Abstract
AIM: To gain new insights into tumor metabolism and to identify possible biomarkers with potential diagnostic values to predict tumor metastasis.
METHODS: Human gastric cancer SGC-7901 cells were implanted into 24 severe combined immune deficiency (SCID) mice, which were randomly divided into metastasis group (n = 8), non-metastasis group (n = 8), and normal group (n = 8). Urinary metabolomic information was obtained by gas chromatography/mass spectrometry (GC/MS).
RESULTS: There were significant metabolic differences among the three groups (t test, P < 0.05). Ten selected metabolites were different between normal and cancer groups (non-metastasis and metastasis groups), and seven metabolites were also different between non-metastasis and metastasis groups. Two diagnostic models for gastric cancer and metastasis were constructed respectively by the principal component analysis (PCA). These PCA models were confirmed by corresponding receiver operating characteristic analysis (area under the curve = 1.00).
CONCLUSION: The urinary metabolomic profile is different, and the selected metabolites might be instructive to clinical diagnosis or screening metastasis for gastric cancer.
Keywords: Metabolomic profile, Gastric cancer, Metastasis, Biomarker, Gas chromatography/mass spectrometry
INTRODUCTION
Gastric cancer is the second leading cause of cancer death worldwide, and in many Asian countries, such as China[1,2]. Until now, there has been no effective treatment for gastric cancer. Even among patients undergoing gastrectomy, because of locoregional relapse and distant metastases, the 5-year survival rates remain disappointing[3]. Early dissemination of the disease through the lymphatic system, blood and peritoneum has limited the therapeutic effects of optimal surgery, except in patients with relatively early-stage tumors[4]. Therefore, it is significant to establish an accurate early diagnosis of gastric cancer. Currently, the diagnosis or screening of gastric cancer or tumor recurrence mainly depends on endoscopy and pathological examinations. The ratio for identifying early gastric cancer with endoscopy is higher than that with X-ray[5], and the diagnosis of gastric cancer using endoscopy is more accurate[6]. Nevertheless, the results of endoscopy are easily affected by artificial factors (e.g. the experience of the endoscopist). Over the past years, epidemiological data have shown that Helicobacter pylori (H. pylori) infection is strongly associated with the development of gastric cancer[1], and H. pylori eradication may be considered as a strategy to prevent gastric cancer[7]. In addition, investigation of gastric cancer tissues and some biomarkers have been used for screening gastric cancer[8-13]. However, compared with tissues and serum, the markers acquired from urine are noninvasive and convenient, especially in the patients with recurrent gastric cancer. The urinary metabolic profiling could be used to get urinary metabolites as gastric cancer or tumor recurrence biomarkers.
Metabolomics is a post-genomic research field for analysis of low molecular weight compounds in biological systems[14], and offers an analysis of metabolite level changes in biological samples[15]. In recent years, studies of metabolomics used in various diseases have been conducted, such as stomach cancer[16], lung cancer[17], renal cancer[18,19], brain tumors[20], and colorectal cancer[21-24]. Nuclear magnetic resonance spectroscopy (NMR) and mass spectrometry (MS) are the most commonly employed techniques for measuring the metabolome[14]. MS-based techniques, including gas chromatography/mass spectrometry (GC/MS), GC-MS/MS, liquid chromatography/mass spectrometry (LC-MS) and LC-MS/MS, are among the most efficient and versatile for quantitative analysis of endogenous and exogenous substances in biological samples[25]. Because of its peak resolution, high sensitivity and reproducibility, GC/MS has been well established and widely utilized in metabolomics[26-28].
In this study, we have established a human gastric cancer non-metastasis model and a metastasis model using severe combined immune deficiency (SCID) mice, and deployed GC/MS following chemical derivatization to profile the mouse model urinary specimens and their matched urine. The metabolic differences among the three groups were characterized by principal components analysis (PCA). On the basis of its results, we expected that the potential metabolic biomarkers could be found in mice for early diagnosis and screening the metastasis or the recurrence of gastric cancer.
MATERIALS AND METHODS
Chemicals and materials
Tetrahydrofuran (THF) and bis-(trimethylsilyl)-trifluoroacetamide (BSTFA) were obtained from Sigma Chemical Co. (St Louis, MO, USA). Vacuum dryer was purchased from Shanghai NOTED Technologies. All other reagents were obtained from Sinopharm Chemical Reagent Co. Ltd.
Animal models
Male SCID mice were acquired from Shanghai Experimental Animal Center of Chinese Academy of Sciences. Animals used were 6-wk old and weighed 20-25 g. Animal and experimental procedures were performed according to the relative ethical regulations for the care and use of laboratory animals of our university. Human gastric cancer SGC-7901 (Shanghai Cancer Institute), a poorly-differentiated adenocarcinoma line, was originally derived from a primary tumor and maintained by passage in the subcutis of nude mice. Tumors were cut out aseptically. Necrotic tissues were cut and the reserved healthy tumor tissues were scissor minced into pieces (about 3 mm × 4 mm in diameter) in Hank’s balanced salt solution. Each tumor piece was weighed and adjusted to be approximately 100 mg. All animals were randomly divided into metastasis group (n = 8), non-metastasis group (n = 8), and normal group (n = 8). Animal models were made using orthotopic implantation of histologically intact tissue of human gastric cancer[29]. Mice were anesthetized with 4.3% trichloraldehyde hydrate. An incision of the metastatic group and the normal group was made through the left upper abdominal pararectal line. Then peritoneal cavity was carefully exposed and a part of serosal membrane in the middle of the greater curvature of stomach was mechanically injured by scissors. A tumor piece of 100 mg was fixed on each injured site of the serosal surface of the metastatic group, while normal control mice received no tumor implantation. The stomach was then returned to the peritoneal cavity, and the abdominal wall and skin were closed. An incision of the non-metastatic group was made at the left oxter. A tumor piece of 100 mg was fixed under the skin. All animals were sent to the breeding room after becoming conscious.
Specimen collection and pathological examination
Six weeks after implantation, all mice were housed in metabolic cages and maintained in an air conditioned room (24 ± 2°C). They were only allowed free access to water during urine sample collection (8:00 pm that day to 8:00 am the next day). All animal urine was collected in frozen tubes at the sixth week after implantation, and immediately stored at -80°C until processing. The specimens were collected at the same time. Then all mice were killed, tumors growing on the stomach wall were resected and fixed in 4% formalin, and processed for routine paraffin embedding after careful macroscopic examination. In order to evaluate histologically for liver metastasis or lymph node metastasis or other organ metastasis under microscope, four-micron-thick sections were stained with hematoxylin and eosin, then observed by a blinded pathologist.
Sample pretreatment and derivatization
Each urinary specimen was transferred to a glass centrifuge tube, subsequently centrifuged at 18 000 × g for 3 min and 50 μL of the supernatant was collected from each sample into a 1-mL EP tube, respectively. The collected supernatant was evaporated to dryness at 60°C for 24 h, using a vacuum dryer. Then 100 μL THF was added to each of the dried urine extracts and vortex-mixed for 2 min, and 50 μL BSTFA was added to the mixture and vortex-mixed for 2 min. The mixture was incubated at 60°C and derivatized for 30 min. After returning to the ambient temperature, samples were prepared for GC/MS analysis.
GC/MS analysis
Each derivatized sample of 1 μL was injected splitless into an Agilent 6980 GC system equipped with an HP5MS capillary column (30 m × 0.25 mm i.d., 0.25 μm), electron impact ionization at 70 eV, and a quadrupole mass spectrometric detector (Agilent Technologies, Palo Alto, CA, USA). The column temperature was initially held at 100°C for 3 min, 10°C/min up to 220°C, then 10°C/min to 280°C, and remained there for 5 min. The injector temperature was 280°C. Carrier gas flow was helium at a constant flow rate of 1.0 mL/min. The interface temperature and the ion source temperature were set at 200°C. Masses were obtained from 100-600 m/z. GC total ion chromatograms (TICs) and fragmentation patterns were acquired using GC/MSD ChemStation Software (Agilent Technologies, Palo Alto, CA, USA). Compound identification was performed by comparing the mass spectrum with a standard mass spectrum in the national institute of standards and technology (NIST) mass spectra library. Peaks with similarity index more than 70% were assigned compound names, while those having less than 70% similarity were listed as unknown metabolites[30]. The chromatograms were subjected to noise reduction prior to peak area integration. Any known artificial peaks, such as peaks due to noise, column bleed and BSTFA derivatization procedure, were excluded from the data set. Integrated peak areas of multiple derivative peaks belonging to the same compound were summed and considered as a single compound. The resulting three dimensional matrix included sample information, peak intensities and peak retention time, and was applied to correlation analysis and pattern recognition.
Data processing and pattern recognition
The relative peak area of each compound would be calculated as the response after the peak areas of compounds were integrated. Each sample was represented by a GC/MS TIC. t test was employed for statistical analysis. Data were expressed as mean ± SD. The differentially expressed compounds with P < 0.05 were considered statistically significant. PCA was used to differentiate the samples and performed using the SPSS 16.0 for Windows.
RESULTS
General state of mice and pathological results
The mean weight of mice was 23.81 ± 0.16 g, 23.87 ± 0.19 g and 23.98 ± 0.19 g for normal group, non-metastasis group and metastasis group, respectively (P > 0.05). All animals from the three groups were alive at the sixth week. The normal group mice had no tumor and metastasis. The non-metastasis group and metastasis group developed localized tumors at the implanted site, which were poorly-differentiated adenocarcinomas under microscope (Figure 1A). The non-metastasis group tumor tissues (4.28 ± 0.20 g) were located at the left oxter, and have no metastasis in regional lymph nodes, liver and other organs. The metastasis group mice had cancer tissues (4.3 ± 0.3 g vs non-metastasis group, P > 0.05) in the stomach, while metastatic tumors were also found in liver (Figure 1B), regional lymph nodes, and other organs. Six mice developed metastatic tumors in regional lymph nodes, four in liver, and two in other organs.
Figure 1.
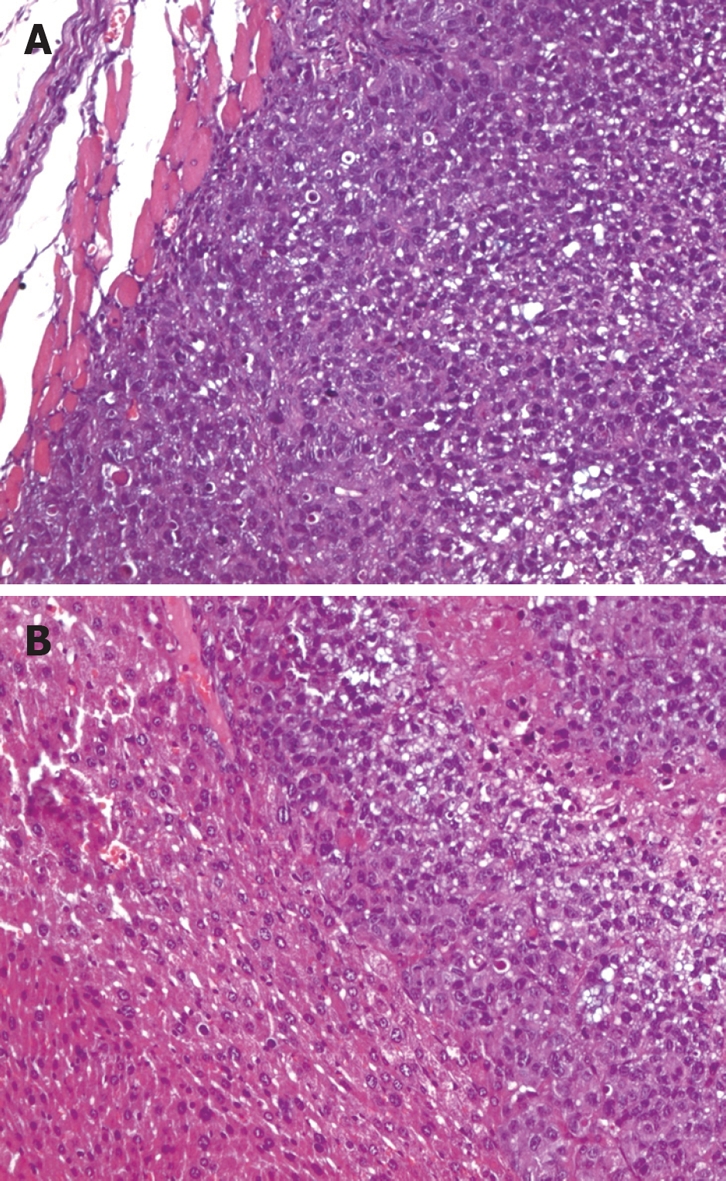
Gastric cancer pathological photographs. A: Gastric cancer cells in mice of the non-metastatic group (HE stain, × 200); B: Gastric cancer metastasis in the liver (HE stain, × 200).
Metabolomic profiling of samples
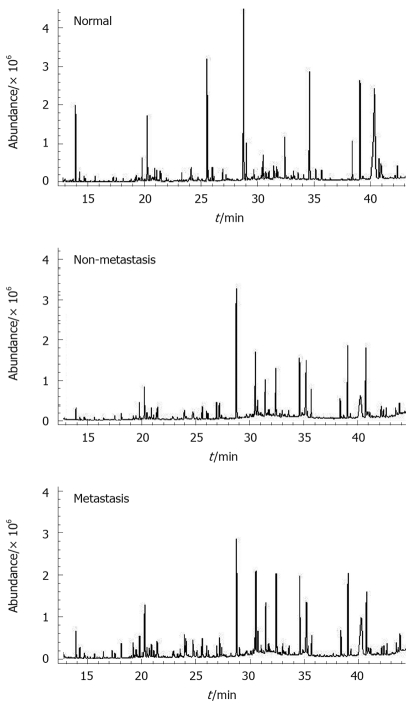
GC/MS TIC chromatograms of urine samples derived from the normal group, the non-metastatic group and the metastatic group are presented in Figure 2. In the GC/MS TICs of urinary samples from the three groups, some peaks were identified based on NIST mass spectra library, and several examples of peaks had statistical significance (Figure 2).
Figure 2.
Representative gas chromatography/mass spectrometry total ion chromatograms of the samples from the three groups (normal group, non-metastasis group and metastasis group) after chemical derivatization.
With GC/MS, around 120 signals were detected per sample using mass spectral deconvolution software for peak detection. However, many of them were not consistently found in other samples or were of too low abundance or too poor spectral quality to be obviously assigned to unique metabolites. Several choline, amino acids, and fatty acids could not be found, which may be associated with the efficiency of chemical derivatization. Table 1 shows that 46 signals could be auto-identified by the NIST library through comparing with a standard mass chromatogram. The remaining peaks which could not be identified were not listed. In addition, the retention time of metabolites and the match percentage to the NIST library are also listed in Table 1.
Table 1.
Urine metabolites of mice in the three groups (normal, non-metastasis and metastasis)
| Peak No. | Retention time | Metabolites | Match percent (%) |
| 1 | 7.196 | Lactic acid | 91 |
| 2 | 7.508 | Acetic acid | 90 |
| 3 | 8.105 | Alanine | 90 |
| 4 | 8.518 | Glycine | 91 |
| 5 | 8.936 | Pentanoic acid, 4-oxo- | 94 |
| 6 | 9.412 | Butanoic acid | 83 |
| 7 | 11.940 | Urea | 95 |
| 8 | 12.511 | Glycerol | 91 |
| 9 | 12.590 | Silanol | 97 |
| 10 | 13.933 | Butanedioic acid | 97 |
| 11 | 14.293 | Propanoic acid | 94 |
| 12 | 14.705 | Pyrimidine | 93 |
| 13 | 14.774 | Triacetin | 83 |
| 14 | 15.657 | 2-Piperidinecarboxylic acid | 90 |
| 15 | 16.070 | L-threonine | 87 |
| 16 | 16.460 | N-(1-oxobutyl)-Glycine | 90 |
| 17 | 17.270 | N-(2-methyl-1-oxopropyl)-Glycine | 91 |
| 18 | 17.530 | (R*,S*)-3,4-Dihydroxybutanoic acid | 94 |
| 19 | 19.301 | Malic acid | 90 |
| 20 | 19.492 | N-(3-methyl-1-oxobutyl)-Glycine | 98 |
| 21 | 19.566 | 2,3,4-oxy-Butanal | 90 |
| 22 | 19.814 | 1,2,3,4-oxy-Butane | 90 |
| 23 | 20.497 | L-proline | 96 |
| 24 | 20.909 | L-threonic acid | 90 |
| 25 | 21.475 | Creatinine | 96 |
| 26 | 25.108 | Hexanedioic acid | 90 |
| 27 | 25.632 | Arabitol | 91 |
| 28 | 25.843 | Nonadecane | 83 |
| 29 | 26.018 | Xylitol | 93 |
| 30 | 26.166 | Ribitol | 91 |
| 31 | 26.838 | 4-Pyrimidinecarboxylic acid | 96 |
| 32 | 26.938 | 1-Propene-1,2,3-tricarboxylic acid | 91 |
| 33 | 27.208 | Phosphoric acid | 90 |
| 34 | 28.768 | Citric acid | 91 |
| 35 | 29.032 | Myo-inositol | 83 |
| 36 | 30.328 | Mannonic acid | 95 |
| 37 | 30.540 | Hydrazone | 96 |
| 38 | 30.730 | N-Phenylacetyl glycine | 93 |
| 39 | 31.037 | Silane | 91 |
| 40 | 31.449 | L-Gluconic acid | 99 |
| 41 | 32.422 | D-Gluconic acid | 91 |
| 42 | 33.025 | Dehydrocholic Acid | 92 |
| 43 | 34.612 | Hexadecanoic acid | 99 |
| 44 | 35.691 | Uric acid | 98 |
| 45 | 38.388 | Retinoic acid, methyl ester | 95 |
| 46 | 39.065 | Octadecanoic acid | 99 |
Peaks in the total ion chromatograms are numbered according to their retention time. The identification of metabolite is based on national institute of standards and technology mass spectra database according to the match of masses (m/z) between the interested peak’s fragmentation pattern and that from the standard database.
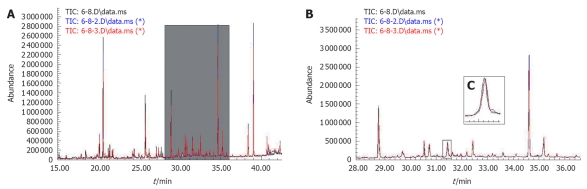
Three TIC profiles of consecutively injected samples of the same aliquot are presented in Figure 3, which showed stable retention time with no drift in all of the peaks. The stable TICs reflected the stability of GC/MS analysis and reliability of the metabolomic data.
Figure 3.
The overlay chromatograms of three parallel samples. A: The total ion chromatograms (TICs) of gas chromatography/mass spectrometry analysis; B: Enlarged part of TIC from 28 to 36 min; C: One peak enlarged.
Urine GC/MS data from the three groups were analyzed. Metabolites selected by t test are listed in Tables 2 and 3 after normalization of data. Lactic acid, butanoic acid, propanoic acid, glycerol, pyrimidine, butanedioic acid, malic acid, citric acid, hexadecanoic acid and uric acid were found at higher levels in the urine of cancer group (non-metastasis group and metastasis group) than in normal control group (Table 2). Furthermore, the decreased levels of alanine, butanoic acid, glycerol, L-proline and L-threonic acid were found in the metastasis group as compared with the non-metastasis group. However, the levels of butanedioic acid and myo-inositol were significantly higher in the metastasis group than in the non-metastasis group (Table 3).
Table 2.
Marker metabolites found in normal and cancer groups
| Metabolites | Retention time | P value1 | A (normal) | B (cancer2) | R3 |
| Lactic acid | 7.196 | 2.4 × 10-5 | 79.24 ± 6.1 | 187.04 ± 71.99 | 1.36 |
| Butanoic acid | 9.412 | 0.000 | 16.79 ± 0.52 | 27.33 ± 4.98 | 0.63 |
| Propanoic acid | 14.293 | 0.000 | 60.58 ± 9.79 | 147.77 ± 15.3 | 1.43 |
| Glycerol | 12.511 | 0.000 | 147 ± 8.98 | 269.13 ± 50.31 | 0.83 |
| Pyrimidine | 14.705 | 0.000 | 61.68 ± 8.05 | 163.11 ± 12.23 | 1.64 |
| Butanedioic acid | 13.933 | 0.1 × 10-5 | 161.51 ± 5.85 | 267.89 ± 54.64 | 0.66 |
| Malic acid | 19.301 | 0.000 | 10.7 ± 1.91 | 32.15 ± 1.16 | 2.00 |
| Citric acid | 28.768 | 1.4 × 10-4 | 1291.89 ± 364.74 | 2164.74 ± 529.58 | 0.68 |
| Hexadecanoic acid | 34.612 | 4.17 × 10-4 | 1347.84 ± 304.67 | 2066.57 ± 437.28 | 0.53 |
| Uric acid | 35.691 | 0.000 | 172.2 ± 17.03 | 214.52 ± 7.74 | 0.25 |
P values were calculated based on Student t test (significance at P < 0.05);
Cancer group included the non-metastasis group and the metastasis group;
R value was calculated from the arithmetic mean values of each group. R = (B-A)/A. R with a positive value indicates a relatively higher concentration in cancer group while a negative value means a relatively lower concentration as compared with the normal group.
Table 3.
Metabolic differences in the two groups
| Metabolites | Retention time | P value1 | A (non-metastasis) | B (metastasis) | R2 |
| Alanine | 8.105 | 0.000 | 173.75 ± 39.59 | 19.28 ± 10.63 | -0.89 |
| Butanoic acid | 9.412 | 0.000 | 32.09 ± 1.00 | 22.58 ± 0.72 | -0.30 |
| Glycerol | 12.511 | 0.003 | 303.23 ± 26.16 | 235.04 ± 45.64 | -0.22 |
| Butanedioic acid | 13.933 | 0.1 × 10-5 | 216.36 ± 2.63 | 319.43 ± 17.89 | 0.48 |
| L-proline | 20.497 | 0.000 | 184.99 ± 10.26 | 117.78 ± 7.05 | -0.36 |
| L-threonic acid | 20.909 | 2.28 × 10-4 | 284.94 ± 46.47 | 181.48 ± 37.25 | -0.36 |
| Myo-inositol | 29.032 | 0.000 | 33.08 ± 3.58 | 114.8 ± 2.20 | 2.47 |
P values were calculated based on Student t test (significance at P < 0.05);
R value was calculated from the arithmetic mean values of each group. R = (B-A)/A. R with a positive value indicates a relatively higher concentration in metastasis group while a negative value means a relatively lower concentration as compared with the non-metastasis group.
Pattern recognition and function analysis
A PCA model for gastric cancer was constructed using the marker metabolite intensities as variables (lactic acid, butanoic acid, propanoic acid, glycerol, pyrimidine, butanedioic acid, malic acid, citric acid, hexadecanoic acid and uric acid). The PCA scores plot showed that the normal group and cancer group (non-metastasis group and metastasis group) samples were scattered into different regions (Figure 4A). ROC analysis, which was performed using the values determined by the first two components of the PCA model, confirmed the robustness of the PCA model. These first two components could present the majority of all significantly different metabolites among the groups (the percentage is 82.7%). Area under the curve (AUC) value of this PCA model was 1.00 (Figure 4B), which demonstrated a good diagnostic value for gastric cancer. In addition, another PCA model for gastric cancer metastasis constructed by seven marker metabolites (alanine, butanoic acid, glycerol, L-threonic acid, L-proline, butanedioic acid and myo-inositol) could differentiate between the non-metastasis group and the metastasis group (Figure 5A). This PCA model was also validated by receiver operating characteristic (ROC) analysis (AUC = 1.00, Figure 5B).
Figure 4.
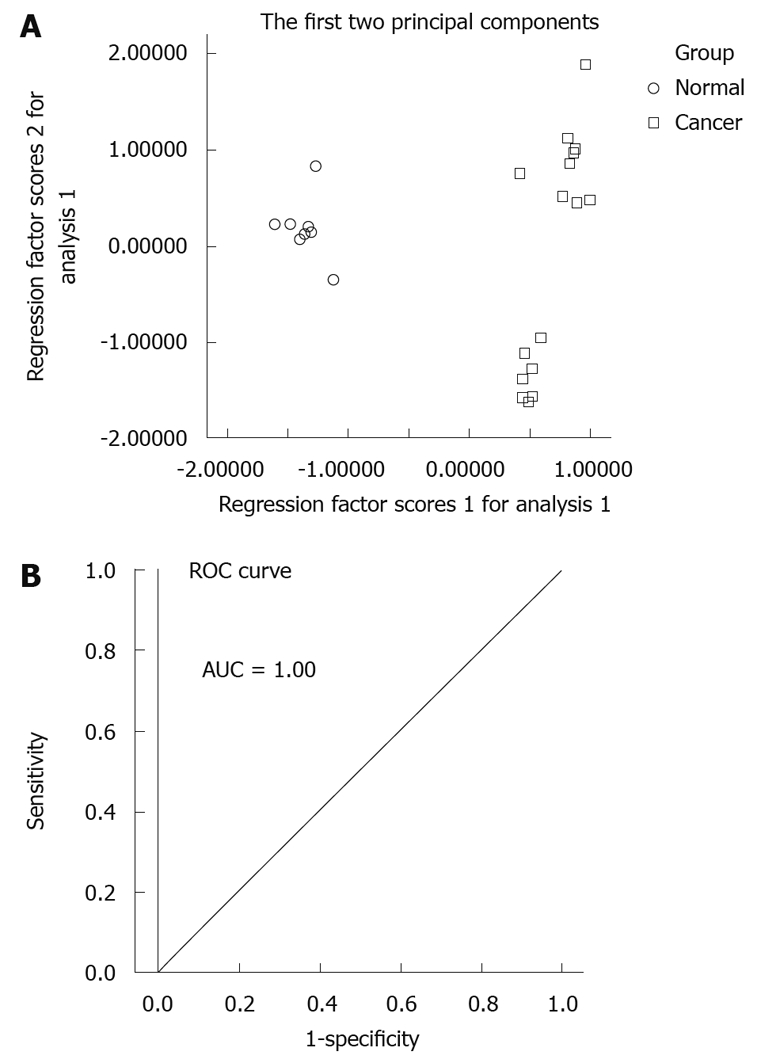
Principal component analysis model and receiver operating characteristic curve for gastric cancer. A: Principal component analysis (PCA) scores plot of gastric tumor specimens from control specimens based on 10 marker metabolites. The PCA scores plot showed different samples (normal group, cancer group including non-metastasis group and metastasis group) were scattered into different regions; B: Receiver operating characteristic (ROC) analysis was performed using the values determined by the first two components. Area under the curve (AUC) = 1.00.
Figure 5.
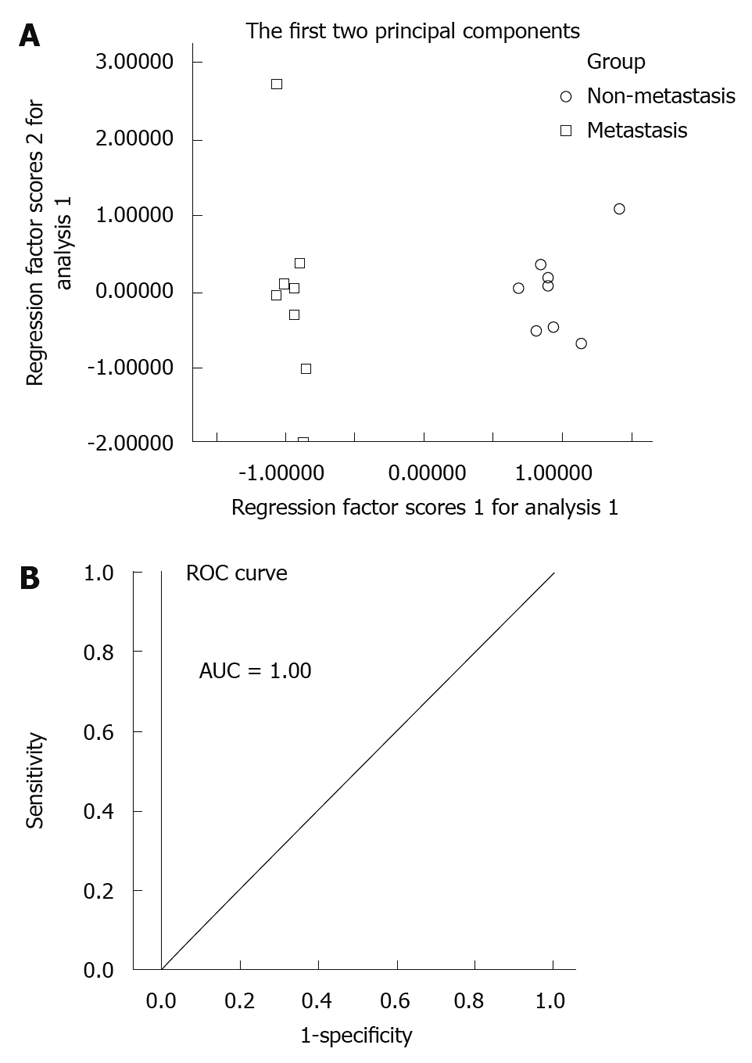
Principal component analysis model and receiver operating characteristic curve for gastric cancer metastasis. A: Principal component analysis (PCA) scores plot of non-metastasis group and metastasis group based on 7 marker metabolites. The PCA scores plot showed the samples from non-metastasis group and metastasis group were scattered into two different regions; B: Receiver operating characteristic (ROC) analysis was performed using the values determined by the first two components. Area under the curve (AUC) = 1.00.
DISCUSSION
In this study, we investigated urinary metabolite profiling using GC/MS. This was assessed non-invasively by measuring two voxels (tumor and healthy controls). We have discriminated the gastric cancer model mice from their healthy controls in a PCA analysis of GC-MS urinary metabolite spectra. Moreover, we could also discriminate the gastric cancer metastasis model mice from the non-metastasis model mice by GC-MS and PCA of urinary metabolites. Some marker metabolites were worth investigating in the future. Compared with the normal group, the level of lactic acid was higher in the cancer group urine. It could be explained that glucose is often converted into lactic acid in cancer cells, which is known as the “Warburg effect”, and cancer cells have a higher rate of aerobic glycolysis[31]. The levels of butanedioic acid, malic acid and citric acid, intermediates of tricarboxylic acid (TCA) cycle, were also found to be higher in the gastric cancer mice. The abnormalities of these metabolite expressions demonstrated a close correlation of TCA cycle with gastric cancer morbidity along with disordered aerobic respiration and mitochondrial functions. The disorder of aerobic respiration (mainly TCA cycle) and the impairment of mitochondrial enzymes have been reported in other malignancies including colorectal cancer, pheochromocytoma and paraganglioma[22,32,33]. Uric acid, the final metabolite of purines, at enhanced level in cancer mice urine, suggests the abnormalities of purine metabolism in gastric cancer[34]. In our study, the significantly higher levels of glycerol and hexadecanoic acid in cancer than in normal groups were interpreted as increased adipocyte lipolysis in cancer and enhanced expression and function of adipocyte hormone-sensitive lipase (HSL)[35].
Cancer metastasis could be considered as an essential prognostic factor[36]. Figure 5A shows the new constructed tumor metastatic model by seven marker metabolites for the non-metastasis group and the metastasis group. This PCA model was also validated by ROC analysis (AUC = 1.00, Figure 5B). Seven metabolites in this model are capable of predicting the gastric cancer metastasis. Compared with the non-metastasis group, levels of alanine and glycerol were found to be lower in the metastasis group. Alanine and glycerol could get into the glycolytic pathway through gluconeogenesis, which produced more energy for the tumor progression and metastasis. The decreased level of L-proline in the metastasis group may be interpreted as increased demand for structural proteins synthesis. These proteins, including receptors, membrane channels and enzymes, play an important role in tumor progression and metastasis[37-39]. Moreover, the higher level of myo-inositol in metastasis group urine, was consistent with the reduction of myo-inositol in lung cancer tissues[40]. The amount of myo-inositol may be a potential indicator for gastric cancer metastasis, as it has been reported that the Gly:Myo-inositol ratio may be a useful index for brain tumor classification[41].
What the difference of metabolite changes of butanoic acid and pyrimidine between the normal and the cancer groups, and the decreased levels of butanoic acid and L-threonic acid in the metastasis group indicates remains unclear.
In conclusion, GC/MS revealed detailed information on the metabolic profile of normal and cancer urine and was found to be suitable, in tandem with the PCA model, for the identification of metabolic variations characteristic of the gastric cancer. Furthermore, seven metabolites have been selected, which constructed a diagnostic model for distinguishing the non-metastatic and the metastatic gastric cancer. To our knowledge, this is the first report on urinary metabolomic investigation of gastric cancer metastasis by GC/MS. On the basis of this research, we believe that urinary metabolomic information obtained by GC/MS might play a significant role in the early diagnosis and screening metastasis or recurrence of gastric cancer.
COMMENTS
Background
Gastric cancer is the second leading cause of cancer death worldwide, and in many Asian countries. Tumor metastasis is one of the leading causes of cancer death. Metabolic alterations play a role in the biology of cancer. The urinary metabolites as gastric cancer or tumor recurrence biomarkers can be obtained by investigating the urinary metabolic profiling.
Research frontiers
Metabolomics is a post-genomic research field for analysis of low molecular weight compounds in biological systems, and its approaches offer an analysis of metabolite level changes in biological samples. Recently, metabolomic method has shown great potentials in identifying the new diagnostic markers and therapeutic targets for cancers. However, metabolomic studies on cancer metastasis remain scarce.
Innovations and breakthroughs
Recently, metabolomic studies on gastric cancer and colon cancer tissues have been conducted. Compared with tissues and serum, markers acquired from urine are noninvasive and convenient, especially in the patients with recurrent gastric cancer. This is the first report on urinary metabolomic investigation in gastric cancer using gas chromatography/mass spectrometry (GC/MS).
Applications
Potential metabolic biomarkers in urine could be used for early diagnosis and screening the metastasis or the recurrence of gastric cancer.
Terminology
Metabolomics is a post-genomic research field for analysis of low molecular weight compounds in biological systems, and its approaches offer an analysis of metabolite level changes in biological samples. Because of its peak resolution, high sensitivity and reproducibility, GC/MS has been widely utilized in metabolomics.
Peer review
This manuscript evaluates tumor metabolism with a goal to identify possible biomarkers with potential diagnostic value and the potential for prediction of tumor metastasis. The authors concluded that the urinary metabolomic profiling of each group is different, and the selected metabolites might be instructive to clinical diagnosis or screening metastasis for gastric cancer. This is a relevant randomized control trial using an animal model to evaluate a non-invasive method for surveillance of gastric cancer.
Footnotes
Supported by The Key Program of Science and Technology Commission of Shanghai Municipality, Project No. 09JC1411600; and Natural Science Foundation of Shanghai, No. 08ZR1411300
Peer reviewers: Dr. Cuneyt Kayaalp, MD, Professor, Department of General Surgery, Staff Surgeon of Gastrointestinal Surgery, Turgut Ozal Medical Center, Inonu University, Malatya 44315, Turkey; Kevin Michael Reavis, MD, Assistant Clinic Professor, Department of Surgery, Division of Gastrointestinal Surgery, University of California, Irvine Medical Center, 333 City Boulevard West, Suite 850, Orange, CA 92868, United States
S- Editor Sun H L- Editor Ma JY E- Editor Ma WH
References
- 1.Leung WK, Wu MS, Kakugawa Y, Kim JJ, Yeoh KG, Goh KL, Wu KC, Wu DC, Sollano J, Kachintorn U, et al. Screening for gastric cancer in Asia: current evidence and practice. Lancet Oncol. 2008;9:279–287. doi: 10.1016/S1470-2045(08)70072-X. [DOI] [PubMed] [Google Scholar]
- 2.Brenner H, Rothenbacher D, Arndt V. Epidemiology of stomach cancer. Methods Mol Biol. 2009;472:467–477. doi: 10.1007/978-1-60327-492-0_23. [DOI] [PubMed] [Google Scholar]
- 3.De Vita F, Vecchione L, Galizia G, Di Martino N, Fabozzi T, Catalano G, Ciardiello F, Orditura M. Perspectives in adjuvant therapy of gastric cancer. Oncology. 2009;77 Suppl 1:38–42. doi: 10.1159/000258494. [DOI] [PubMed] [Google Scholar]
- 4.Cunningham D, Chua YJ. East meets west in the treatment of gastric cancer. N Engl J Med. 2007;357:1863–1865. doi: 10.1056/NEJMe078182. [DOI] [PubMed] [Google Scholar]
- 5.Tashiro A, Sano M, Kinameri K, Fujita K, Takeuchi Y. Comparing mass screening techniques for gastric cancer in Japan. World J Gastroenterol. 2006;12:4873–4874. doi: 10.3748/wjg.v12.i30.4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Catalano V, Labianca R, Beretta GD, Gatta G, de Braud F, Van Cutsem E. Gastric cancer. Crit Rev Oncol Hematol. 2009;71:127–164. doi: 10.1016/j.critrevonc.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 7.Shin CM, Kim N, Yang HJ, Cho SI, Lee HS, Kim JS, Jung HC, Song IS. Stomach cancer risk in gastric cancer relatives: interaction between Helicobacter pylori infection and family history of gastric cancer for the risk of stomach cancer. J Clin Gastroenterol. 2010;44:e34–e39. doi: 10.1097/MCG.0b013e3181a159c4. [DOI] [PubMed] [Google Scholar]
- 8.Hao Y, Yu Y, Wang L, Yan M, Ji J, Qu Y, Zhang J, Liu B, Zhu Z. IPO-38 is identified as a novel serum biomarker of gastric cancer based on clinical proteomics technology. J Proteome Res. 2008;7:3668–3677. doi: 10.1021/pr700638k. [DOI] [PubMed] [Google Scholar]
- 9.Chan DC, Chen CJ, Chu HC, Chang WK, Yu JC, Chen YJ, Wen LL, Huang SC, Ku CH, Liu YC, et al. Evaluation of serum amyloid A as a biomarker for gastric cancer. Ann Surg Oncol. 2007;14:84–93. doi: 10.1245/s10434-006-9091-z. [DOI] [PubMed] [Google Scholar]
- 10.Kang X, Zhang L, Sun J, Ni Z, Ma Y, Chen X, Sheng X, Chen T. Prohibitin: a potential biomarker for tissue-based detection of gastric cancer. J Gastroenterol. 2008;43:618–625. doi: 10.1007/s00535-008-2208-3. [DOI] [PubMed] [Google Scholar]
- 11.Al-Moundhri MS, Al-Shukaili A, Al-Nabhani M, Al-Bahrani B, Burney IA, Rizivi A, Ganguly SS. Measurement of circulating levels of VEGF-A, -C, and -D and their receptors, VEGFR-1 and -2 in gastric adenocarcinoma. World J Gastroenterol. 2008;14:3879–3883. doi: 10.3748/wjg.14.3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abbaszadegan MR, Moaven O, Sima HR, Ghafarzadegan K, A'rabi A, Forghani MN, Raziee HR, Mashhadinejad A, Jafarzadeh M, Esmaili-Shandiz E, et al. p16 promoter hypermethylation: a useful serum marker for early detection of gastric cancer. World J Gastroenterol. 2008;14:2055–2060. doi: 10.3748/wjg.14.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu H, Xue R, Tang Z, Deng C, Liu T, Zeng H, Sun Y, Shen X. Metabolomic investigation of gastric cancer tissue using gas chromatography/mass spectrometry. Anal Bioanal Chem. 2010;396:1385–1395. doi: 10.1007/s00216-009-3317-4. [DOI] [PubMed] [Google Scholar]
- 14.Madsen R, Lundstedt T, Trygg J. Chemometrics in metabolomics--a review in human disease diagnosis. Anal Chim Acta. 2010;659:23–33. doi: 10.1016/j.aca.2009.11.042. [DOI] [PubMed] [Google Scholar]
- 15.Vinayavekhin N, Homan EA, Saghatelian A. Exploring disease through metabolomics. ACS Chem Biol. 2010;5:91–103. doi: 10.1021/cb900271r. [DOI] [PubMed] [Google Scholar]
- 16.Hirayama A, Kami K, Sugimoto M, Sugawara M, Toki N, Onozuka H, Kinoshita T, Saito N, Ochiai A, Tomita M, et al. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res. 2009;69:4918–4925. doi: 10.1158/0008-5472.CAN-08-4806. [DOI] [PubMed] [Google Scholar]
- 17.Tan C, Chen H, Xia C. Early prediction of lung cancer based on the combination of trace element analysis in urine and an Adaboost algorithm. J Pharm Biomed Anal. 2009;49:746–752. doi: 10.1016/j.jpba.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 18.Kim K, Aronov P, Zakharkin SO, Anderson D, Perroud B, Thompson IM, Weiss RH. Urine metabolomics analysis for kidney cancer detection and biomarker discovery. Mol Cell Proteomics. 2009;8:558–570. doi: 10.1074/mcp.M800165-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kind T, Tolstikov V, Fiehn O, Weiss RH. A comprehensive urinary metabolomic approach for identifying kidney cancerr. Anal Biochem. 2007;363:185–195. doi: 10.1016/j.ab.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 20.Monleón D, Morales JM, Gonzalez-Darder J, Talamantes F, Cortés O, Gil-Benso R, López-Ginés C, Cerdá-Nicolás M, Celda B. Benign and atypical meningioma metabolic signatures by high-resolution magic-angle spinning molecular profiling. J Proteome Res. 2008;7:2882–2888. doi: 10.1021/pr800110a. [DOI] [PubMed] [Google Scholar]
- 21.Monleón D, Morales JM, Barrasa A, López JA, Vázquez C, Celda B. Metabolite profiling of fecal water extracts from human colorectal cancer. NMR Biomed. 2009;22:342–348. doi: 10.1002/nbm.1345. [DOI] [PubMed] [Google Scholar]
- 22.Mal M, Koh PK, Cheah PY, Chan EC. Development and validation of a gas chromatography/mass spectrometry method for the metabolic profiling of human colon tissue. Rapid Commun Mass Spectrom. 2009;23:487–494. doi: 10.1002/rcm.3898. [DOI] [PubMed] [Google Scholar]
- 23.Qiu Y, Cai G, Su M, Chen T, Liu Y, Xu Y, Ni Y, Zhao A, Cai S, Xu LX, et al. Urinary metabonomic study on colorectal cancer. J Proteome Res. 2010;9:1627–1634. doi: 10.1021/pr901081y. [DOI] [PubMed] [Google Scholar]
- 24.Qiu Y, Cai G, Su M, Chen T, Zheng X, Xu Y, Ni Y, Zhao A, Xu LX, Cai S, et al. Serum metabolite profiling of human colorectal cancer using GC-TOFMS and UPLC-QTOFMS. J Proteome Res. 2009;8:4844–4850. doi: 10.1021/pr9004162. [DOI] [PubMed] [Google Scholar]
- 25.Tsikas D. Quantitative analysis of biomarkers, drugs and toxins in biological samples by immunoaffinity chromatography coupled to mass spectrometry or tandem mass spectrometry: A focused review of recent applications. J Chromatogr B Analyt Technol Biomed Life Sci. 2010;878:133–148. doi: 10.1016/j.jchromb.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 26.Wu H, Xue R, Lu C, Deng C, Liu T, Zeng H, Wang Q, Shen X. Metabolomic study for diagnostic model of oesophageal cancer using gas chromatography/mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:3111–3117. doi: 10.1016/j.jchromb.2009.07.039. [DOI] [PubMed] [Google Scholar]
- 27.Jonsson P, Johansson AI, Gullberg J, Trygg J, A J, Grung B, Marklund S, Sjöström M, Antti H, Moritz T. High-throughput data analysis for detecting and identifying differences between samples in GC/MS-based metabolomic analyses. Anal Chem. 2005;77:5635–5642. doi: 10.1021/ac050601e. [DOI] [PubMed] [Google Scholar]
- 28.Jonsson P, Gullberg J, Nordström A, Kusano M, Kowalczyk M, Sjöström M, Moritz T. A strategy for identifying differences in large series of metabolomic samples analyzed by GC/MS. Anal Chem. 2004;76:1738–1745. doi: 10.1021/ac0352427. [DOI] [PubMed] [Google Scholar]
- 29.Chen JL, Hong J, Lu JL, Chen MX, Chen WX, Zhu JS, Chen NW, Chen GQ, Geng JG. Effect of non-anticoagulant N-desulfated heparin on expression of vascular endothelial growth factor, angiogenesis and metastasis of orthotopic implantation of human gastric carcinoma. World J Gastroenterol. 2007;13:457–461. doi: 10.3748/wjg.v13.i3.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chan EC, Koh PK, Mal M, Cheah PY, Eu KW, Backshall A, Cavill R, Nicholson JK, Keun HC. Metabolic profiling of human colorectal cancer using high-resolution magic angle spinning nuclear magnetic resonance (HR-MAS NMR) spectroscopy and gas chromatography mass spectrometry (GC/MS) J Proteome Res. 2009;8:352–361. doi: 10.1021/pr8006232. [DOI] [PubMed] [Google Scholar]
- 31.Liu X, Wang X, Zhang J, Lam EK, Shin VY, Cheng AS, Yu J, Chan FK, Sung JJ, Jin HC. Warburg effect revisited: an epigenetic link between glycolysis and gastric carcinogenesis. Oncogene. 2010;29:442–450. doi: 10.1038/onc.2009.332. [DOI] [PubMed] [Google Scholar]
- 32.Goncalves S, Paupe V, Dassa EP, Brière JJ, Favier J, Gimenez-Roqueplo AP, Bénit P, Rustin P. Rapid determination of tricarboxylic acid cycle enzyme activities in biological samples. BMC Biochem. 2010;11:5. doi: 10.1186/1471-2091-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, Pan Y, Simon MC, Thompson CB, Gottlieb E. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell. 2005;7:77–85. doi: 10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 34.Gocmen E, Tez M, Ozturk S, Koc M, Demirci S. Activities of adenosine deaminase and 5'-nucleotidase in cancereous and non-cancereous human gastric tissues. Bratisl Lek Listy. 2009;110:416–418. [PubMed] [Google Scholar]
- 35.Agustsson T, Rydén M, Hoffstedt J, van Harmelen V, Dicker A, Laurencikiene J, Isaksson B, Permert J, Arner P. Mechanism of increased lipolysis in cancer cachexia. Cancer Res. 2007;67:5531–5537. doi: 10.1158/0008-5472.CAN-06-4585. [DOI] [PubMed] [Google Scholar]
- 36.Bilici A, Ustaalioglu BB, Gumus M, Seker M, Yilmaz B, Kefeli U, Yildirim E, Sonmez B, Salepci T, Kement M, et al. Is metastatic lymph node ratio superior to the number of metastatic lymph nodes to assess outcome and survival of gastric cancer? Onkologie. 2010;33:101–105. doi: 10.1159/000277927. [DOI] [PubMed] [Google Scholar]
- 37.Matsuura I, Chiang KN, Lai CY, He D, Wang G, Ramkumar R, Uchida T, Ryo A, Lu K, Liu F. Pin1 promotes transforming growth factor-beta-induced migration and invasion. J Biol Chem. 2010;285:1754–1764. doi: 10.1074/jbc.M109.063826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song IS, Oh NS, Kim HT, Ha GH, Jeong SY, Kim JM, Kim DI, Yoo HS, Kim CH, Kim NS. Human ZNF312b promotes the progression of gastric cancer by transcriptional activation of the K-ras gene. Cancer Res. 2009;69:3131–3139. doi: 10.1158/0008-5472.CAN-08-2240. [DOI] [PubMed] [Google Scholar]
- 39.Surazynski A, Liu Y, Miltyk W, Phang JM. Nitric oxide regulates prolidase activity by serine/threonine phosphorylation. J Cell Biochem. 2005;96:1086–1094. doi: 10.1002/jcb.20631. [DOI] [PubMed] [Google Scholar]
- 40.Rocha CM, Barros AS, Gil AM, Goodfellow BJ, Humpfer E, Spraul M, Carreira IM, Melo JB, Bernardo J, Gomes A, et al. Metabolic profiling of human lung cancer tissue by 1H high resolution magic angle spinning (HRMAS) NMR spectroscopy. J Proteome Res. 2010;9:319–332. doi: 10.1021/pr9006574. [DOI] [PubMed] [Google Scholar]
- 41.Righi V, Andronesi OC, Mintzopoulos D, Black PM, Tzika AA. High-resolution magic angle spinning magnetic resonance spectroscopy detects glycine as a biomarker in brain tumors. Int J Oncol. 2010;36:301–306. doi: 10.3892/ijo_00000500. [DOI] [PMC free article] [PubMed] [Google Scholar]