigley D.B. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- 10.Lohman T.M., Bjornson K.P. Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 11.Delagoutte E., von Hippel P.H. Helicase mechanisms and the coupling of helicases within macromolecular machines. Part I: Structures and properties of isolated helicases. Q. Rev. Biophys. 2002;35:431–478. doi: 10.1017/s0033583502003852. [DOI] [PubMed] [Google Scholar]
- 12.Betterton M.D., Jülicher F. A motor that makes its own track: helicase unwinding of DNA. Phys. Rev. Lett. 2003;91:258103. doi: 10.1103/PhysRevLett.91.258103. [DOI] [PubMed] [Google Scholar]
- 13.Betterton M.D., Jülicher F. Opening of nucleic-acid double strands by helicases: active versus passive opening. Phys. Rev. E. 2005;71:011904. doi: 10.1103/PhysRevE.71.011904. [DOI] [PubMed] [Google Scholar]
- 14.Johnson D.S., Bai L., Wang M.D. Single-molecule studies reveal dynamics of DNA unwinding by the ring-shaped T7 helicase. Cell. 2007;129:1299–1309. doi: 10.1016/j.cell.2007.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lionnet T., Spiering M.M., Croquette V. Real-time observation of bacteriophage T4 gp41 helicase reveals an unwinding mechanism. Proc. Natl. Acad. Sci. USA. 2007;104:19790–19795. doi: 10.1073/pnas.0709793104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng W., Dumont S., Bustamante C. NS3 helicase actively separates RNA strands and senses sequence barriers ahead of the opening fork. Proc. Natl. Acad. Sci. USA. 2007;104:13954–13959. doi: 10.1073/pnas.0702315104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soultanas P., Dillingham M.S., Wigley D.B. Uncoupling DNA translocation and helicase activity in PcrA: direct evidence for an active mechanism. EMBO J. 2000;19:3799–3810. doi: 10.1093/emboj/19.14.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amaratunga M., Lohman T.M. Escherichia coli Rep helicase unwinds DNA by an active mechanism. Biochemistry. 1993;32:6815–6820. doi: 10.1021/bi00078a003. [DOI] [PubMed] [Google Scholar]
- 19.Dessinges M.N., Lionnet T., Croquette V. Single-molecule assay reveals strand switching and enhanced processivity of UvrD. Proc. Natl. Acad. Sci. USA. 2004;101:6439–6444. doi: 10.1073/pnas.0306713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischer C.J., Maluf N.K., Lohman T.M. Mechanism of ATP-dependent translocation of E. coli UvrD monomers along single-stranded DNA. J. Mol. Biol. 2004;344:1287–1309. doi: 10.1016/j.jmb.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 21.Yuzhakov A., Turner J., O'Donnell M. Replisome assembly reveals the basis for asymmetric function in leading and lagging strand replication. Cell. 1996;86:877–886. doi: 10.1016/s0092-8674(00)80163-4. [DOI] [PubMed] [Google Scholar]
- 22.Kobori J.A., Kornberg A. The Escherichia coli DnaC gene product. II. Purification, physical properties, and role in replication. J. Biol. Chem. 1982;257:13763–13769. [PubMed] [Google Scholar]
- 23.Kaplan D.L., O'Donnell M. DnaB drives DNA branch migration and dislodges proteins while encircling two DNA strands. Mol. Cell. 2002;10:647–657. doi: 10.1016/s1097-2765(02)00642-1. [DOI] [PubMed] [Google Scholar]
- 24.Ribeck N., Saleh O.A. Multiplexed single-molecule measurements with magnetic tweezers. Rev. Sci. Instrum. 2008;79:094301. doi: 10.1063/1.2981687. [DOI] [PubMed] [Google Scholar]
- 25.Gosse C., Croquette V. Magnetic tweezers: micromanipulation and force measurement at the molecular level. Biophys. J. 2002;82:3314–3329. doi: 10.1016/S0006-3495(02)75672-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berg-Sorensen K., Flyvbjerg H. Power spectrum analysis for optical tweezers. Rev. Sci. Instrum. 2004;75:594–612. [Google Scholar]
- 27.Klaue D., Seidel R. Torsional stiffness of single superparamagnetic microspheres in an external magnetic field. Phys. Rev. Lett. 2009;102:028302. doi: 10.1103/PhysRevLett.102.028302. [DOI] [PubMed] [Google Scholar]
- 28.Efron B., Tibshirani R. Statistical data analysis in the computer age. Science. 1991;253:390–395. doi: 10.1126/science.253.5018.390. [DOI] [PubMed] [Google Scholar]
- 29.Dumont S., Cheng W., Bustamante C. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature. 2006;439:105–108. doi: 10.1038/nature04331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bujalowski W., Jezewska M.J. Interactions of Escherichia coli primary replicative helicase DnaB protein with single-stranded DNA. The nucleic acid does not wrap around the protein hexamer. Biochemistry. 1995;34:8513–8519. doi: 10.1021/bi00027a001. [DOI] [PubMed] [Google Scholar]
- 31.Bujalowski W., Jezewska M.J. Kinetic mechanism of the single-stranded DNA recognition by Escherichia coli replicative helicase DnaB protein. Application of the matrix projection operator technique to analyze stopped-flow kinetics. J. Mol. Biol. 2000;295:831–852. doi: 10.1006/jmbi.1999.3378. [DOI] [PubMed] [Google Scholar]
- 32.Rouzina I., Bloomfield V.A. Force-induced melting of the DNA double helix. 1. Thermodynamic analysis. Biophys. J. 2001;80:882–893. doi: 10.1016/S0006-3495(01)76067-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cocco S., Yan J., Marko J.F. Overstretching and force-driven strand separation of double-helix DNA. Phys. Rev. E. 2004;70:011910. doi: 10.1103/PhysRevE.70.011910. [DOI] [PubMed] [Google Scholar]
- 34.Smith S.B., Cui Y.J., Bustamante C. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 35.Cluzel P., Lebrun A., Caron F. DNA: an extensible molecule. Science. 1996;271:792–794. doi: 10.1126/science.271.5250.792. [DOI] [PubMed] [Google Scholar]
- 36.van Mameren J., Gross P., Peterman E.J. Unraveling the structure of DNA during overstretching by using multicolor, single-molecule fluorescence imaging. Proc. Natl. Acad. Sci. USA. 2009;106:18231–18236. doi: 10.1073/pnas.0904322106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bailey S., Eliason W.K., Steitz T.A. Structure of hexameric DnaB helicase and its complex with a domain of DnaG primase. Science. 2007;318:459–463. doi: 10.1126/science.1147353. [DOI] [PubMed] [Google Scholar]
- 38.Bustamante C., Chemla Y.R., Izhaky D. Mechanical processes in biochemistry. Annu. Rev. Biochem. 2004;73:705–748. doi: 10.1146/annurev.biochem.72.121801.161542. [DOI] [PubMed] [Google Scholar]
- 39.Wang M.D., Schnitzer M.J., Block S.M. Force and velocity measured for single molecules of RNA polymerase. Science. 1998;282:902–907. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- 40.Galletto R., Jezewska M.J., Bujalowski W. Unzipping mechanism of the double-stranded DNA unwinding by a hexameric helicase: the effect of the 3′ arm and the stability of the dsDNA on the unwinding activity of the Escherichia coli DnaB helicase. J. Mol. Biol. 2004;343:101–114. doi: 10.1016/j.jmb.2004.07.056. [DOI] [PubMed] [Google Scholar]
- 41.Rothenberg E., Trakselis M.A., Ha T. MCM forked substrate specificity involves dynamic interaction with the 5′-tail. J. Biol. Chem. 2007;282:34229–34234. doi: 10.1074/jbc.M706300200. [DOI] [PubMed] [Google Scholar]
- 42.Sawaya M.R., Guo S.Y., Ellenberger T. Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7. Cell. 1999;99:167–177. doi: 10.1016/s0092-8674(00)81648-7. [DOI] [PubMed] [Google Scholar]
- 43.Stano N.M., Jeong Y.J., Patel S.S. DNA synthesis provides the driving force to accelerate DNA unwinding by a helicase. Nature. 2005;435:370–373. doi: 10.1038/nature03615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Egelman E.H., Yu X., Patel S.S. Bacteriophage T7 helicase/primase proteins form rings around single-stranded DNA that suggest a general structure for hexameric helicases. Proc. Natl. Acad. Sci. USA. 1995;92:3869–3873. doi: 10.1073/pnas.92.9.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lo Y.H., Tsai K.L., Hsiao C.D. The crystal structure of a replicative hexameric helicase DnaC and its complex with single-stranded DNA. Nucleic Acids Res. 2009;37:804–814. doi: 10.1093/nar/gkn999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


