Abstract
We simulated spontaneous fusion of small unilamellar vesicles mediated by lung surfactant protein B (SP-B) using the MARTINI force field. An SP-B monomer triggers fusion events by anchoring two vesicles and facilitating the formation of a lipid bridge between the proximal leaflets. Once a lipid bridge is formed, fusion proceeds via a previously described stalk – hemifusion diaphragm – pore-opening pathway. In the absence of protein, fusion of vesicles was not observed in either unbiased simulations or upon application of a restraining potential to maintain the vesicles in close proximity. The shape of SP-B appears to enable it to bind to two vesicles at once, forcing their proximity, and to facilitate the initial transfer of lipids to form a high-energy hemifusion intermediate. Our results may provide insight into more general mechanisms of protein-mediated membrane fusion, and a possible role of SP-B in the secretory pathway and transfer of lung surfactant to the gas exchange interface.
Introduction
Cells and their organelles are surrounded by membranes. Efficient exchange of their contents can be achieved via membrane fusion. Membrane fusion is a required step in intracellular trafficking, exocytosis (secretion of signaling molecules), development, and fertilization in eukaryotes, as well as in viral infection (1–6). In the fusion pathway, two membranes first approach at close distance and then merge into a hemifusion structure through which a fusion pore opens (7–9) (Fig. 1). This pathway involves overcoming intermembrane repulsion and strong deformations of the lipid bilayers constituting the membranes. Transitions between fusion intermediates are associated with high-energy barriers and are generally driven by proteins.
Figure 1.
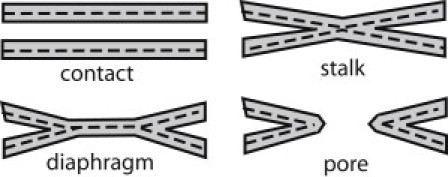
Schematic representation of intermediate steps of the fusion reaction.
Fusion proteins are believed to mediate most steps of fusion reactions, starting from recognition of the target membrane (10). The structure of fusion proteins and their interactions with lipids are as diverse as the biological processes involving fusion. Fusion machinery can vary from a single protein (e.g., FAST proteins of reoviruses (11)) to assemblies of protein complexes (e.g., homotrimeric class 1 fusion proteins in enveloped viruses (12) or heterotetrameric SNARE complexes (13)). Many proteins from unrelated families share common structural motifs, such as transmembrane domains anchoring the complex to the membrane and fusion peptides partially embedded in the membrane, and modes of action, including membrane local bending and membrane merger driven by refolding of oligomer ectodomains (1,14). However, the mechanisms of coupling between the protein and bilayer rearrangements that promote fusion are still poorly understood. All known fusion reactions appear to converge to the hemifusion and fusion pore formation. The exact sequence of hemifusion intermediates and the detailed structure of the fusion pore remain to be determined, and although a large number of fusion reactions have been studied to date, many have yet to be identified. Given the diversity of biological fusion events, variations along the fusion pathway or other mechanisms for fusion are possible.
In this work, we simulated fusion of lipid vesicles mediated by surfactant-associated protein B (SP-B). In contrast to experimental methods or theoretical models, computer simulations make it possible to directly follow nanoscale transformations along the fusion pathway without any assumptions regarding the intermediate steps. SP-B is a small hydrophobic protein that is crucial for regulating surface tension in the lungs during breathing (15,16). The fusogenic and lytic properties of SP-B demonstrated in vitro (17–19) are believed to play an important role in the secretory pathway and transfer of lung surfactant to the gas exchange interface (20–22). Although its high-resolution structure is not known, a reasonable model can be constructed based on homology with the saposin-like protein family (23,24) and NMR structures of several related fragments (20,25).
We employ molecular-dynamics (MD) simulations to study small unilamellar lipid vesicles and SP-B proteins with the coarse-grained MARTINI model (26,27). In this model, approximately four heavy atoms are grouped to represent a particle, and the particles interact via soft potentials. This allows a significant computational speedup compared to atomistic simulations while retaining a substantial amount of molecular detail. The model has been successfully applied to study the phase behavior of lipids, and the properties and transformations of protein-lipid aggregates (28–37). Earlier large-scale simulations with coarse-grained models demonstrated membrane remodeling by curvature-inducing proteins, including N-BAR domains (38–42). Fusion of lipid vesicles in the absence of proteins was previously simulated using coarse-grained and atomistic models (43–52). The vesicles were either constrained in close contact or pushed together to initiate stalk formation. Spontaneous fusion of vesicles/membranes was observed only in more coarse-grained systems, likely due to their softness and the high flip-flop rates of strongly simplified lipids, leading to low-energy barriers for fusion intermediates. Our simulations are not biased by any external force or constraint.
Materials and Methods
To study protein-mediated fusion reactions, we simulated lipid vesicles and SP-B proteins in water. We employed the Gromacs (version 4.0.5) (53) software package with the MARTINI coarse-grained force field for lipids and proteins (26,27). In this model, the molecules are represented by grouping four heavy atoms (two to three in the case of ring structures) into a particle. All lipids used are standard components of this force field, except for palmitoyloleoyl phosphatidylglycerol (POPG), for which the glycerol group in the headgroup was represented by a polar particle (P4). In dioleoyl phosphatidylcholine (DOPC) lipids, the angle potential in the hydrocarbon chains between particles C1, C2, and D3 was modified analogously to polyunsaturated chains in this model. The protein topology was built with the use of scripts downloaded from the MARTINI Web site (http://md.chem.rug.nl/∼marrink/MARTINI/). In the protein, selected bonds were constrained using the LINCS algorithm (54,55). For nonbonded interactions, the standard cutoffs for the MARTINI force field were used: the Lennard-Jones potential was shifted to zero between 0.9 and 1.2 nm, and the Coulomb potential was shifted to zero between 0 and 1.2 nm, with a relative dielectric constant of 15 (26). The time step was 20 fs with the neighbor list updated every 10 steps. The system was coupled to an isotropic pressure of 1 bar using the Berendsen barostat (56) with a time constant of 4 ps. Lipids, water, and proteins were coupled separately to a temperature of 310 K using the velocity rescaling thermostat (57) with a time constant of 1 ps.
The simulation box contained two lipid vesicles and ∼200,000 water particles with Na+ ions to neutralize the negative charge of the POPG lipids. SP-B proteins were added to this system as described below. Each vesicle was composed of 1152 lipids, giving a diameter of ∼14 nm. We simulated lipid mixtures of DPPC, DOPC, POPG, and PA. Two lipid compositions were tested: DPPC/POPG in a ratio of 3:1 (mixture 1) and DPPC/POPG/DOPC/PA in a ratio of 5:2:2:1 (mixture 2). In these mixtures, the anionic lipid POPG can interact specifically with positively charged SP-B proteins (total charge of one SP-B protein: +7).
To study the fusogenic activity of SP-B, we randomly distributed eight proteins in water. We performed six independent, unbiased simulations for each lipid composition (each 10 μs long, 120 μs in total). The actual simulation time is given in the text. To characterize the times required for transformation between the hemifusion intermediates, we used vesicles anchored by a single protein as an additional starting configuration, without any further constraints. We ran five independent simulations for each system (each 2 μs long). To assess the fusogenic properties of other proteins in the MARTINI force field, we tested surfactant-associated protein C (SP-C). The SP-C topology was built based on the known NMR structure (58), and an analogous setup with eight SP-C proteins (without SP-B) was simulated (five independent runs for each composition, each 5 μs long).
To study the ability of lipid vesicles to fuse without proteins, we performed a set of protein-free simulations for each lipid composition. We ran unbiased simulations (four runs, each 5 μs long) as well as simulations with a harmonic potential (with a force constant of 100 kJ/mol nm2) restraining the vesicles at distances ranging from 10 to 14 nm with a 1 nm interval between their center of mass (10 runs, each 1 μs long). As a control, all simulations with restrained vesicles were repeated using a previous version of the MARTINI force field. In this version, the nonbonded interactions between the C1- and Q-type particles are changed back from the superrepulsive to the repulsive level, and the van der Waals radius is reduced to 0.47. This modification facilitates protrusions of lipids and favors formation of the hemifusion stalk, making fusion more likely.
Results
The vesicles were preformed upon spontaneous bicelle bending and closing. Two lipid compositions were simulated: DPPC and POPG in a ratio of 3:1 (mixture 1), and DPPC, POPG, DOPC, and PA in a ratio of 5:2:2:1 (mixture 2). In both mixtures, lipids were distributed roughly homogeneously on the vesicle surface and between the leaflets; no demixing along the fusion reaction was observed. The vesicles had a diameter of ∼14 nm. Employing homology modeling (based on saposin C and NK-lysin) (59–61) and data on SP-B synthetic peptides (mini-B: residues 8–25, 63–78) (20,25), we obtained a structure characteristic of a saposin fold (Fig. 2). The full-length (79 residues) SP-B protein (62) contains four α-helices connected by unstructured loops and three internal disulfide bridges, forming a hairpin-shape (63). This secondary structure is incorporated in simulations with the MARTINI force field by defining bonded interaction parameters for the backbone. Spatial rearrangements between the secondary structure elements remain possible, however, and side-chain partitioning between water and lipids is well reproduced (27).
Figure 2.
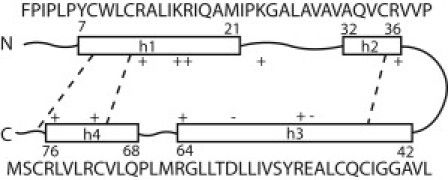
The secondary structure of SP-B protein contains four α-helices (h1–h4) connected by unstructured loops (solid lines) and linked by disulfide bridges (dashed lines) forming a hairpin shape.
The vesicles and SP-B proteins are initially placed randomly in water. The proteins diffuse toward the vesicle surface and insert into the headgroup region of the outer lipid leaflet. Because of the large fraction of hydrophobic residues on both faces of SP-B (Fig. 3), it can adopt two conformations on the surface of the vesicle: extended (Fig. 4 a) or bent (Fig. 4 b). The density distributions for these conformations are shown in Fig. S1 of the Supporting Material. In both conformations, helices 1 and 4 at the N- and C-termini partition into the headgroup region. In the bent conformation, helices 2 and 3 are exposed to solvent, with negatively charged residues in helix 3 interacting with positively charged residues in helix 1. This bent-hairpin conformation readily triggers fusion events.
Figure 3.
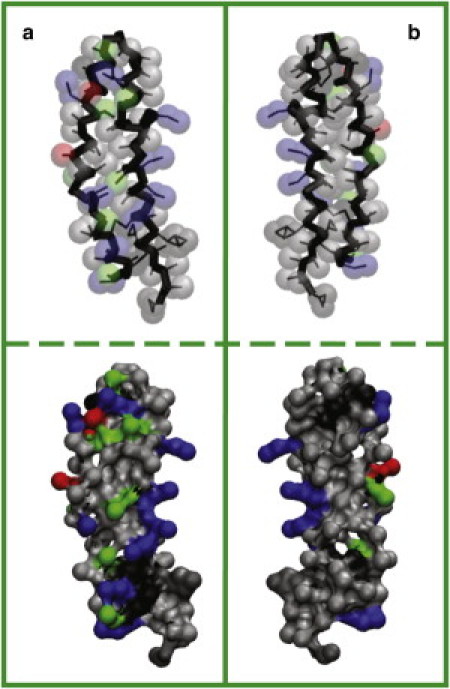
Distribution of residues of (a) water- and (b) membrane-facing sides of SP-B. In the upper panel, bonds are shown as thin black lines, backbone as thick black lines, and side chains as semitransparent spheres. Apolar residues are colored in gray, polar in green, positively charged in blue, and negatively charged in red. The solvent-accessible surface with the same color scheme is given in the lower panel.
Figure 4.
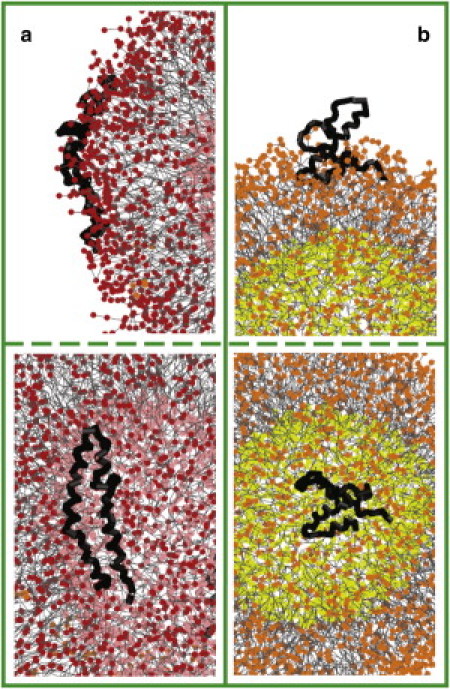
Extended (a) and bent (b) conformations of SPB on the surface of the vesicles, side (upper panel) and top (lower panel) views. Apolar groups of lipids are colored in gray, and polar groups belonging to the outer and lower leaflets are shown in red (a)/orange (b) and pink (a)/yellow (b), respectively. Protein backbone is shown as thick black lines. For clarity, side chains and water/ions are not shown.
Fusion begins when the vesicles diffuse to a close distance, such that the solvent-exposed part of the protein on one vesicle can reach the second vesicle (Movie S1). Upon contact with the surface, the protein inserts into the headgroup region, anchoring the two vesicles together. The protein then rearranges as a wedge between the two vesicles (Fig. 5 a). Near the vertices of the wedge, lipids protrude out of the membrane and tilt their molecular axes along the vesicle surface, with the hydrocarbon chains facing the hydrophobic part of the protein. Thus a lipid bridge is formed connecting the outer leaflets of the vesicles (Fig. 5 b). As the flow of lipids continues (Fig. 5 c), these proximal leaflets merge to form a hemifusion stalk (Fig. 5 d). At the same time, the conformation of SP-B changes from bent to extended. The stalk is elongated in shape, with the protein lining a positively curved side of the stalk. The stalk bends and expands anisotropically into a hemifusion diaphragm (Fig. 5 e), which then ruptures (Fig. 5 f) to complete the fusion process (Fig. 5 g). Fusion events were observed in six cases (three for each mixture). In mixture 1 the reaction did not proceed beyond the stalk on the simulation timescale, whereas in mixture 2, fusion was always completed.
Figure 5.
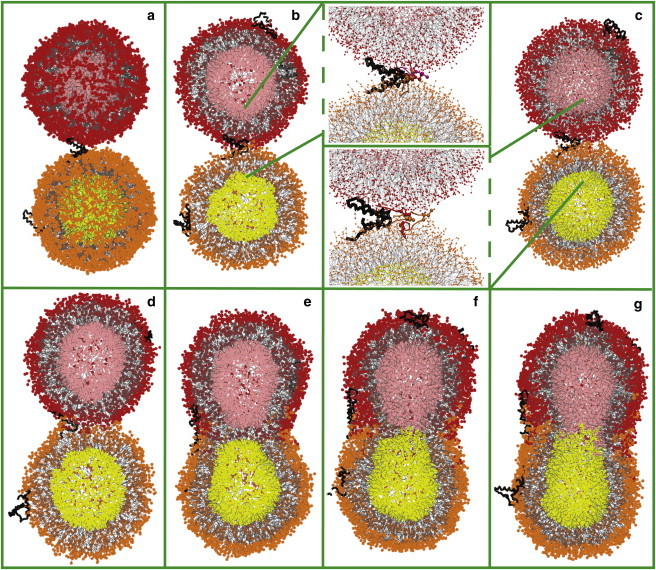
Intermediate steps for fusion of lipid vesicles mediated by SP-B: vesicles anchored by protein (a), lipid bridge (b), lipid flow (c), hemifusion stalk (d), hemifusion diaphragm (e), and fusion pore (f). (g) Fusion is complete. Vesicles are shown in cross-section in b–g. Molecular representations are as in Fig. 4.
The limiting step for fusion is establishment of close contact between the vesicles. This step requires insertion of SP-B into both vesicles, and is controlled by diffusion of the vesicles in solution and of the proteins in solution and on the vesicle surface. The time required for this depends on the vesicle and protein concentration or the simulation box size; in our simulations, it took hundreds of nanoseconds to microseconds. Once SP-B reaches a close distance to the membrane (∼3 nm for mixture 1 and ∼2.5 nm for mixture 2), its insertion proceeds rapidly (∼20 ns; see Fig. S2). The fusion reaction always continues after the protein in the bent conformation anchors two vesicles (including in 10 control simulations). Formation of a lipid bridge between the anchored vesicles took between 20 and 950 ns; such variation may originate from adjustments in protein insertion. Transformation of the lipid bridge into a hemifusion stalk occurred within 27 ± 5 ns. Formation of a hemifusion diaphragm in mixture 1 was not observed on the simulation timescale (10 μs). Transformation of the stalk into a hemifusion diaphragm in mixture 2 was completed between 28 and 416 ns, and the diaphragm ruptured in 16 ± 11 ns. The hemifusion diaphragm was formed from inner leaflets of both vesicles in five cases. In three other cases, one of the vesicles ruptured at the end of stalk expansion, and the lipids belonging to the other vesicle lined the diaphragm. In earlier simulations of tethered/restrained vesicles using the MARTINI model (43,48), stalk development and diaphragm rupture also proceeded rapidly, whereas diaphragm formation diverged for the same system and had faster or slower kinetics depending on the lipid composition.
In an alternative pathway, a lipid bridge is formed inside the protein hairpin in the extended conformation (Fig. 6 a), and the fusion reaction is trapped on the microsecond timescale (two and three cases in mixtures 1 and 2, respectively). Another protein in the bent conformation releases the trapped protein (one case in mixture 2) by pushing it away from the connection (Fig. 6 b). This trapped configuration may result form the small radii of the vesicles (which stretches the protein for optimal insertion to the surface), and may be less likely to occur between planar bilayers or vesicles of larger radii (data not shown). Depending on their local concentration, the proteins also form aggregates in water and on the vesicle's surface. However, an SP-B monomer in the bent-hairpin conformation is sufficient to initiate vesicle fusion.
Figure 6.
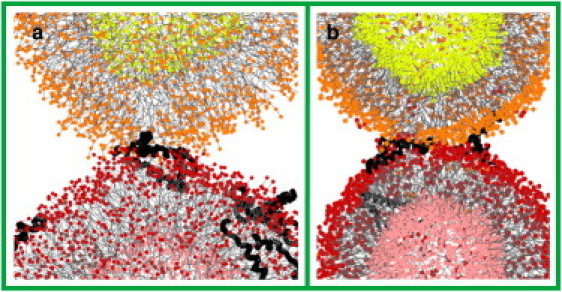
Lipid bridge through SP-B protein in extended conformation. Fusion reaction is trapped (a) and released by another SP-B protein in the bent conformation (b). Color scheme as in Fig. 4.
Unlike SP-B, SP-C did not result in vesicle fusion in our simulations (five attempts for each mixture). SP-C partitions deeper into the membrane interior as compared to SP-B, with its helix oriented nearly parallel to the membrane surface and palmitoyl chains aligned with the lipid chains. This insertion prevents the protein from reaching the second vesicle, which hinders vesicle anchoring and establishment of close contact (in nine out of 10 cases). In one simulation (mixture 1), an SP-C dimer protruding out of the membrane into the water region (with the helices oriented antiparallel) connected two vesicles. However, the dimer remained in the center of the connection, with a lipid bridge on both its sides, and thus the hemifusion stalk could not be fully formed.
In the absence of proteins, no fusion events were observed. The vesicles approached each other in a random manner but did not remain at close distances, and diffused away. Restraining the vesicles in close contact by a harmonic potential also did not lead to their fusion on the microsecond timescale (five attempts for each mixture). A reduction of repulsion between the lipid tails and the charged particles (as in the previous version of the force field; see Materials and Methods), which favors lipid protrusions, also did not lead to fusion (10 additional attempts). Even if the imposed distance was smaller then the sum of their radii, the vesicles deformed to obey the restraint but failed to fuse (Fig. 7).
Figure 7.

Vesicles restrained at close distance (10 nm between their centers of mass) in the absence of SP-B protein. Cross-section view, color scheme as in Fig. 4.
Discussion
The simulation results suggest that SP-B proteins promote early stages of vesicle fusion. At close distances, interactions between the membranes are net repulsive due to hydration forces and bilayer undulations (64,65). Here, thermal undulations likely have a weak effect as the vesicles are of small radius and the headgroup region of the outer leaflet is stretched. Insertion of the protein into the vesicle surface allows burying of its hydrophobic residues and positioning of the positively charge residues near the headgroups of anionic lipids. Insertion of the protein into the second vesicle anchors the vesicles together and establishes a close contact between the membranes. Favorable protein-lipid interactions counteract intermembrane repulsion.
Approach in close contact can be associated with the formation of strongly bent areas (bulges or dimples) in biological membranes (7,66). In particular, insertion of fusion peptides is believed to produce membrane distortion/local curvature (10,14,67). For example, the fusion peptide of influenza hemagglutinin (HA) has a helix-hinge-helix amphipathic structure arranged into a V-shape with a hydrophobic pocket in the cavity of the V that disrupts the bilayer (68,69). Here, the lipid layers are deformed significantly due to the small radius of the vesicle. SP-B in the bent conformation does not create noticeable curvature. Of interest, SP-B in the extended conformation, which resembles the V-shape of the HA fusion peptide, similarly to the latter induces local curvature in bilayers (data not shown).
Maintaining the vesicles in close contact is, however, not sufficient for fusion. This is shown in protein-free simulations with the two vesicles kept in close proximity by a restraining potential (Fig. 7). Transition to the hemifusion stalk requires the creation of a high negative curvature in the proximal leaflets of the vesicles and exposure of lipid hydrocarbon chains to the polar environment. This step is facilitated in membranes with lower bending rigidity and increased lipid protrusions, which is typical for more coarse-grained lipid models (e.g., lipids represented by three beads (44,50)), and with more polar and flexible lipid tails (as manifested by back folding (49)). Formation of the stalk is also promoted by cone-shaped lipids (such as unsaturated phosphatidylethanolamines (70)). In earlier simulations, fusion between restrained vesicles was observed for different lipid compositions (including cone-shaped lipids) using a previous version of the MARTINI model (43,47). In our simulations, restoring the interactions between the lipid tails and charged particles to the level of the previous version favoring lipid protrusions did not produce fusion. This suggests that the chosen lipid mixtures are not strongly fusogenic, and a significant external driving force is required to lower the energy barrier for stalk formation. SP-B in the bent conformation acts as a scaffold (71,72), providing both a positive charge and a hydrophobic surface along which the lipids protrude out of the membrane and tilt to establish a lipid bridge. Once a lipid bridge is formed, the flow of lipids continues and leads to the hemifusion stalk. Establishment of a lipid bridge is thus the limiting step for stalk formation.
SP-B plays no additional role in later stages of fusion reaction in our simulations. Transformation of the stalk into a hemifusion diaphragm and opening of the pore through the diaphragm are controlled by the properties of lipid mixtures. Release of the energy accumulated due to intermembrane repulsion and elastic stresses trigger these transformations (7,67,73,74). Here, the elastic stresses originate from the small radii of the vesicles and the high curvature of the stalk. Transformation into a hemifusion diaphragm and pore opening can, however, require higher energies than the stalk formation (66,70,75). The accumulated energy is not sufficient or activation energies are higher for these steps in mixture 1. The stalk represents a long-lived hemifusion intermediate that may decay toward fusion on a timescale longer than the simulation times (48). In mixture 2, the higher content of unsaturated lipids reduces the elastic moduli of the leaflets, which in turn lowers the energy barriers, and fusion is completed. Tension due to stretching of the headgroup region of outer vesicle leaflets may also facilitate formation of the diaphragm and its rupture (8,50,74,76).
The fusion events in our simulations proceed via a contact – hemifusion stalk – hemifusion diaphragm – fusion pore pathway. All intermediates of this pathway are lipid-based (76,77) and are generally consistent with theoretical continuum models (8,75,78–80). Lipid tilting to produce stalk, radial, or anisotropic (banana-shaped) stalk expansion, and a diaphragm composed either of inner leaflets of both vesicles or of one vesicle were observed in previous simulations in the absence of proteins (43–45,49,51). Earlier simulations also demonstrated a theoretically predicted (66,81) alternative evolution by a direct stalk-pore transition (44,47,48), as well as a transition between an adhesion zone and a diaphragm without stalk formation (50,82). Of interest, fusion appears to follow different pathways for the same lipid mixture under the same conditions. The kinetics and intermediates along the fusion reaction depend on many factors, including lipid composition, simulation parameters/force field, system size and geometry (vesicles/bilayers), and membrane tension. Of importance, all simulated fusion events converge to the contact – hemifusion – pore pattern.
We have shown that an SP-B monomer in the bent-hairpin conformation is sufficient to initiate vesicle fusion. SP-B proteins were observed to induce fusion of liposomes (DPPC/POPG lipids in a 7:3 ratio) in earlier experimental studies (17–19). These studies also reported a minor ability of SP-C to induce lipid mixing, which is in agreement with our simulation results. Fusogenic activity of SP-B does not appear to require other proteins and does not involve significant structural rearrangements. This may be similar to the action of small FAST proteins in reoviruses (11,83), and is distinct from enveloped viruses (6,12) or SNARE-mediated fusion (5,13,84), where conformational changes of the fusion protein complex, including refolding (zippering) of ectodomains, are thought to drive fusion reactions. It is worth mentioning that the secondary structure of the protein is fixed by parameters of the employed coarse-grained model. Hence, it was not possible to capture changes in the secondary structure of SP-B along the fusion pathway. However, changes of structure upon binding to lipid aggregates are expected to be small (85). In addition, SP-B is expected to form a homodimer connected by a disulfide bridge (86,87). Dimer conformations or aggregation may mediate later stages of fusion: transformation of the stalk into a hemifusion diaphragm and fusion pore opening. This offers a subject for future studies, which could perhaps incorporate a more detailed model and larger vesicles or bilayers. Previous simulations have already demonstrated promotion of pores in the stalk by scramblases (40), stabilization of the stalk-pore complex in a bicontinuous cubic phase by HA fusion peptides (32), and membrane perturbation by pulling on the SNARE complex (88).
The fusogenic activity of SP-B is important for the physiological function of the lung. The protein is believed to play a role in several steps in the lung surfactant secretory pathway (20), including fusion of a late endosome/multivesicular body with the lamellar body, organization of surfactant lipids as concentric bilayers in the lamellar body (89,90), and formation of tubular myelin from the lamellar bodies (91,92). In addition, SP-B provides rapid transfer of lung surfactant to the gas exchange interface, which is required for lung function. Rapid transfer is likely achieved via a hemifusion stalk connection between the interface and surfactant vesicles/reservoirs (93,94). SP-B induces/stabilizes the highly curved stalk, which allows collective adsorption/flow of lipids. However, the exact mechanism of stalk formation and the role of SP-B in the function of lung surfactant are still not fully understood. Our simulations provide insights into SP-B-facilitated stalk formation. The simulations also show that the proteins line positively curved edges of the stalk. We hypothesize that SP-B stabilizes the stalk by reducing the line tension at its edges.
In summary, we simulated fusion of small unilamellar vesicles mediated by lung surfactant protein SP-B. The simulation results suggest that SP-B initiates fusion by anchoring the vesicles together and facilitating formation of a lipid bridge, which converts into a hemifusion stalk.
Acknowledgments
We thank Megan O'Mara for help with the homology models. Simulations were performed in part on WestGrid/Compute Canada facilities.
S.B. was supported by postdoctoral fellowships from the Alberta Heritage Foundation for Medical Research, and the Canadian Institutes for Health Research. D.P.T. is an Alberta Heritage Foundation for Medical Research Scientist. This work was supported by the Natural Sciences and Engineering Research Council.
Supporting Material
References
- 1.Jahn R., Lang T., Südhof T.C. Membrane fusion. Cell. 2003;112:519–533. doi: 10.1016/s0092-8674(03)00112-0. [DOI] [PubMed] [Google Scholar]
- 2.Sapir A., Avinoam O., Chernomordik L.V. Viral and developmental cell fusion mechanisms: conservation and divergence. Dev. Cell. 2008;14:11–21. doi: 10.1016/j.devcel.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen E.H., Grote E., Vignery A. Cell-cell fusion. FEBS Lett. 2007;581:2181–2193. doi: 10.1016/j.febslet.2007.03.033. [DOI] [PubMed] [Google Scholar]
- 4.Wickner W., Schekman R. Membrane fusion. Nat. Struct. Mol. Biol. 2008;15:658–664. doi: 10.1038/nsmb.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rizo J., Rosenmund C. Synaptic vesicle fusion. Nat. Struct. Mol. Biol. 2008;15:665–674. doi: 10.1038/nsmb.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harrison S.C. Viral membrane fusion. Nat. Struct. Mol. Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kozlov M.M., Chernomordik L.V. A mechanism of protein-mediated fusion: coupling between refolding of the influenza hemagglutinin and lipid rearrangements. Biophys. J. 1998;75:1384–1396. doi: 10.1016/S0006-3495(98)74056-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chernomordik L.V., Melikyan G.B., Chizmadzhev Y.A. Biomembrane fusion: a new concept derived from model studies using two interacting planar lipid bilayers. Biochim. Biophys. Acta. 1987;906:309–352. doi: 10.1016/0304-4157(87)90016-5. [DOI] [PubMed] [Google Scholar]
- 9.Lee J., Lentz B.R. Evolution of lipidic structures during model membrane fusion and the relation of this process to cell membrane fusion. Biochemistry. 1997;36:6251–6259. doi: 10.1021/bi970404c. [DOI] [PubMed] [Google Scholar]
- 10.Chernomordik L.V., Zimmerberg J., Kozlov M.M. Membranes of the world unite! J. Cell Biol. 2006;175:201–207. doi: 10.1083/jcb.200607083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Top D., de Antueno R., Duncan R. Liposome reconstitution of a minimal protein-mediated membrane fusion machine. EMBO J. 2005;24:2980–2988. doi: 10.1038/sj.emboj.7600767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kielian M., Rey F.A. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat. Rev. Microbiol. 2006;4:67–76. doi: 10.1038/nrmicro1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jahn R., Scheller R.H. SNAREs—engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 14.Chernomordik L.V., Kozlov M.M. Mechanics of membrane fusion. Nat. Struct. Mol. Biol. 2008;15:675–683. doi: 10.1038/nsmb.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clark J.C., Wert S.E., Whitsett J.A. Targeted disruption of the surfactant protein B gene disrupts surfactant homeostasis, causing respiratory failure in newborn mice. Proc. Natl. Acad. Sci. USA. 1995;92:7794–7798. doi: 10.1073/pnas.92.17.7794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nogee L.M., Garnier G., Colten H.R. A mutation in the surfactant protein B gene responsible for fatal neonatal respiratory disease in multiple kindreds. J. Clin. Invest. 1994;93:1860–1863. doi: 10.1172/JCI117173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oosterlaken-Dijksterhuis M.A., van Eijk M., Haagsman H.P. Lipid mixing is mediated by the hydrophobic surfactant protein SP-B but not by SP-C. Biochim. Biophys. Acta. 1992;1110:45–50. doi: 10.1016/0005-2736(92)90292-t. [DOI] [PubMed] [Google Scholar]
- 18.Poulain F.R., Allen L., Hawgood S. Effects of surfactant apolipoproteins on liposome structure: implications for tubular myelin formation. Am. J. Physiol. 1992;262:L730–L739. doi: 10.1152/ajplung.1992.262.6.L730. [DOI] [PubMed] [Google Scholar]
- 19.Shiffer K., Hawgood S., Goerke J. Interactions of the low molecular weight group of surfactant-associated proteins (SP 5-18) with pulmonary surfactant lipids. Biochemistry. 1988;27:2689–2695. doi: 10.1021/bi00408a008. [DOI] [PubMed] [Google Scholar]
- 20.Ryan M.A., Qi X.Y., Weaver T.E. Mapping and analysis of the lytic and fusogenic domains of surfactant protein B. Biochemistry. 2005;44:861–872. doi: 10.1021/bi0485575. [DOI] [PubMed] [Google Scholar]
- 21.Whitsett J.A., Weaver T.E. Hydrophobic surfactant proteins in lung function and disease. N. Engl. J. Med. 2002;347:2141–2148. doi: 10.1056/NEJMra022387. [DOI] [PubMed] [Google Scholar]
- 22.Zuo Y.Y., Veldhuizen R.A.W., Possmayer F. Current perspectives in pulmonary surfactant—inhibition, enhancement and evaluation. Biochim. Biophys. Acta. 2008;1778:1947–1977. doi: 10.1016/j.bbamem.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 23.Bruhn H. A short guided tour through functional and structural features of saposin-like proteins. Biochem. J. 2005;389:249–257. doi: 10.1042/BJ20050051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patthy L. Homology of the precursor of pulmonary surfactant-associated protein SP-B with prosaposin and sulfated glycoprotein 1. J. Biol. Chem. 1991;266:6035–6037. [PubMed] [Google Scholar]
- 25.Sarker M., Waring A.J., Booth V. Structure of mini-B, a functional fragment of surfactant protein B, in detergent micelles. Biochemistry. 2007;46:11047–11056. doi: 10.1021/bi7011756. [DOI] [PubMed] [Google Scholar]
- 26.Marrink S.J., Risselada H.J., de Vries A.H. The MARTINI force field: coarse grained model for biomolecular simulations. J. Phys. Chem. B. 2007;111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 27.Monticelli L., Kandasamy S.K., Marrink S.J. The MARTINI coarse-grained force field: Extension to proteins. J. Chem. Theory Comput. 2008;4:819–834. doi: 10.1021/ct700324x. [DOI] [PubMed] [Google Scholar]
- 28.Risselada H.J., Marrink S.J. The molecular face of lipid rafts in model membranes. Proc. Natl. Acad. Sci. USA. 2008;105:17367–17372. doi: 10.1073/pnas.0807527105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marrink S.J., Risselada J., Mark A.E. Simulation of gel phase formation and melting in lipid bilayers using a coarse grained model. Chem. Phys. Lipids. 2005;135:223–244. doi: 10.1016/j.chemphyslip.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 30.Baoukina S., Monticelli L., Tieleman D.P. Pressure-area isotherm of a lipid monolayer from molecular dynamics simulations. Langmuir. 2007;23:12617–12623. doi: 10.1021/la702286h. [DOI] [PubMed] [Google Scholar]
- 31.Marrink S.J., Mark A.E. Molecular view of hexagonal phase formation in phospholipid membranes. Biophys. J. 2004;87:3894–3900. doi: 10.1529/biophysj.104.048710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fuhrmans M., Knecht V., Marrink S.J. A single bicontinuous cubic phase induced by fusion peptides. J. Am. Chem. Soc. 2009;131:9166–9167. doi: 10.1021/ja903224q. [DOI] [PubMed] [Google Scholar]
- 33.Catte A., Patterson J.C., Segrest J.P. Structure of spheroidal HDL particles revealed by combined atomistic and coarse-grained simulations. Biophys. J. 2008;94:2306–2319. doi: 10.1529/biophysj.107.115857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yefimov S., van der Giessen E., Marrink S.J. Mechanosensitive membrane channels in action. Biophys. J. 2008;94:2994–3002. doi: 10.1529/biophysj.107.119966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scott K.A., Bond P.J., Sansom M.S. Coarse-grained MD simulations of membrane protein-bilayer self-assembly. Structure. 2008;16:621–630. doi: 10.1016/j.str.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 36.Periole X., Huber T., Sakmar T.P. G protein-coupled receptors self-assemble in dynamics simulations of model bilayers. J. Am. Chem. Soc. 2007;129:10126–10132. doi: 10.1021/ja0706246. [DOI] [PubMed] [Google Scholar]
- 37.Baoukina S., Monticelli L., Tieleman D.P. The molecular mechanism of lipid monolayer collapse. Proc. Natl. Acad. Sci. USA. 2008;105:10803–10808. doi: 10.1073/pnas.0711563105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ayton G.S., Lyman E., Voth G.A. New insights into BAR domain-induced membrane remodeling. Biophys. J. 2009;97:1616–1625. doi: 10.1016/j.bpj.2009.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reynwar B.J., Illya G., Deserno M. Aggregation and vesiculation of membrane proteins by curvature-mediated interactions. Nature. 2007;447:461–464. doi: 10.1038/nature05840. [DOI] [PubMed] [Google Scholar]
- 40.Smeijers A.F., Pieterse K., Hilbers P.A. Coarse-grained transmembrane proteins: hydrophobic matching, aggregation, and their effect on fusion. J. Phys. Chem. B. 2006;110:13614–13623. doi: 10.1021/jp062012y. [DOI] [PubMed] [Google Scholar]
- 41.Yin Y., Arkhipov A., Schulten K. Simulations of membrane tubulation by lattices of amphiphysin N-BAR domains. Structure. 2009;17:882–892. doi: 10.1016/j.str.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marrink S.J., de Vries A.H., Tieleman D.P. Lipids on the move: simulations of membrane pores, domains, stalks and curves. Biochim. Biophys. Acta. 2009;1788:149–168. doi: 10.1016/j.bbamem.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 43.Marrink S.J., Mark A.E. The mechanism of vesicle fusion as revealed by molecular dynamics simulations. J. Am. Chem. Soc. 2003;125:11144–11145. doi: 10.1021/ja036138+. [DOI] [PubMed] [Google Scholar]
- 44.Noguchi H., Takasu M. Fusion pathways of vesicles: a Brownian dynamics simulation. J. Chem. Phys. 2001;115:9547–9551. [Google Scholar]
- 45.Stevens M.J., Hoh J.H., Woolf T.B. Insights into the molecular mechanism of membrane fusion from simulation: evidence for the association of splayed tails. Phys. Rev. Lett. 2003;91:188102. doi: 10.1103/PhysRevLett.91.188102. [DOI] [PubMed] [Google Scholar]
- 46.Müller M., Katsov K., Schick M. A new mechanism of model membrane fusion determined from Monte Carlo simulation. Biophys. J. 2003;85:1611–1623. doi: 10.1016/S0006-3495(03)74592-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kasson P.M., Pande V.S. Control of membrane fusion mechanism by lipid composition: predictions from ensemble molecular dynamics. PLOS Comput. Biol. 2007;3:e220. doi: 10.1371/journal.pcbi.0030220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kasson P.M., Kelley N.W., Pande V.S. Ensemble molecular dynamics yields submillisecond kinetics and intermediates of membrane fusion. Proc. Natl. Acad. Sci. USA. 2006;103:11916–11921. doi: 10.1073/pnas.0601597103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smeijers A.F., Markvoort A.J., Hilbers P.A. A detailed look at vesicle fusion. J. Phys. Chem. B. 2006;110:13212–13219. doi: 10.1021/jp060824o. [DOI] [PubMed] [Google Scholar]
- 50.Shillcock J.C., Lipowsky R. Tension-induced fusion of bilayer membranes and vesicles. Nat. Mater. 2005;4:225–228. doi: 10.1038/nmat1333. [DOI] [PubMed] [Google Scholar]
- 51.Knecht V., Marrink S.J. Molecular dynamics simulations of lipid vesicle fusion in atomic detail. Biophys. J. 2007;92:4254–4261. doi: 10.1529/biophysj.106.103572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marrink S.J., Tieleman D.P. Molecular dynamics simulation of spontaneous membrane fusion during a cubic-hexagonal phase transition. Biophys. J. 2002;83:2386–2392. doi: 10.1016/s0006-3495(02)75252-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hess B., Kutzner C., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 54.Hess B. P-LINCS: a parallel linear constraint solver for molecular simulation. J. Chem. Theory Comput. 2008;4:116–122. doi: 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- 55.Hess B., Bekker H., Fraaije J. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. doi: 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- 56.Berendsen H.J.C., Postma J.P.M., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 57.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 58.Johansson J., Szyperski T., Wüthrich K. The NMR structure of the pulmonary surfactant-associated polypeptide SP-C in an apolar solvent contains a valyl-rich α-helix. Biochemistry. 1994;33:6015–6023. doi: 10.1021/bi00185a042. [DOI] [PubMed] [Google Scholar]
- 59.Liepinsh E., Andersson M., Otting G. Saposin fold revealed by the NMR structure of NK-lysin. Nat. Struct. Biol. 1997;4:793–795. doi: 10.1038/nsb1097-793. [DOI] [PubMed] [Google Scholar]
- 60.Rossmann M., Schultz-Heienbrok R., Maier T. Crystal structures of human saposins C and D: implications for lipid recognition and membrane interactions. Structure. 2008;16:809–817. doi: 10.1016/j.str.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 61.Ahn V.E., Leyko P., Privé G.G. Crystal structures of saposins A and C. Protein Sci. 2006;15:1849–1857. doi: 10.1110/ps.062256606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hawgood S., Derrick M., Poulain F. Structure and properties of surfactant protein B. Biochim. Biophys. Acta. 1998;1408:150–160. doi: 10.1016/s0925-4439(98)00064-7. [DOI] [PubMed] [Google Scholar]
- 63.Andersson M., Curstedt T., Johansson J. An amphipathic helical motif common to tumourolytic polypeptide NK-lysin and pulmonary surfactant polypeptide SP-B. FEBS Lett. 1995;362:328–332. doi: 10.1016/0014-5793(95)00268-e. [DOI] [PubMed] [Google Scholar]
- 64.Helfrich W. Elastic properties of lipid bilayers: theory and possible experiments. Z. Naturforsch. C. 1973;28(11, C):693–703. doi: 10.1515/znc-1973-11-1209. [DOI] [PubMed] [Google Scholar]
- 65.Rand R.P., Parsegian V.A. Hydration forces between phospholipid-bilayers. Biochim. Biophys. Acta. 1989;988:351–376. [Google Scholar]
- 66.Kuzmin P.I., Zimmerberg J., Cohen F.S. A quantitative model for membrane fusion based on low-energy intermediates. Proc. Natl. Acad. Sci. USA. 2001;98:7235–7240. doi: 10.1073/pnas.121191898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martens S., Kozlov M.M., McMahon H.T. How synaptotagmin promotes membrane fusion. Science. 2007;316:1205–1208. doi: 10.1126/science.1142614. [DOI] [PubMed] [Google Scholar]
- 68.Tamm L.K. Hypothesis: spring-loaded boomerang mechanism of influenza hemagglutinin-mediated membrane fusion. Biochim. Biophys. Acta. 2003;1614:14–23. doi: 10.1016/s0005-2736(03)00159-7. [DOI] [PubMed] [Google Scholar]
- 69.Han X., Bushweller J.H., Tamm L.K. Membrane structure and fusion-triggering conformational change of the fusion domain from influenza hemagglutinin. Nat. Struct. Biol. 2001;8:715–720. doi: 10.1038/90434. [DOI] [PubMed] [Google Scholar]
- 70.Chernomordik L.V., Kozlov M.M. Protein-lipid interplay in fusion and fission of biological membranes. Annu. Rev. Biochem. 2003;72:175–207. doi: 10.1146/annurev.biochem.72.121801.161504. [DOI] [PubMed] [Google Scholar]
- 71.Jackson M.B., Chapman E.R. Fusion pores and fusion machines in Ca2+-triggered exocytosis. Annu. Rev. Biophys. Biomol. Struct. 2006;35:135–160. doi: 10.1146/annurev.biophys.35.040405.101958. [DOI] [PubMed] [Google Scholar]
- 72.Rizo J., Chen X.C., Araç D. Unraveling the mechanisms of synaptotagmin and SNARE function in neurotransmitter release. Trends Cell Biol. 2006;16:339–350. doi: 10.1016/j.tcb.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 73.Lee J.Y., Schick M. Calculation of free energy barriers to the fusion of small vesicles. Biophys. J. 2008;94:1699–1706. doi: 10.1529/biophysj.107.119511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Katsov K., Müller M., Schick M. Field theoretic study of bilayer membrane fusion. I. Hemifusion mechanism. Biophys. J. 2004;87:3277–3290. doi: 10.1529/biophysj.103.038943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kozlovsky Y., Chernomordik L.V., Kozlov M.M. Lipid intermediates in membrane fusion: formation, structure, and decay of hemifusion diaphragm. Biophys. J. 2002;83:2634–2651. doi: 10.1016/S0006-3495(02)75274-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cohen F.S., Zimmerberg J., Finkelstein A. Fusion of phospholipid vesicles with planar phospholipid bilayer membranes. II. Incorporation of a vesicular membrane marker into the planar membrane. J. Gen. Physiol. 1980;75:251–270. doi: 10.1085/jgp.75.3.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zimmerberg J., Cohen F.S., Finkelstein A. Fusion of phospholipid vesicles with planar phospholipid bilayer membranes. I. Discharge of vesicular contents across the planar membrane. J. Gen. Physiol. 1980;75:241–250. doi: 10.1085/jgp.75.3.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kozlov M.M., Markin V.S. [Possible mechanism of membrane fusion] Biofizika. 1983;28:242–247. [PubMed] [Google Scholar]
- 79.Kozlovsky Y., Kozlov M.M. Stalk model of membrane fusion: solution of energy crisis. Biophys. J. 2002;82:882–895. doi: 10.1016/S0006-3495(02)75450-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zimmerberg J., Chernomordik L.V. Membrane fusion. Adv. Drug Deliv. Rev. 1999;38:197–205. doi: 10.1016/s0169-409x(99)00029-0. [DOI] [PubMed] [Google Scholar]
- 81.Siegel D.P. Energetics of intermediates in membrane fusion: comparison of stalk and inverted micellar intermediate mechanisms. Biophys. J. 1993;65:2124–2140. doi: 10.1016/S0006-3495(93)81256-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gao L.H., Lipowsky R., Shillcock J. Tension-induced vesicle fusion: pathways and pore dynamics. Soft Matter. 2008;4:1208–1214. doi: 10.1039/b801407h. [DOI] [PubMed] [Google Scholar]
- 83.Shmulevitz M., Duncan R. A new class of fusion-associated small transmembrane (FAST) proteins encoded by the non-enveloped fusogenic reoviruses. EMBO J. 2000;19:902–912. doi: 10.1093/emboj/19.5.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stein A., Weber G., Jahn R. Helical extension of the neuronal SNARE complex into the membrane. Nature. 2009;460:525–528. doi: 10.1038/nature08156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cruz A., Casals C., Perez-Gil J. Conformational flexibility of pulmonary surfactant proteins SP-B and SP-C, studied in aqueous organic solvents. Biochim. Biophys. Acta. 1995;1255:68–76. doi: 10.1016/0005-2760(94)00210-p. [DOI] [PubMed] [Google Scholar]
- 86.Possmayer F. A proposed nomenclature for pulmonary surfactant-associated proteins. Am. Rev. Respir. Dis. 1988;138:990–998. doi: 10.1164/ajrccm/138.4.990. [DOI] [PubMed] [Google Scholar]
- 87.Johansson J., Curstedt T., Jörnvall H. Size and structure of the hydrophobic low molecular weight surfactant-associated polypeptide. Biochemistry. 1988;27:3544–3547. doi: 10.1021/bi00410a002. [DOI] [PubMed] [Google Scholar]
- 88.Delalande O., Férey N., Baaden M. Complex molecular assemblies at hand via interactive simulations. J. Comput. Chem. 2009;30:2375–2387. doi: 10.1002/jcc.21235. [DOI] [PubMed] [Google Scholar]
- 89.Voorhout W.F., Veenendaal T., Geuze H.J. Intracellular processing of pulmonary surfactant protein B in an endosomal/lysosomal compartment. Am. J. Physiol. 1992;263:L479–L486. doi: 10.1152/ajplung.1992.263.4.L479. [DOI] [PubMed] [Google Scholar]
- 90.Stahlman M.T., Gray M.P., Weaver T.E. Lamellar body formation in normal and surfactant protein B-deficient fetal mice. Lab. Invest. 2000;80:395–403. doi: 10.1038/labinvest.3780044. [DOI] [PubMed] [Google Scholar]
- 91.Suzuki Y., Fujita Y., Kogishi K. Reconstitution of tubular myelin from synthetic lipids and proteins associated with pig pulmonary surfactant. Am. Rev. Respir. Dis. 1989;140:75–81. doi: 10.1164/ajrccm/140.1.75. [DOI] [PubMed] [Google Scholar]
- 92.deMello D.E., Heyman S., Colten H.R. Ultrastructure of lung in surfactant protein B deficiency. Am. J. Respir. Cell Mol. Biol. 1994;11:230–239. doi: 10.1165/ajrcmb.11.2.8049084. [DOI] [PubMed] [Google Scholar]
- 93.Rugonyi S., Biswas S.C., Hall S.B. The biophysical function of pulmonary surfactant. Respir. Physiol. Neurobiol. 2008;163:244–255. doi: 10.1016/j.resp.2008.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chavarha M., Khoojinian H., Hall S.B. Hydrophobic surfactant proteins induce a phosphatidylethanolamine to form cubic phases. Biophys. J. 2010;98:1549–1557. doi: 10.1016/j.bpj.2009.12.4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


