Abstract
AIM: To investigate the effects of four probiotic bacteria and their combination on human mast cell gene expression using microarray analysis.
METHODS: Human peripheral-blood-derived mast cells were stimulated with Lactobacillus rhamnosus (L. rhamnosus) GG (LGG®), L. rhamnosus Lc705 (Lc705), Propionibacterium freudenreichii ssp. shermanii JS (PJS) and Bifidobacterium animalis ssp. lactis Bb12 (Bb12) and their combination for 3 or 24 h, and were subjected to global microarray analysis using an Affymetrix GeneChip® Human Genome U133 Plus 2.0 Array. The gene expression differences between unstimulated and bacteria-stimulated samples were further analyzed with GOrilla Gene Enrichment Analysis and Visualization Tool and MeV Multiexperiment Viewer-tool.
RESULTS: LGG and Lc705 were observed to suppress genes that encoded allergy-related high-affinity IgE receptor subunits α and γ (FCER1A and FCER1G, respectively) and histamine H4 receptor. LGG, Lc705 and the combination of four probiotics had the strongest effect on the expression of genes involved in mast cell immune system regulation, and on several genes that encoded proteins with a pro-inflammatory impact, such as interleukin (IL)-8 and tumour necrosis factor alpha. Also genes that encoded proteins with anti-inflammatory functions, such as IL-10, were upregulated.
CONCLUSION: Certain probiotic bacteria might diminish mast cell allergy-related activation by downregulation of the expression of high-affinity IgE and histamine receptor genes, and by inducing a pro-inflammatory response.
Keywords: Probiotic bacteria, Mast cells, Microarray, Allergy, IgE receptor
INTRODUCTION
Mast cells are multifunctional regulator cells that are located at strategic host-environment interfaces including skin, vascular barriers and gastrointestinal tract, where they encounter antigens and pathogens, as well as commensal microbes. In the healthy intestinal mucosa, mast cells constitute 2%-3% of the cells of the lamina propria[1]. Mast cells are very heterogeneous cells that are traditionally classified by the content of their specific proteases tryptase and chymase. A mast cell subtype that contains only tryptase (MCT) is predominant in the lung and intestine, whereas mast cells that contain tryptase and chymase (MCTC) are prevalent in the skin and conjunctiva[2]. Mast cell subtypes can even vary between different parts of the same organ. Thus, MCT cells are enriched in the mucosal layer of the intestine but MCTC cells outnumber them in the intestinal submucosa. However, this classification is thought to be interchangeable and can be shaped according to the microenvironment of the cells[3].
Mast cells participate in a variety of physiological functions, such as epithelial secretion and permeability, blood flow, peristalsis, neuroimmune interactions, and wound healing. One significant task for mast cells is host defence against pathogenic microbes. By secreting several mediators including histamine, proteases, lipid mediators and pro- and anti-inflammatory cytokines, mast cells regulate the immune system and interact with other immune cells[4]. The multifunctionality of mast cells can explain why they are also involved in the pathogenesis of many inflammatory diseases, such as allergy. The number of mast cells and the amount of mast-cell-derived mediators, such as histamine, which is the key mediator in allergy, are increased at sites of allergic inflammation. The released mediators induce mucus and electrolyte secretion, smooth muscle contraction, nerve-cell activation and other symptoms common in allergic reactions[5]. The regulation of mast-cell mediators is complex. The best characterized mechanism of mast cell activation is high-affinity IgE receptor (FcεR1)-mediated activation[6]. IgE-receptor aggregation induces multiple signaling pathways that control the secretion of allergy-related mediators, such as histamine and leukotrienes, and the induction of T helper cell 2 (Th2) type cytokine and tumor necrosis factor (TNF) gene transcription[7]. The inflammatory effects of the released histamine are mediated by histamine receptors H1-H4[8]. However, some of the mast cell mediators, including interleukin (IL)-10 and histamine[9] can have anti-inflammatory effects and decrease inflammation[10].
Probiotics are defined as live microbes that have beneficial effects on the host’s health when administered in adequate amounts[11]. In clinical intervention studies, certain probiotics have been documented to be effective in the prevention and treatment of various clinical conditions. The most promising results of the health effects of probiotics have been discovered in studies of diarrhea[12], allergy[13], irritable bowel syndrome (IBS)[14,15], and respiratory infections[16,17]. The effects of probiotics are suggested to be strain-specific, although one strain can have multiple influences[18]. In the treatment of a complex and heterogeneous condition such as IBS, the use of combinations of different strains of probiotics can have advantages over using a single strain[14]. The most investigated and used probiotic genera are Lactobacillus and Bifidobacterium. Lactobacillus strains have been effective in beneficially modulating commensal microbes and inhibiting pathogen adhesion to gut mucosa. Lactobacillus and Bifidobacterium have been shown to produce antimicrobial agents and to alleviate symptoms of allergy[19]. Dairy Propionibacterium, which has been observed to exclude pathogenic microbes from gut mucosa[20], has also been used as a probiotic.
Evidence from two clinical trials performed with the combination of four probiotic bacteria, i.e. Lactobacillus rhamnosus (L. rhamnosus) GG (LGG), L. rhamnosus Lc705 (Lc705), Propionibacterium freudenreichii ssp. shermanii JS (PJS) and Bifidobacterium animalis ssp. lactis Bb12 (Bb12), suggests that the consumption of such combination alleviates the symptoms in IBS patients[14,21]. As mast cells are believed to be important in regulating intestinal immunity and perhaps also intestinal sensory functions, we chose to study the effects of the above probiotic combination and each bacterium alone on the global gene expression of primary peripheral-blood-derived human mast cells.
MATERIALS AND METHODS
Cell culture
Freshly collected buffy coats from healthy adult blood donors were provided by the Finnish Red Cross Blood Transfusion Service (Helsinki, Finland). The health of the subjects for blood donation is strictly controlled. The donors must be 18-65 years of age, free of infections including HIV, hepatitis B and C, and free of allergic symptoms and most chronic illnesses including autoimmune diseases. Mononuclear cells were purified from heparinized blood by Ficoll-Paque™ Plus (GE Healthcare, Uppsala, Sweden) density gradient centrifugation. Mast cell precursor cells were isolated by positive immunomagnetic selection using indirect CD34 MicroBead Kit and MACS® separation columns (Miltenyi Biotec, Bergish Gladbach, Germany) according to the manufacturer’s instructions. After selection, the CD34+ cells were cultured for 9-11 wk in serum-free Stem Span™ cell culture medium (Stem Cell Technologies, Vancouver, Canada) supplemented with penicillin and streptomycin (GIBCO BRL, Grand Island, NY, USA), human recombinant stem cell factor (SCF; Peprotech, Rocky Hill, NJ, USA), IL-3 (Peprotech), IL-9 (Peprotech), IL-6 (Peprotech) and human low-density lipoprotein, as previously described[22]. Cultured mast cells were phenotypically and functionally similar to mature MCTC cells as measured by c-kit and IgE receptor (FcεR1) expression, and the presence of chymase, tryptase, heparin and histamine in their granules, as described previously in detail[23].
Bacterial strains
L. rhamnosus GG (ATCC 53103), L. rhamnosus Lc705 (DSM 7061), P. freudenreichii ssp. shermanii JS (DSM 7067) and B. animalis ssp. lactis Bb12 (DSM 15954) were provided by Valio Research Centre (Helsinki, Finland). LGG and Lc705 were grown as previously described[24]. PJS was grown under optimized aerobic conditions at 30°C in whey broth (Valio) twice for 2 d at a concentration of 2%. Bb12 was grown under anaerobic conditions at 37°C in de Man, Rogosa and Sharpe (MRS) broth enriched with 5 g/L L-cysteine hydrochloride monohydrate (Merck, Darmstadt, Germany) three times for 17-18 h at a concentration of 2%[25]. LGG, Lc705, PJS and Bb12 were grown to logarithmic growth phase and the number of bacteria was determined by counting in a Petroff-Hauser counting chamber. Chlamydia pneumoniae isolate Kajaani 6 (Cpn) was used as a reference strain, and was obtained from the National Institute for Health and Welfare (Helsinki, Finland) and was propagated as described previously[22].
Mast-cell stimulation with bacteria
After differentiation, mast cells were collected and re-suspended in fresh Stem Span™ medium that contained antibiotics and SCF as described above. Single live bacterial strains were added to the cell culture in a bacterium-to-cell ratio of 5:1 based on preliminary experiments (data not shown). When the stimulation was performed with the combination of bacteria, each of the four strains was dosed in a bacterium-to-cell ratio of 1.25:1, thus the total bacterium-to-cell ratio of the combination was 5:1. The cells were incubated with bacteria for 3 or 24 h at 37°C in 5% CO2. After incubation, mast cells were separated from the medium by centrifugation, and the medium was aliquoted and stored at -20°C. The cells were washed free of bacteria, lysed and homogenized in RLT buffer (Qiagen, Valencia, CA, USA) and stored at -70°C before RNA isolation. All experiments were performed with mast cells obtained from three different blood donors. For analysis, the cells from different donors of each experiment were pooled.
RNA isolation and microarray
Total RNA was isolated from the cell lysates using RNeasy Mini Kit (Qiagen) according to the protocol provided by the manufacturer. Microarray experiments were performed at Biomedicum Genomics (Helsinki, Finland) using an Affymetrix GeneChip® Human Genome U133 Plus 2.0 Array (Affymetrix, Santa Clara, CA, USA). Integrity and purity of the RNA were verified with Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). Total RNA was prepared and hybridized using the two-cycle protocol of the GeneChip® Expression Analysis kit (Affymetrix) according to the manufacturer’s recommendations. Double-stranded cDNA was synthesized from total RNA. Next, biotin-labeled cRNA was transcribed from the cDNA, and the cRNA was fragmented and hybridized. The hybridization reactions were scanned using a GeneChip Scanner 3000 (Affymetrix).
Microarray analysis
The robust multiarray averaging algorithm[26] in the Bioconductor simpleaffy package[27,28] was used to calculate expression estimates from GeneChip signal intensity data. To provide better precision and accuracy and to overcome interpretation problems related to conflicting id gene references[27], an updated probe set definition was used[29], based on Ensemble gene information. In contrast to the default Affymetrix chip description file with 54 675 probe sets, the used custom chip description file (version 11.0.1) contained 17 492 unique Ensemble gene probe sets. The significance of differential expression was assessed using the empirical Bayes moderated paired t statistics (eBayes function) in the limma package, followed by intensity-based hierarchical Bayes analysis[30,31]. In the analysis, a moderated paired t test was computed by constructing cell line effects in the linear model. All P values were adjusted for multiple hypotheses testing using the bootstrapped q value approach in the qvalue package[32]. Genes with P values ≤ 0.05 were identified as significantly differentially expressed.
The GOrilla Gene Enrichment Analysis and Visualization Tool[33] was used to discover functional categories that were enriched at either end of a gene list sorted by the moderated t test score, which was calculated using limma. The input gene set was used as a background. Clustering and visualization of the gene expression differences were done using MeV Multiexperiment Viewer tool and hierarchical clustering with Euclidean as a distance, and average linkage clustering as the linkage method[34,35].
Quantitative reverse transcriptase-polymerase chain reaction
To validate the microarray data, TaqMan® real-time reverse transcriptase-polymerase chain reaction (RT-PCR) was performed as previously described[22] for selected genes. Total RNA from the same samples used for the microarray experiments was reverse transcribed to cDNA using random hexamers (Invitrogen, Paisley, UK) and Moloney murine leukemia virus reverse transcriptase (Invitrogen). TaqMan® Gene Expression assays (Applied Biosystems, Foster City, CA, USA) were chosen for detection of IL8 (Hs00174103_m1), CCL2 (Hs00234140_m1), IL10 (Hs00174086_m1), HRH4 (Hs00222094_m1), FCER1A (Hs00758600_m1) and FCER1G (Hs00610227_m1). Transcripts for TNF-α were detected by using sense primer 5'-GCTGCACTTTGGAGTGATCG-3', antisense primer 5'-GTTTGCTACAACATGGGCTACAG-3' and probe 5'-FAM-CCCAGGCAGTCAGATCATCTTCTCGA-BHQ1-3'. Samples were analyzed in triplicate. β-Actin was used as an endogenous normalization control. Relative quantification was determined by standard 2-ΔΔCT calculations[36].
RESULTS
Gene expression profiling
In order to explore the effects of different probiotic bacteria or their combination on mast cells, transcriptional changes of the bacteria-stimulated mast cells were studied using a whole genome microarray. A total of 42 Affymetrix GeneChip® Human Genome U133 Plus 2.0 Arrays were used to analyze gene expression profiles of unstimulated mast cells or those stimulated with live LGG, Lc705, PJS, Bb12, or the combination of these four bacteria. Cpn was included in the analysis as a reference bacterium, which represented a non-probiotic, pathogenic microbe with certain known transcriptional effects on mast cells[22]. Results are representative of three independent experiments, each performed with cells from three donors at two different time points (3 and 24 h). After 3 h bacterial stimulation, the differences in the levels of mast-cell gene expression were so low that no statistical significance was observed as compared to unstimulated samples (data not shown). At 24 h, however, a statistically significant change was observed in 698 genes. Numbers of differentially expressed genes are illustrated in Figure 1. Lc705, LGG and the combination of the four probiotic strains were the most effective stimulators. Bb12 affected the expression of only one gene, and PJS and Cpn failed to change mast-cell gene expression significantly. Lc705 affected mast-cell gene expression the most by changing the expression of 629 genes significantly. Of these genes, 288 were upregulated and 341 were downregulated. LGG altered the expression of 345 genes (160 upregulated and 185 downregulated), and the combination of the four probiotic strains changed the expression of 91 genes (57 upregulated and 34 downregulated). Raw data of the microarray analysis are available at http://ekhidna.biocenter.helsinki.fi/poxo/download_data.
Figure 1.
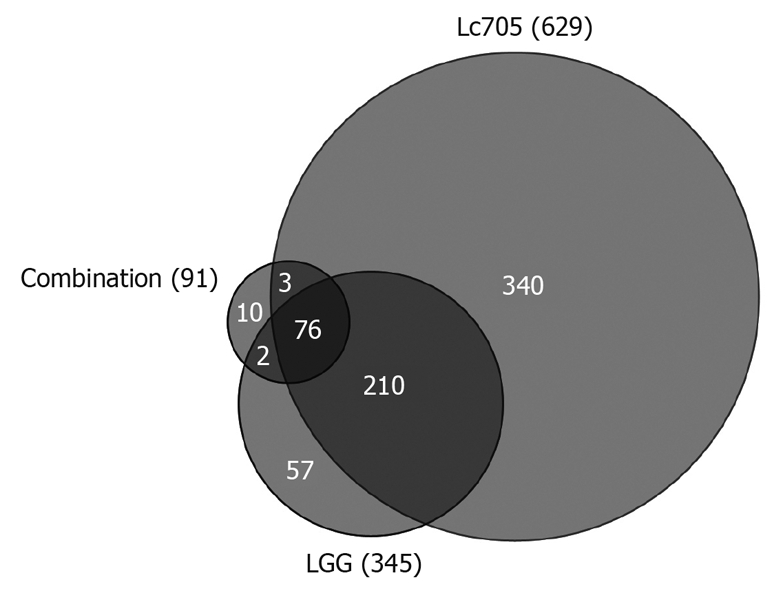
Schematic representation of statistically significant changes (P < 0.05) in mast-cell gene expression after 24 h stimulation with Lactobacillus rhamnosus Lc705, Lactobacillus rhamnosus GG and the combination of four probiotic bacteria. LGG: Lactobacillus rhamnosus GG; Lc705: Lactobacillus rhamnosus Lc705.
Gene functional category analysis and hierarchical clustering
To characterize the biological significance of the differentially expressed mast-cell genes, a gene ontology category analysis was performed. GOrilla Gene Enrichment Analysis and Visualization Tool[33] was used to discover whether some functional categories showed statistically significant, concordant differences between the stimulated sample and the unstimulated sample. The enrichment analyzes were carried out for the whole array gene set, including the genes that did not reach statistical significance in the array analysis, that were ranked based on their moderated t values. For the analysis, the ranked genes were sorted in the order of the highest and lowest t value of each sample. The analysis software used the sorted gene lists to classify the genes of each sample into gene ontology categories[37] by their biological function. Selected functional groups and examples of genes that represented each category are depicted in Table 1.
Table 1.
Representative subsets of differentially expressed mast-cell genes after bacterial stimulation classified into functional categories
| Go class | Description |
P value |
Gene examples | |||||
| LGG | Lc705 | PJS | Bb12 | Combination | Cpn | |||
| Upregulation | ||||||||
| GO:0002376 | Immune system process | 3.48E-16 | 2.95E-13 | - | 5.25E-7 | 5.66E-12 | 1.64E-4 | TNF, IL1B, IL8, IL10, CCL2, TLR1, TLR6, NOD2 |
| GO:0043067 | Regulation of programmed cell death | 2.77E-8 | 9.63E-8 | - | 7.18E-4 | 3.49E-6 | - | CASP3, CASP8, CDKN1 |
| GO:0045321 | Leukocyte activation | 8.97E-9 | 2.17E-4 | - | - | 2.23E-4 | - | CD8A, LCP2 |
| Downregulation | ||||||||
| GO:0007049 | Cell cycle | 4.76E-5 | 9.61E-7 | - | - | 5.81E-4 | - | CCNA2, MAPK12 |
| GO:0033033 | Regulation of mast cell activation | - | 7.51E-4 | - | - | - | - | FCER1A, FCER1G |
| GO:0008610 | Lipid biosynthetic process | - | - | - | 2.02E-5 | - | - | LPGAT1 |
The three selected highly upregulated or downregulated functional categories from the enrichment analysis performed with GOrilla Gene Enrichment Analysis and Visualization Tool. P value is the enrichment P value reported by GOrilla. The resultant categories reflect the gene expression differences of the bacteria-stimulated sample compared to unstimulated sample in 24 h time point. LGG: Lactobacillus rhamnosus GG; Lc705: Lactobacillus rhamnosus Lc705; PJS: Propionibacterium freudenreichii ssp. shermanii JS; Bb12: Bifidobacterium animalis ssp. lactis Bb12; Cpn: Chlamydia pneumoniae isolate Kajaani 6; TNF: Tumor necrosis factor; IL-1B: Interleukin-1β; IL-8: Interleukin-8; IL-10: Interleukin-10; CCL2: Chemokine (C-C motif) 2; TLR1: Toll-Like receptor 1; TLR6: Toll-Like receptor 6; NOD2: Nucleotide-binding oligomerization domain-containing protein 2; CASP3: Caspase 3; CASP8: Caspase 8; CDKN1: Cyclin-dependent kinase inhibitor 1; CD8A: Cluster of differentiation 8 A (T cell surface glycoprotein); LCP2: Lymphocyte cytosolic protein 2; CCNA2: Cyclin A2; MAPK12: Mitogen-activated protein kinase 12; FCER1A: Fc fragment of IgE high affinity I receptor for α polypeptide; FCER1G: Fc fragment of IgE high affinity I receptor for γ polypeptide; LPGAT1: Lysophosphatidylglycerol acyltransferase 1.
Stimulation of mast cells with LGG, Lc705, Bb12, combination of probiotics, or Cpn resulted in upregulation of genes that belong to categories that involve immune system processes, regulation of programmed cell death, and leukocyte activation. Stimulation with LGG, Lc705, Bb12 or the combination of probiotics suppressed mast-cell genes that are involved in general cell activities and metabolism such as cell cycle and lipid biosynthetic processes. Lc705 was found to suppress genes that are related to mast-cell activation.
Representative genes from the functional categorization (Table 1) were selected to compare the expression patterns of different samples. Mast cells have a central role in many inflammatory responses as well as in allergy, therefore, immunologically relevant genes and genes involved in mast cell activation and mediator release were selected. In order to compare the expression of the selected genes, a hierarchical comparison analysis with the MeV two-way hierarchical clustering method was performed. As expected, LGG and Lc705 but also the combination of four probiotics showed similar expression patterns in the clustering analysis, and were therefore considered to alter mast-cell gene expression in a similar manner (Figure 2). Genes that are involved in similar processes were also grouped: genes that encode Toll-like receptor (TLR) 1, nucleotide-binding oligomerization domain containing 2 (NOD2), FCER1A and FCER1G (high-affinity IgE receptor 1 gene subunits α and γ, respectively) grouped together; and genes that encode proteins with inflammatory functions, such as IL-1β and IL-8 constituted another distinct cluster.
Figure 2.
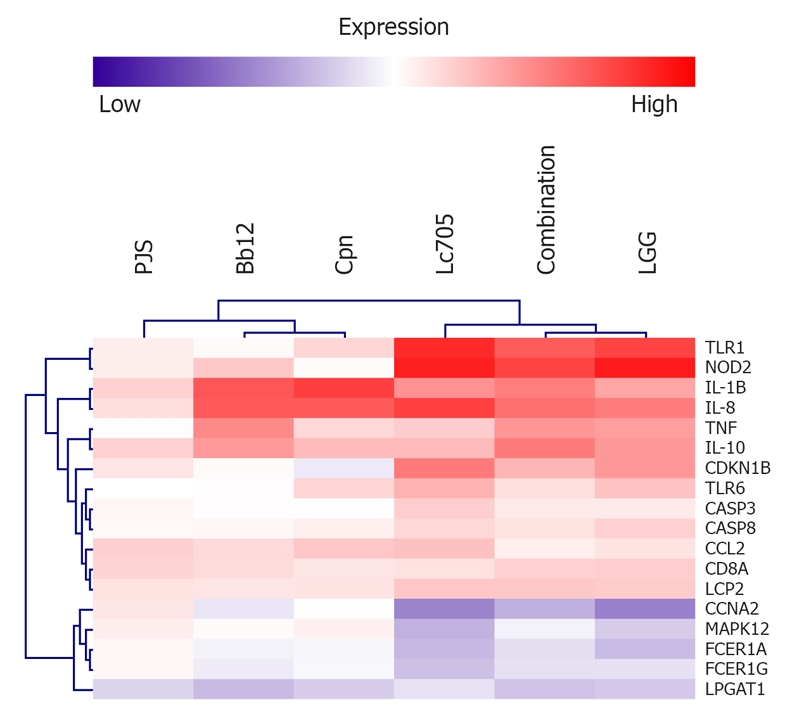
Two-way hierarchical clustering of representative genes selected from the functional category analysis (Table 1). Data are average expression differences between unstimulated and bacteria-stimulated mast cells from three independent experiments at the 24-h time point. Analysis was performed using MeV software. PJS: Propionibacterium freudenreichii ssp. shermanii JS; Bb12: Bifidobacterium animalis ssp. lactis Bb12; Cpn: Chlamydia pneumoniae isolate Kajaani 6; Lc705: Lactobacillus rhamnosus Lc705; LGG: Lactobacillus rhamnosus GG; TLR1: Toll-Like receptor 1; NOD2: Nucleotide-binding oligomerization domain-containing protein 2; IL-1B: Interleukin-1β; IL-8: Interleukin-8; TNF: Tumor necrosis factor; IL-10: Interleukin-10; CDKN1B: Cyclin-dependent kinase inhibitor 1B; TLR6: Toll-Like receptor 6; CASP3: Caspase 3; CASP8: Caspase 8; CCL2: Chemokine (C-C motif) 2; CD8A: Cluster of differentiation 8 A (T cell surface glycoprotein); LCP2: Lymphocyte cytosolic protein 2; CCNA2: Cyclin A2; MAPK12: Mitogen-activated protein kinase 12; FCER1A: Fc fragment of IgE high affinity I receptor for α polypeptide; FCER1G: Fc fragment of IgE high affinity I receptor for γ polypeptide; LPGAT1: Lysophosphatidylglycerol acyltransferase 1.
Although there were differences in the intensity of the expression levels, all bacteria except PJS induced upregulation of mast-cell genes included in the functional group of immune system regulation. Examples of upregulated genes in this category were TLR1, TLR6, NOD2, IL1B, IL8, chemokine (C-C motif) 2 (CCL2), TNF and IL10. Genes that are involved in regulation of programmed cell death, such as caspases 3 and 8 (CASP3 and CASP8) and cyclin-dependent kinase inhibitor 1B (CDKN1B), were observed to be upregulated in LGG, Lc705, and the combination-stimulated cells. LGG, Lc705 and the combination also significantly enhanced the expression of the cluster of differentiation 8 A (CD8A) and lymphocyte cytosolic protein 2 (LPC2) genes, which are involved in leukocyte activation.
Cyclin A2 (CCNA2) and mitogen-activated protein kinase 12 (MAPK12) genes involved in the regulation of cell cycle, and the latter also in IgE receptor signaling, were observed to be downregulated in cells stimulated with LGG, Lc705, and the combination of four probiotics. Stimulation of mast cells with Lc705 significantly suppressed the expression of FCER1A and FCER1G genes. The same trend was also observed after stimulation with LGG and the combination, although these changes in the analysis failed to reach statistical significance. Bb12 was found to suppress the expression of LPGAT1 (lysophosphatidylglycerol acyltransferase 1), a mast cell gene that is related to lipid biosynthesis, and also the other bacteria studied had a similar impact on the expression of this gene.
Manual screening of the array data
To gain further insight into the probiotic-induced changes in mast-cell activation, the array data set was screened for additional genes that were related to the IgE receptor signaling pathway and mast-cell activation and immunomodulation. The screening was performed manually using the list of genes that reached statistical difference in the array analysis at the 24-h time point (http://ekhidna.biocenter.helsinki.fi/poxo/download_data). The gene that encodes phospholipase C (PLC) that is involved in the mast-cell IgE receptor signaling pathway was observed to be significantly downregulated by LGG and Lc705. The gene that encodes prostaglandin E2 receptor (PTGER), which mediates the effects of this inflammatory prostaglandin, was also downregulated by LGG and Lc705. Although only Lc705 was categorized in the functional group as downregulating mast-cell activation, LGG-stimulated cells also reached statistical significance in suppressing FCER1A expression in the microarray analysis. Both Lc705 and LGG also suppressed the expression of the gene that encodes mast-cell histamine H4 receptor (HRH4). Expression alterations of the genes that encode the end-products of the IgE receptor signaling pathway were also screened, but no changes were observed in the mast-cell expression of leukotrienes, heparin or Th2 type cytokines, such as IL-3, IL-4, IL-5 and IL-15 after bacterial stimulation.
Quantitative RT-PCR
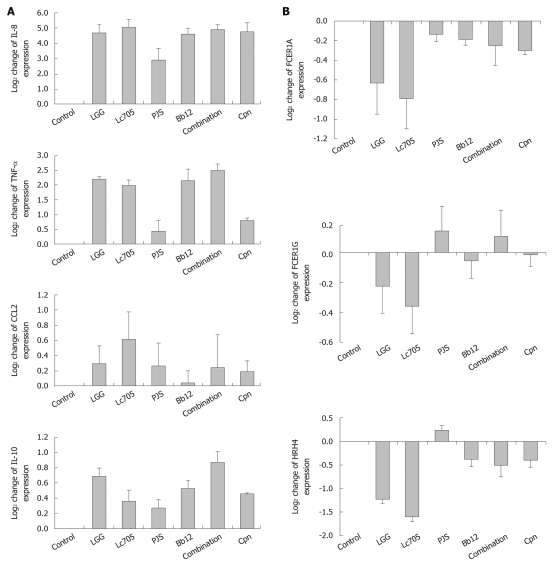
Findings from the microarray analysis were verified by quantitative RT-PCR. Seven genes with statistical significance after 24 h stimulation with at least one of the bacteria in the array analysis or in the functional categorization analysis were selected. Four of the genes encoded proteins that are involved in mast-cell regulation of immunological events: IL8, TNF-α, CCL2 and IL10 (Figure 3A). Three of the genes encoded proteins that are involved in mast-cell activation and in allergy: FCER1A, FCER1G and HRH4 (Figure 3B). With minor differences in the levels of intensities, all of the selected genes followed a similar expression pattern detected in the microarray analysis, and thus confirmed the results of the microarray data.
Figure 3.
Verification of mast-cell microarray results by quantitative reverse transcriptase-polymerase chain reaction with seven selected genes that are involved in mast-cell immune system regulation (A) and mast-cell activation (B). Gene expression was quantified after 24 h stimulation with four probiotic bacteria; Lactobacillus rhamnosus GG (LGG), Lactobacillus rhamnosus Lc705 (Lc705), Propionibacterium freudenreichii ssp. shermanii JS (PJS), Bifidobacterium animalis ssp. lactis Bb12 (Bb12) and their combination or with Chlamydia pneumoniae isolate Kajaani 6 (Cpn). Data are mean values ± SE of three independent experiments. IL: Interleukin; TNF-α: Tumor necrosis factor-α; CCL2: Chemokine (C-C motif) 2; FCER1A: Fc fragment of IgE high affinity I receptor for α polypeptide; FCER1G: Fc fragment of IgE high affinity I receptor for γ polypeptide; HRH4: Histamine H4 receptor.
DISCUSSION
In the present study, microarray analysis was used to gain a broad understanding of interactions between probiotic bacteria and human primary mast cells. We report here, that live probiotic bacteria have species-specific effects on human mast cells. The most significant changes in mast-cell gene expression were observed in the regulation of genes related to mast-cell activation and mediator release, including FCER1A, FCER1G and HRH4, and immunological responses such as IL8, TNF, CCL2 and IL10.
In the microarray analysis, lactobacilli affected mast-cell gene expression more than the other bacteria. Stimulation of mast cells with both LGG and Lc705 significantly downregulated the expression of the high-affinity IgE receptor subtype α (FCER1A) and HRH4 (HRH4) genes after 24 h stimulation. In addition, Lc705 stimulation downregulated the gene expression of FCεR1 receptor subtype γ (FCER1G). PJS, Bb12, the combination, or Cpn did not have an effect on FCER1 and HRH4 genes. FCεR1 plays a key role in mediating the allergy-related IgE-dependent activation and degranulation of mast cells[38] as demonstrated in FCεR1-deficient mice that fail to show allergic reactions after sensitization[39]. After FCεR1 aggregation, mast cells release inflammatory mediators, such as histamine. Histamine is the key mediator in causing the symptoms of allergy, and it also has a potent role as a modulator of immune responses[40]. The effects of histamine are mediated through histamine receptors H1-H4 that are expressed on the surface on many cell types, including mast cells and other inflammatory cells as well as epithelial cells[41]. The most recently discovered HRH4 has been shown to have mainly immunomodulatory effects[41]. The expression of histamine receptors is suggested to be influenced by inflammatory stimuli[40]. In addition, modification of histamine receptor gene expression is suggested to play a role in the pathogenesis of allergy, atherosclerosis and rheumatoid arthritis[7]. By suppressing the expression of FCER1 and HRH4 genes, probiotic lactobacilli could attenuate mast-cell activation and release of allergy-related mediators.
To obtain more evidence of the possible ability of probiotics to suppress mast-cell activation, the array gene set was manually screened for other genes involved in IgE receptor signaling. Expression of the gene that encodes PLC, which is involved in the release of intracellular calcium and in mast-cell degranulation[39], was suppressed significantly after Lc705 and LGG stimulation. Additionally, the gene that encodes expression of the member of the mitogen-activated protein kinase (MAPK) family, MAPK12, that participates in the signaling events that lead to the production of Th2 type cytokines IL-3, IL-4, IL-5 and IL-13, and in the generation of eicosanoids[38], was also downregulated in Lc705 and LGG-stimulated mast cells. These findings suggest that Lc705 and LGG have an inhibitory effect on mast-cell genes that are involved in the IgE signaling cascade, beyond inhibition of IgE receptor gene expression.
Our results with human primary mast cells are in line with those of two previous studies of the effects of non-pathogenic bacteria on human or mouse mast cells. Escherichia coli (E. coli) K12 strain has been found to downregulate FCER1A in the malignant human mast cell line (LAD3)[42]. The other study has reported that E. coli strain DSM 17252 inhibits mast-cell degranulation in mouse peritoneal mast cells[43]. Commensal non-pathogenic E. coli and probiotic LGG and Lc705 seem to downregulate mast-cell IgE responses similarly. These results suggest that commensal and probiotic bacteria do not stimulate mast cells but rather diminish their activation. However, it is worth noticing that this effect is not universal response to bacteria, because not all probiotic bacteria or pathogenic Cpn affected the gene expression of high-affinity IgE receptor similarly.
Probiotic bacteria were also observed to alter the expression of genes that have inflammatory functions. In the functional categorization analysis, the expression of a category of immune system process that contains, for example, a gene that encodes an inflammatory mediator that is also regulated by the MAPK pathway, TNFα[44], was upregulated in mast cells stimulated with LGG, Lc705, Bb12 and the combination. The categorization analysis also highlighted upregulated functions in known inflammatory genes such as IL8, CCL2, and IL1B in cells stimulated with LGG, Lc705, Bb12 and the combination. The same genes were also upregulated in mast cells stimulated with Cpn, which is in line with our previous study in which Cpn elicited pro-inflammatory effects in mast cells[22]. However, no upregulation in any of the Th2 type cytokine genes in mast cells after probiotic stimulation in the gene microarray was observed. In the prevention of the symptoms in allergic diseases, probiotics have been suggested to elicit low-grade inflammation and thus shift the immune response away from the allergy-related Th2 type inflammation[45,46]. In in vitro studies, probiotic bacteria have been observed to induce the secretion and expression of Th1 type cytokines in monocytes, macrophages and dendritic cells[47-49]. Additionally, in a clinical study, low-grade inflammation induced by LGG in allergy-prone children has been proposed to be one mechanism to prevent atopic diseases[45]. The ability of LGG, Lc705, Bb12 and the combination to induce in mast cells pro-inflammatory rather than Th2 type cytokine expression could have contributed to the observed beneficial low-grade inflammatory response in the above-cited study. Our findings in mast cells support the idea that some probiotic strains shift the mast-cell-mediated immunological response from the Th2 to Th1 type, by inducing expression of pro-inflammatory mediators.
Mast cells stimulated with LGG, Lc705, Bb12, the combination, or with Cpn also induced the expression of a gene that encodes anti-inflammatory IL-10. Mast-cell-derived IL-10 has been observed to be crucial in restriction of chronic skin inflammation in hypersensitivity reactions[50]. Probiotic Bifidobacterium strains have been observed to stimulate IL-10 production in immune cells[51]. IL-10 induction in mast cells could participate in balancing the inflammatory impact. In vivo, the combination of affected genes after bacterial stimulation is likely to be more important than a change in the expression of any single gene. Thus, the downregulation of FCER1 and HRH4 genes combined with the upregulation of IL10 after stimulation with probiotic Lactobacillus might be involved in downregulation of inflammatory responses in allergy and other inflammatory diseases in which mast cells are known to a play role, such as atherosclerosis, rheumatoid arthritis, inflammatory bowel disease and IBS[5,52-54]. As Cpn upregulated only IL10 without affecting FCER1 and HRH4, mast cells stimulated with pathogenic Cpn might not have the same clinical effects as those stimulated with probiotic bacteria.
In addition to FCεR1 and HRH4, the function of mast cells is regulated through a variety of other receptors, such as TLRs. In our previous study, we have shown that TLR2 is a receptor for LGG, which triggers the nuclear factor-κB signaling cascade, which leads to expression of different cytokines in human primary macrophages[55]. In addition, probiotic bacteria have been shown to suppress mast-cell degranulation by interrupting FCεR1-mediated signaling through TLR2[56]. The paradox of the ability of probiotic LGG and Lc705 to diminish mast-cell activation, but enhance the mast-cell immune response, could be regulated through the same receptor, TLR2.
PJS failed to change mast-cell gene expression significantly at either time point. Previously, PJS has been observed to induce TNFα expression after 3 h stimulation, and IL-10 expression after 3 and 24 h stimulation in human peripheral blood mononuclear cells (PBMCs)[24]. It could be that mast cells are unresponsive to PJS unlike the PBMC population. It could also be that the quantitative spectrum of microarray is a limiting factor, especially if the expression differences between unstimulated and stimulated samples are low or if the gene is expressed in low quantities[57]. In our experiments, the differences in the gene expression levels of the microarrays were relatively low, which is in accordance with other microarray studies performed with probiotic bacteria[58].
Crosstalk between probiotic bacteria and intestinal mast cells is likely to occur mainly through gut epithelial cells[59]. Probiotic bacteria or their products could incidentally translocate to the lamina propria through intestinal M cells and make direct contact with immunological cells such as mast cells. Additionally, mast cells are suggested to have the possibility to be in direct contact with bacteria or bacterial fragments in atherosclerotic plaques[53,60]. Fragments of Firmicutes and Proteobacteria, which are probably of intestinal origin, as well as Cpn have been detected from atherosclerotic plaques[60]. The intestinal barrier is believed to leak also in healthy humans, which would allow probiotic bacteria to access spaces that contain macrophages, dendritic cells and mast cells. Even though in vivo evidence of direct contact between mast cells and probiotic bacteria in the human intestine is missing, in vitro interaction studies performed with direct contact between bacteria and host cells, such as mast cells, are required for better understanding of the molecular basis of the immunomodulative properties of probiotic bacteria.
The mast cells differentiated by the peripheral-blood-derived isolation method comprise mainly the MCTC phenotype[23], whereas the gut contains both MCTC and MCT phenotypes; the latter being the predominant mucosal mast-cell phenotype[2]. However, the MCT and MCTC phenotypes both express high-affinity IgE receptors and produce a variety of inflammatory mediators. The responses observed in this study could be further evaluated in a more physiological context, e.g. in ex vivo culture models using mast cells derived from human intestinal tissues, or in in vivo animal or clinical studies.
The present study is believed to be the first to describe the effects of probiotic bacteria on human mast cells. Our data suggest that especially probiotic L. rhamnosus Lc705 and L. rhamnosus GG could diminish mast-cell activation and the effects of allergy-related mediators by downregulating expression of the high-affinity IgE and HRH4 receptors, and by stimulating mast-cell immune responses. Mast cells are important mediators of allergic responses on host surfaces including the intestine, therefore, we propose that mast cells participate in regulating the beneficial immunological responses to probiotic bacteria.
COMMENTS
Background
Probiotic bacteria are widely used to prevent or relieve symptoms of various clinical conditions, such as intestinal disorders and allergy. However, it is not fully understood what make probiotics effective.
Research frontiers
Mast cells are important immunological cells that have many functions. Mast cells also participate in the pathogenesis of many inflammatory diseases, with allergy being the best-known example. In the prevention of the symptoms in allergic diseases, probiotics have been suggested to elicit low-grade inflammation. In the present study, the authors explored for the first time the role of human mast cells in contributing to the beneficial effects of probiotic bacteria.
Innovations and breakthroughs
In the present study, the authors found that probiotic lactobacilli strains Lactobacillus rhamnosus (L. rhamnosus) GG and L. rhamnosus Lc705, but not propionibacteria or bifidobacteria, downregulated expression of high-affinity IgE and histamine H4 receptors, and enhanced mast-cell immune activity, and thus, might diminish the impact of the allergenic response.
Applications
Understanding the mechanisms by which probiotic bacteria elicit their health effects is crucial when designing and using different probiotic strains for specific preventive or therapeutic purposes.
Terminology
Probiotic bacteria are defined as live microorganisms that have beneficial effects on human health. High-affinity IgE receptor mediates mast-cell activation and histamine production in allergy. Histamine H4 receptor is a protein on mast cells that mediates the effects of histamine.
Peer review
In this well-designed study, the authors explored the action of four probiotic strains and their combination on human mast-cell global gene expression. The analysis revealed changes in genes involved in immune responses and mast-cell activation, which are especially involved in the pathogenesis of inflammatory diseases, such as allergy. The authors suggest that lactobacilli are able to diminish the impact of allergenic responses by downregulating expression of high-affinity IgE and histamine H4 receptors, and by enhancing mast-cell immune activity. The study addresses an interesting and important question.
Acknowledgments
Professor Heikki Vapaatalo is acknowledged for his advice and critical comments on the study and the manuscript. The authors thank Mirja Puolakkainen in the National Institute for Health and Welfare for kindly providing Cpn for this study. The authors acknowledge the excellent technical assistance of Anna-Maija Ahonen in growing bacteria, and Mari Jokinen and Maija Atuegwu for their help with culturing mast cells.
Footnotes
Supported by Valio Research Centre, the Foundation for Nutrition Research, Academy of Finland Research Council for Biosciences and Environment, Grant No. 129954; and Finnish Funding Agency for Technology and Innovation (TEKES) grant No. 2243/31/05. Wihuri Research Institute is maintained by Jenny and Antti Wihuri Foundation
Peer reviewer: Dr. William R Parker, PhD, Assistant Professor, Department of Surgery, Duke University Medical Center, Box 2605, Durham, NC 27710, United States
S- Editor Sun H L- Editor Kerr C E- Editor Lin YP
References
- 1.Bischoff SC, Wedemeyer J, Herrmann A, Meier PN, Trautwein C, Cetin Y, Maschek H, Stolte M, Gebel M, Manns MP. Quantitative assessment of intestinal eosinophils and mast cells in inflammatory bowel disease. Histopathology. 1996;28:1–13. doi: 10.1046/j.1365-2559.1996.262309.x. [DOI] [PubMed] [Google Scholar]
- 2.Welle M. Development, significance, and heterogeneity of mast cells with particular regard to the mast cell-specific proteases chymase and tryptase. J Leukoc Biol. 1997;61:233–245. doi: 10.1002/jlb.61.3.233. [DOI] [PubMed] [Google Scholar]
- 3.Rao KN, Brown MA. Mast cells: multifaceted immune cells with diverse roles in health and disease. Ann N Y Acad Sci. 2008;1143:83–104. doi: 10.1196/annals.1443.023. [DOI] [PubMed] [Google Scholar]
- 4.Bischoff SC, Krämer S. Human mast cells, bacteria, and intestinal immunity. Immunol Rev. 2007;217:329–337. doi: 10.1111/j.1600-065X.2007.00523.x. [DOI] [PubMed] [Google Scholar]
- 5.Bischoff SC. Role of mast cells in allergic and non-allergic immune responses: comparison of human and murine data. Nat Rev Immunol. 2007;7:93–104. doi: 10.1038/nri2018. [DOI] [PubMed] [Google Scholar]
- 6.Daëron M, Malbec O, Latour S, Arock M, Fridman WH. Regulation of high-affinity IgE receptor-mediated mast cell activation by murine low-affinity IgG receptors. J Clin Invest. 1995;95:577–585. doi: 10.1172/JCI117701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turner H, Kinet JP. Signalling through the high-affinity IgE receptor Fc epsilonRI. Nature. 1999;402:B24–B30. doi: 10.1038/35037021. [DOI] [PubMed] [Google Scholar]
- 8.Zampeli E, Tiligada E. The role of histamine H4 receptor in immune and inflammatory disorders. Br J Pharmacol. 2009;157:24–33. doi: 10.1111/j.1476-5381.2009.00151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Metz M, Grimbaldeston MA, Nakae S, Piliponsky AM, Tsai M, Galli SJ. Mast cells in the promotion and limitation of chronic inflammation. Immunol Rev. 2007;217:304–328. doi: 10.1111/j.1600-065X.2007.00520.x. [DOI] [PubMed] [Google Scholar]
- 10.Galli SJ, Grimbaldeston M, Tsai M. Immunomodulatory mast cells: negative, as well as positive, regulators of immunity. Nat Rev Immunol. 2008;8:478–486. doi: 10.1038/nri2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guides for the Evaluation of Probiotics in Food. London, Ontario, Canada: Food and Agriculture Organization of the United Nations and World Health Organization; 2002. [Google Scholar]
- 12.Guarino A, Lo Vecchio A, Canani RB. Probiotics as prevention and treatment for diarrhea. Curr Opin Gastroenterol. 2009;25:18–23. doi: 10.1097/MOG.0b013e32831b4455. [DOI] [PubMed] [Google Scholar]
- 13.Kopp MV, Salfeld P. Probiotics and prevention of allergic disease. Curr Opin Clin Nutr Metab Care. 2009;12:298–303. doi: 10.1097/MCO.0b013e32832989a3. [DOI] [PubMed] [Google Scholar]
- 14.Kajander K, Hatakka K, Poussa T, Färkkilä M, Korpela R. A probiotic mixture alleviates symptoms in irritable bowel syndrome patients: a controlled 6-month intervention. Aliment Pharmacol Ther. 2005;22:387–394. doi: 10.1111/j.1365-2036.2005.02579.x. [DOI] [PubMed] [Google Scholar]
- 15.Agrawal A, Houghton LA, Morris J, Reilly B, Guyonnet D, Goupil Feuillerat N, Schlumberger A, Jakob S, Whorwell PJ. Clinical trial: the effects of a fermented milk product containing Bifidobacterium lactis DN-173-010 on abdominal distension and gastrointestinal transit in irritable bowel syndrome with constipation. Aliment Pharmacol Ther. 2008:Epub ahead of print. doi: 10.1111/j.1365-2036.2008.03853.x. [DOI] [PubMed] [Google Scholar]
- 16.Hatakka K, Savilahti E, Pönkä A, Meurman JH, Poussa T, Näse L, Saxelin M, Korpela R. Effect of long term consumption of probiotic milk on infections in children attending day care centres: double blind, randomised trial. BMJ. 2001;322:1327. doi: 10.1136/bmj.322.7298.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turchet P, Laurenzano M, Auboiron S, Antoine JM. Effect of fermented milk containing the probiotic Lactobacillus casei DN-114001 on winter infections in free-living elderly subjects: a randomised, controlled pilot study. J Nutr Health Aging. 2003;7:75–77. [PubMed] [Google Scholar]
- 18.Timmerman HM, Koning CJ, Mulder L, Rombouts FM, Beynen AC. Monostrain, multistrain and multispecies probiotics--A comparison of functionality and efficacy. Int J Food Microbiol. 2004;96:219–233. doi: 10.1016/j.ijfoodmicro.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 19.Ouwehand AC. Antiallergic effects of probiotics. J Nutr. 2007;137:794S–797S. doi: 10.1093/jn/137.3.794S. [DOI] [PubMed] [Google Scholar]
- 20.Collado MC, Meriluoto J, Salminen S. Role of commercial probiotic strains against human pathogen adhesion to intestinal mucus. Lett Appl Microbiol. 2007;45:454–460. doi: 10.1111/j.1472-765X.2007.02212.x. [DOI] [PubMed] [Google Scholar]
- 21.Kajander K, Myllyluoma E, Rajilić-Stojanović M, Kyrönpalo S, Rasmussen M, Järvenpää S, Zoetendal EG, de Vos WM, Vapaatalo H, Korpela R. Clinical trial: multispecies probiotic supplementation alleviates the symptoms of irritable bowel syndrome and stabilizes intestinal microbiota. Aliment Pharmacol Ther. 2008;27:48–57. doi: 10.1111/j.1365-2036.2007.03542.x. [DOI] [PubMed] [Google Scholar]
- 22.Oksaharju A, Lappalainen J, Tuomainen AM, Pussinen PJ, Puolakkainen M, Kovanen PT, Lindstedt KA. Pro-atherogenic lung and oral pathogens induce an inflammatory response in human and mouse mast cells. J Cell Mol Med. 2009;13:103–113. doi: 10.1111/j.1582-4934.2008.00285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lappalainen J, Lindstedt KA, Kovanen PT. A protocol for generating high numbers of mature and functional human mast cells from peripheral blood. Clin Exp Allergy. 2007;37:1404–1414. doi: 10.1111/j.1365-2222.2007.02778.x. [DOI] [PubMed] [Google Scholar]
- 24.Kekkonen RA, Kajasto E, Miettinen M, Veckman V, Korpela R, Julkunen I. Probiotic Leuconostoc mesenteroides ssp. cremoris and Streptococcus thermophilus induce IL-12 and IFN-gamma production. World J Gastroenterol. 2008;14:1192–1203. doi: 10.3748/wjg.14.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Latvala S, Pietila TE, Veckman V, Kekkonen RA, Tynkkynen S, Korpela R, Julkunen I. Potentially probiotic bacteria induce efficient maturation but differential cytokine production in human monocyte-derived dendritic cells. World J Gastroenterol. 2008;14:5570–5583; discussion 5581-5582. doi: 10.3748/wjg.14.5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson CL, Miller CJ. Simpleaffy: a BioConductor package for Affymetrix Quality Control and data analysis. Bioinformatics. 2005;21:3683–3685. doi: 10.1093/bioinformatics/bti605. [DOI] [PubMed] [Google Scholar]
- 29.Sandberg R, Larsson O. Improved precision and accuracy for microarrays using updated probe set definitions. BMC Bioinformatics. 2007;8:48. doi: 10.1186/1471-2105-8-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 31.Sartor MA, Tomlinson CR, Wesselkamper SC, Sivaganesan S, Leikauf GD, Medvedovic M. Intensity-based hierarchical Bayes method improves testing for differentially expressed genes in microarray experiments. BMC Bioinformatics. 2006;7:538. doi: 10.1186/1471-2105-7-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Storey J, Taylor J, Siegmund D. Strong control, conservative point estimation, and simultaneous conservative consistency of false discovery rates: A unified approach. J Roy Stat Soc (Series B) 2004;66:187–205. [Google Scholar]
- 33.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dudoit S, Gentleman RC, Quackenbush J. Open source software for the analysis of microarray data. Biotechniques. 2003;Suppl:45–51. [PubMed] [Google Scholar]
- 35.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 37.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Galli SJ, Kalesnikoff J, Grimbaldeston MA, Piliponsky AM, Williams CM, Tsai M. Mast cells as "tunable" effector and immunoregulatory cells: recent advances. Annu Rev Immunol. 2005;23:749–786. doi: 10.1146/annurev.immunol.21.120601.141025. [DOI] [PubMed] [Google Scholar]
- 39.Kinet JP. The high-affinity IgE receptor (Fc epsilon RI): from physiology to pathology. Annu Rev Immunol. 1999;17:931–972. doi: 10.1146/annurev.immunol.17.1.931. [DOI] [PubMed] [Google Scholar]
- 40.Jutel M, Watanabe T, Akdis M, Blaser K, Akdis CA. Immune regulation by histamine. Curr Opin Immunol. 2002;14:735–740. doi: 10.1016/s0952-7915(02)00395-3. [DOI] [PubMed] [Google Scholar]
- 41.Thurmond RL, Gelfand EW, Dunford PJ. The role of histamine H1 and H4 receptors in allergic inflammation: the search for new antihistamines. Nat Rev Drug Discov. 2008;7:41–53. doi: 10.1038/nrd2465. [DOI] [PubMed] [Google Scholar]
- 42.Kulka M, Fukuishi N, Rottem M, Mekori YA, Metcalfe DD. Mast cells, which interact with Escherichia coli, up-regulate genes associated with innate immunity and become less responsive to Fc(epsilon)RI-mediated activation. J Leukoc Biol. 2006;79:339–350. doi: 10.1189/jlb.1004600. [DOI] [PubMed] [Google Scholar]
- 43.Magerl M, Lammel V, Siebenhaar F, Zuberbier T, Metz M, Maurer M. Non-pathogenic commensal Escherichia coli bacteria can inhibit degranulation of mast cells. Exp Dermatol. 2008;17:427–435. doi: 10.1111/j.1600-0625.2008.00704.x. [DOI] [PubMed] [Google Scholar]
- 44.Okayama Y, Tkaczyk C, Metcalfe DD, Gilfillan AM. Comparison of Fc epsilon RI- and Fc gamma RI-mediated degranulation and TNF-alpha synthesis in human mast cells: selective utilization of phosphatidylinositol-3-kinase for Fc gamma RI-induced degranulation. Eur J Immunol. 2003;33:1450–1459. doi: 10.1002/eji.200323563. [DOI] [PubMed] [Google Scholar]
- 45.Marschan E, Kuitunen M, Kukkonen K, Poussa T, Sarnesto A, Haahtela T, Korpela R, Savilahti E, Vaarala O. Probiotics in infancy induce protective immune profiles that are characteristic for chronic low-grade inflammation. Clin Exp Allergy. 2008;38:611–618. doi: 10.1111/j.1365-2222.2008.02942.x. [DOI] [PubMed] [Google Scholar]
- 46.Viljanen M, Pohjavuori E, Haahtela T, Korpela R, Kuitunen M, Sarnesto A, Vaarala O, Savilahti E. Induction of inflammation as a possible mechanism of probiotic effect in atopic eczema-dermatitis syndrome. J Allergy Clin Immunol. 2005;115:1254–1259. doi: 10.1016/j.jaci.2005.03.047. [DOI] [PubMed] [Google Scholar]
- 47.Miettinen M, Vuopio-Varkila J, Varkila K. Production of human tumor necrosis factor alpha, interleukin-6, and interleukin-10 is induced by lactic acid bacteria. Infect Immun. 1996;64:5403–5405. doi: 10.1128/iai.64.12.5403-5405.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miettinen M, Matikainen S, Vuopio-Varkila J, Pirhonen J, Varkila K, Kurimoto M, Julkunen I. Lactobacilli and streptococci induce interleukin-12 (IL-12), IL-18, and gamma interferon production in human peripheral blood mononuclear cells. Infect Immun. 1998;66:6058–6062. doi: 10.1128/iai.66.12.6058-6062.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Veckman V, Miettinen M, Pirhonen J, Sirén J, Matikainen S, Julkunen I. Streptococcus pyogenes and Lactobacillus rhamnosus differentially induce maturation and production of Th1-type cytokines and chemokines in human monocyte-derived dendritic cells. J Leukoc Biol. 2004;75:764–771. doi: 10.1189/jlb.1003461. [DOI] [PubMed] [Google Scholar]
- 50.Grimbaldeston MA, Nakae S, Kalesnikoff J, Tsai M, Galli SJ. Mast cell-derived interleukin 10 limits skin pathology in contact dermatitis and chronic irradiation with ultraviolet B. Nat Immunol. 2007;8:1095–1104. doi: 10.1038/ni1503. [DOI] [PubMed] [Google Scholar]
- 51.Young SL, Simon MA, Baird MA, Tannock GW, Bibiloni R, Spencely K, Lane JM, Fitzharris P, Crane J, Town I, et al. Bifidobacterial species differentially affect expression of cell surface markers and cytokines of dendritic cells harvested from cord blood. Clin Diagn Lab Immunol. 2004;11:686–690. doi: 10.1128/CDLI.11.4.686-690.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barbara G, Stanghellini V, De Giorgio R, Cremon C, Cottrell GS, Santini D, Pasquinelli G, Morselli-Labate AM, Grady EF, Bunnett NW, et al. Activated mast cells in proximity to colonic nerves correlate with abdominal pain in irritable bowel syndrome. Gastroenterology. 2004;126:693–702. doi: 10.1053/j.gastro.2003.11.055. [DOI] [PubMed] [Google Scholar]
- 53.Lindstedt KA, Mäyränpää MI, Kovanen PT. Mast cells in vulnerable atherosclerotic plaques--a view to a kill. J Cell Mol Med. 2007;11:739–758. doi: 10.1111/j.1582-4934.2007.00052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eklund KK. Mast cells in the pathogenesis of rheumatic diseases and as potential targets for anti-rheumatic therapy. Immunol Rev. 2007;217:38–52. doi: 10.1111/j.1600-065X.2007.00504.x. [DOI] [PubMed] [Google Scholar]
- 55.Miettinen M, Veckman V, Latvala S, Sareneva T, Matikainen S, Julkunen I. Live Lactobacillus rhamnosus and Streptococcus pyogenes differentially regulate Toll-like receptor (TLR) gene expression in human primary macrophages. J Leukoc Biol. 2008;84:1092–1100. doi: 10.1189/jlb.1206737. [DOI] [PubMed] [Google Scholar]
- 56.Kasakura K, Takahashi K, Aizawa T, Hosono A, Kaminogawa S. A TLR2 ligand suppresses allergic inflammatory reactions by acting directly on mast cells. Int Arch Allergy Immunol. 2009;150:359–369. doi: 10.1159/000226237. [DOI] [PubMed] [Google Scholar]
- 57.Allanach K, Mengel M, Einecke G, Sis B, Hidalgo LG, Mueller T, Halloran PF. Comparing microarray versus RT-PCR assessment of renal allograft biopsies: similar performance despite different dynamic ranges. Am J Transplant. 2008;8:1006–1015. doi: 10.1111/j.1600-6143.2008.02199.x. [DOI] [PubMed] [Google Scholar]
- 58.Shima T, Fukushima K, Setoyama H, Imaoka A, Matsumoto S, Hara T, Suda K, Umesaki Y. Differential effects of two probiotic strains with different bacteriological properties on intestinal gene expression, with special reference to indigenous bacteria. FEMS Immunol Med Microbiol. 2008;52:69–77. doi: 10.1111/j.1574-695X.2007.00344.x. [DOI] [PubMed] [Google Scholar]
- 59.Corthésy B, Gaskins HR, Mercenier A. Cross-talk between probiotic bacteria and the host immune system. J Nutr. 2007;137:781S–790S. doi: 10.1093/jn/137.3.781S. [DOI] [PubMed] [Google Scholar]
- 60.Ott SJ, El Mokhtari NE, Musfeldt M, Hellmig S, Freitag S, Rehman A, Kühbacher T, Nikolaus S, Namsolleck P, Blaut M, et al. Detection of diverse bacterial signatures in atherosclerotic lesions of patients with coronary heart disease. Circulation. 2006;113:929–937. doi: 10.1161/CIRCULATIONAHA.105.579979. [DOI] [PubMed] [Google Scholar]