Abstract
The global evolution and spread of Mycobacterium tuberculosis (MTB), one of the most successful bacterial pathogens, remain a mystery. Advances in molecular technology in the past decade now make it possible to understand MTB strain evolution and transmission in the context of human population migration. Taiwan is a relatively isolated island, serving as a mixing vessel over the past four centuries as colonization by different waves of ethnic groups occurred. By using mycobacterial tandem repeat sequences as genetic markers, the prevalence of MTB strains in Taiwan revealed an interesting association with historical migrations of different ethnic populations, thus providing a good model to explore the global evolution and spread of MTB.
1. Introduction
Tuberculosis (TB) remains a major worldwide health concern and has been characterized as one of three epidemics by the World Health Organization [1]. In 2006, more than 1.5 million people died of TB, an estimated 9.1 million new cases appeared, and the number of total TB cases worldwide reached about 14 million [2]. Findings from sites representative of Neolithic Europe, ancient Egypt, and the Greek and Roman empires revealed that TB is an ancient human disease [3]. Population migration due to wars and New World expedition accounts for the major transmission patterns of microbial pathogens, including Mycobacterium tuberculosis (MTB). In the past decade, the prevalence of MTB strains in different geographic regions and ethnic populations has been explored by molecular methods [4–6]. The reports revealed interesting patterns of strain distribution in different ethnic populations, which matched well to historical population migrations [5, 6]. Therefore, strain variations in different populations may be used to elucidate the transmission patterns of MTB.
The distribution of TB in different geographic regions is characterized by the prevalence of different MTB strains with varied virulence and drug resistance. Both environmental and host factors are responsible for the transmission and prevalence of different MTB strains. Because MTB has no detectable horizontal gene transfer [7, 8], large sequence polymorphisms (LPSs) can be used as phylogenetic markers to trace the evolutionary relationships of different strain families. Hirsh et al. presented a phylogenetic analysis of genomic deletions or LSPs, which were identified by comparative genome hybridization using DNA microarrays [7]. Mycobacterial interspersed repetitive units (MIRUs) loci comprise variable numbers of tandem repeat (VNTR) sequences, which allow them to be used as powerful genotyping markers [9]. In terms of genetic diversity and mutation rates, they resemble human microsatellites, which are widely used in human population genetics studies. By conducting MIRU-VNTR typing, Supply et al. were able to detect strong linkage disequilibrium between allele variants at these loci, indicative of a predominant clonal evolution in the MTB complex [8].
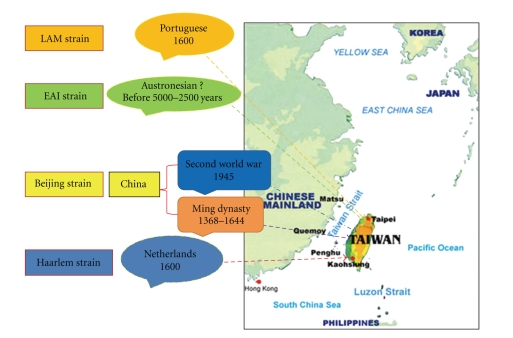
Taiwan is a relatively isolated island situated to the southeast of mainland China. The ethnic populations of Taiwan include Han Chinese who migrated to the island in two major waves: the first during the Ming dynasty around 1600 and the second between 1945 and 1950, when members of the military, veterans, and some civilians emigrated from mainland China due to the civil war there [4]; in total, about two million mainland Chinese have migrated to Taiwan to date. Taiwan was occupied by the Dutch beginning in 1660 for 40 years, and by the Japanese from 1895 until 1945. There are 12 tribes of aboriginals on this island, which are presumed to represent the ethnics who have inhabited the island for at least four thousand years (Figure 1).
Figure 1.
Proposed origins and routes of spread of four strains of MTB to Taiwan.
Although both the incidence and mortality rate of TB have shown steadily declined since 1950, TB is still a leading notifiable infectious disease on Taiwan. The populations of Taiwan that have tuberculosis among them include aborigines, veterans, and Taiwanese (Hoklo). Therefore, the heterogeneous components of ethnic populations constitute a good model with which to study MTB transmission and host-pathogen relationships. An important question to be answered here is whether distinct genotypes or lineages of MTB are distributed differently according to their hosts' ethnic origins and birthplaces. In the past years, we applied MIRU-VNTR sequences as genetic markers and discovered interesting findings on the origin and evolution of MTB in Taiwan, as described below.
2. Associations of Mycobacterium tuberculosis Genotypes with Different Ethnic and Migratory Populations in Taiwan
Some epidemiologic studies have revealed that MTB genotype distribution is closely associated with geography, ethnicity, and population migrations [4, 5, 7]. Similar phylogeographical population structures have been reported for other human pathogens [10–13], some of which have been linked to ancient human migrations [11, 12, 14].
In Taiwan, TB is a major disease with an annual incidence of about 16,000 confirmed cases. The proportion of ethnic populations on the island is about 2% native aborigines and 98% Han Chinese (Council of Indigenous Peoples, Executive Yuan Taiwan, 2007). Previous studies in Taiwan have demonstrated a fivefold higher incidence of TB among aborigines compared to Han Chinese [15]. In addition, polymorphism at the NRAMP1 gene appears to be associated with susceptibility to TB among aborigines but not among the Han Chinese population [15]. Preliminary studies on Beijing family MTB strains reveal differential distributions by geographic region in Taiwan [16]. These multifactor influences, including waves of immigration, allow us to trace the evolutionary history of pulmonary TB in Taiwan. Accordingly, we investigated TB evolution or transmission in (1) the aborigines of Austronesian ethnicity, whose ancestors came to Taiwan more than 500 years ago; (2) the veterans of Han Chinese origin, first-generation immigrants who moved to Taiwan 55–60 years ago; (3) the general Taiwanese population of Han Chinese, most of whose ancestors migrated to Taiwan around 200–400 years ago [4].
Based on spoligotyping classification, six distinct clades of MTB isolates among three Taiwanese subpopulations were identified: Beijing, Haarlem, East-African Indian (EAI), Latin American and Mediterranean (LAM), U, and the ill-defined T clade. Of the six known clades, the Beijing genotype overall was the most prevalent, being found in 40% of TB-positive aborigines, 72% of TB-positive veterans, and 56% of the TB-positive general population [4]. This result coincides with the global situation, with the most prevalent MTB strain worldwide being the Beijing genotype. Because Beijing strains are rapidly spreading worldwide, major TB outbreaks are most often associated with this strain [6, 17–19]. The second most frequent clade was that of the Haarlem family, which was present in 27% of aborigines and 13% of the general population, but in only 7% of veterans [4]. The third most frequent type was the T family, which was present in 5% of aborigines, 10% of veterans, and 6% of the general population. The remaining types were, in descending order of frequency, LAM, EAI, and U [4].
The Beijing family, which has the highest prevalence in the three Taiwanese subpopulations, can be further grouped into ancestral, modern, and recent strains by NTF locus analysis and RD deletion analysis. The NTF region and RD deletion are associated with the length of time since an MTB strain emerged in the human population; thus, they can be used to estimate the relative age of Beijing family clusters. Results of NTF and RD analyses revealed that ancient Beijing strains are prevalent among the aborigines, and modern Beijing strains predominate among veterans and the general population [4]. The retention of ancient characteristics of MTB among aborigines may be due to the historical tendency of Taiwan aborigines to live separately from the general population and thus have relatively little intermingling with Han Chinese.
The Haarlem genotype is the second prevalent type of TB in Taiwan. The Haarlem strain was first isolated from a patient living in Holland [20, 21] and is found mainly in Central America, the Caribbean, Europe, and West Africa, suggesting a link between Haarlem and post-Columbus Europeans [19]. ogt and mgtC gene analyses for the Haarlem lineage demonstrated that Haarlem strains circulating among aborigines in Taiwan are wild-type strains, whereas most Haarlem strains currently isolated in Europe contain single nucleotide polymorphisms (SNPs) and are comparatively modern. These results are similar to those of the Beijing strains. Given Taiwan aborigines' geographic isolation, the first transmission or exchange of Haarlem strains between the Dutch and the aborigines in Taiwan may have occurred in the 16th century during the Dutch colonization period. The late 16th century of Ming Dynasty was also the period in which Han Chinese began to migrate from mainland China to Taiwan. Thus, the Han Chinese may have introduced Beijing ancient strains into the MTB gene pool in Taiwan at that time.
3. Molecular Epidemiology and Evolutionary Genetics of Mycobacterium tuberculosis in Taipei
We then turn to study the strain distribution of MTB in Taipei, which is located in northern Taiwan and is the island's capital city. The strain distribution of MTB in Taipei provides us with the transmission pattern in this metropolitan city against the background described above. The city proper occupies 272 square km and has a population of 2.6 million, with an additional 4.3 million inhabitants in the surrounding metropolitan area. The population of Taipei includes the same ethnicities as described above for the entire island: Han Chinese, veterans, and Taiwanese aborigines [22]. The prevalence of TB in large urban areas such as Taipei is complicated by the close human-to-human contacts and potential multiple sources of MTB strains from different ethnic and migratory populations.
In a molecular epidemiologic analysis undertaken to investigate the prevalence of genotypes, cluster pattern, and drug resistance of MTB isolates in metropolitan Taipei, 356 MTB isolates from patients presenting with pulmonary TB were studied; the major spoligotypes found were Beijing lineages (52.5%), followed by Haarlem lineages (13.5%) and EAI plus EAI-like lineages (11%) [1]. Based on NTF and RD analyses, as well as on drug-resistance testing, strains of the Beijing family were more likely to be modern strains and have a higher percentage of multiple drug resistance than all of the other families combined. Because Han Chinese make up almost all of the general population of Taipei City, Beijing isolates found there were overwhelmingly modern strains (96%). The predominance of the Beijing strain in Taipei city constitutes a big challenge for TB control. Another important observation was that patients infected with the Beijing family were statistically younger than those infected with other genotypes (Table 1). These results suggest a possible recent spread of the Beijing genotype among younger individuals in this area. Thus, even though Taiwan has had comprehensive BCG vaccinations for more than 40 years, the predominance of the Beijing family strain in the younger cohort in our study suggests that BCG may not adequately protect young people from the Beijing strain of MTB.
Table 1.
Association of Beijing MTB genotype and different age groups of patientsa.
| Age group (yr) | No. (%) isolates | No. (%) of Beijing isolates | Odds ratio | 95% C.I. | P value |
|---|---|---|---|---|---|
| 356 | 187 (52.53) | ||||
| ≤25 | 34 (9.55) | 29 (85.29) | 5.80 | 2.11–15.98 | .0002 |
| ≤30 | 54 (15.17) | 37 (68.52) | 2.18 | 1.11–4.28 | .02 |
| 31–60 | 95 (26.69) | 50 (52.63) | 1.11 | 0.65–1.90 | .7 |
| 61–75 | 85 (23.88) | 39 (45.88) | 0.85 | 0.49–1.48 | .56 |
| ≥76 | 122 (34.27) | 61 (50.00) | 1 | reference group |
aAdapted from [1].
This situation warrants closer attention to control policy and suggests that a better BCG vaccine is needed.
Of the 356 strains in this study, 281 isolates (79%) were sensitive to all four of the first-line agents tested and 75 (21%) were resistant to at least one drug; 2.8% were multidrug resistant (MDR) (Table 2). Analysis of the association between MDR and genotypes (as determined by spoligotyping) showed that the Beijing genotype is more likely to be MDR than all other genotypes (Haarlem, T, EAI, others, and orphan combined) (P = .08, OR = 3.73, and 95% C.I. = (0.78–17.83)). The EAI family is significantly more likely to be sensitive to all drugs compared to other genotypes (P = .02, OR = 3.64, and 95% C.I. = (1.09–12.15)). EAI belongs to a branch in the early evolution of MTB and shows more antibiotic-sensitive properties, perhaps due to a lack of drug selection pressure. Interestingly among the orphan strains, 5% were MDR and 20% were resistant to one drug, showing a distribution similar to that of the Beijing family.
Table 2.
Association between MTB genotype and drug resistance in patientse.
| Genotype family | No. of isolates (%) | MDR (%) | Any one drug (%) | All sensitivity (%) |
|---|---|---|---|---|
| Beijinga | 187 (52.5) | 8(4.2) | 36 (19.4) | 143 (76.4) |
| Haarlem | 48 (13.5) | 0 | 9 (18.8) | 39 (81.2) |
| EAIb | 40 (11.2) | 0 | 3 (7.5) | 37 (92.5) |
| T | 25 (7.1) | 0 | 8 (32.0) | 17 (68.0) |
| “Others”c (LAM, U, MANU, Bovis1) | 16 (4.5) | 0 | 1 (6.3) | 15 (93.7) |
| Unclassifiedd | 40 (11.2) | 2 (5) | 8 (20) | 30 (75) |
| Total | 356 | 10 (2.8) | 65 (18.2) | 281 (79) |
aIncluding Beijing-like strains.
bIncluding EAI-like strains.
c“Others”, all genotype families with a frequency of less than 10 cases.
dUnclassified, no internationally recognized genotype family assigned, based on the SpolDB4 spoligotype database.
eAdapted from [1].
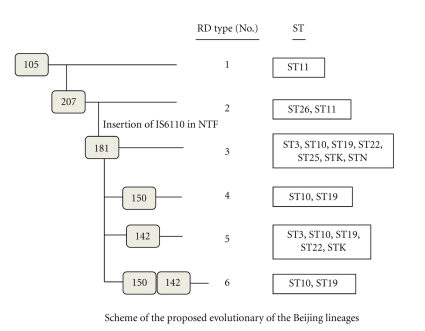
Taken together, our data summarized in Figure 2 show the evolutionary relationships within the Beijing family of strains in Taipei city. RD group 1 sublineage: 1 isolate of ST11; this isolate shows a deletion of the RD105 region. RD group 2 sublineages include ST11 and ST26; these isolates show deletion of the RD105 and RD207 regions. RD group 3 sublineages include ST3, ST10, ST19, ST22, STK, and STN; these isolates show deletion of the RD105, RD207, and RD181 regions. RD group 4 sublineages include ST10 and ST19; these isolates show deletion of the RD105, RD207, RD181, and RD150 regions. RD group 5 sublineages include ST3, ST10 ST19, and ST22; these isolates show deletion of the RD105, RD207, RD181, and RD142 regions. RD group 6 sublineages include ST10 and ST19; these isolates show deletion of the RD105, RD207, RD181, RD142, and RD150 regions. It has been suggested that insertion sequence- (IS-) mediated deletion events are an important mechanism driving mycobacterial genome variation. Based on our results (Figure 2), the RD105 and RD207 deletions appear to have been early events in the evolutionary history of Beijing strains; however, the IS6110 insertion occurred after the RD181 deletion but has not always persisted in later sublineage evolution. Thus, neither of the RD type 1 and type 2 groups (which include ST26 and ST11) have an IS6110 insert in the NTF region (N family). We still found some characteristics of ancient Beijing strains (N family) in ST19, ST10, and ST22.
Figure 2.
Scheme of the proposed evolution of Beijing lineages. The scheme is based on the deletion of genomic regions (RD: region of difference, shown in gray rectangles), and types of sequence (ST) designations from the studies of Filliol et al. [23] and Iwamoto et al. [24].
Figure 1 illustrates the proposed origins and routes of spread of four strains of MTB in Taiwan.
Route 1 —
The Beijing strain may have migrated to Taiwan through two separate historic events: the first during the Ming dynasty and the second wave shortly after World War II. Through these two migrations, the ancient Beijing strain has evolved into the modern Beijing strain.
Route 2 —
Haarlem originated in the Netherlands. It migrated to Taiwan during the Dutch reign over the island in the 16th century and continues to be a major strain here. It is also important to note that there has been no observed genetic mutation in the strain that was passed onto the natives of Taiwan. The Haarlem strain that remained in the Netherlands, however, has mutations in the ogt and mgtC genes, thus, resulting in SNP variants.
Route 3 —
LAM originated in both Europe and the Americas. It may have migrated to Taiwan during the Portuguese reign in the 16th century and been passed on to the natives of Taiwan.
Route 4 —
EAI originated in Taiwanese aborigines, entering Taiwan four thousand years ago. It may be closely associated with the Austronesian culture. The Austronesian peoples are a population in Oceania and Southeast Asia who speak languages of the Austronesian family. They include Taiwanese aborigines; the majority ethnic groups of East Timor, Indonesia, Malaysia, the Philippines, Brunei, Madagascar, Micronesia, and Polynesia; the Polynesian peoples of New Zealand and Hawaii and the Austronesian peoples of Melanesia.
Problems are remaining to be solved. Molecular genetic analysis of clinical MTB strains delineates relationships among closely related strains of pathogenic microbes and allows construction of genetic frameworks for examining the distribution of biomedically relevant traits such as virulence, transmissibility, and host range. Based on the strain distribution in different ethnic populations, we will attempt to identify factors that determine the disease transmission. Comparative genomic hybridization (CGH) microarray chips will be designed based on the genomic sequence to conduct the population genetic study efficiently. The information we provided in this paper will help us to better understand the dynamics of TB transmission in Taiwan and hence is a good model to understand the global distribution of MTB strains among different geographic regions and ethnic populations.
Conflict of Interests
The authors declare no conflict of interests.
Acknowledgments
This project was supported by grants from the National Health Research Institutes. All participants of this consortium are acknowledged for valuable discussions.
References
- 1.Dou HY, Tseng FC, Lin CW, et al. Molecular epidemiology and evolutionary genetics of Mycobacterium tuberculosis in Taipei. BMC Infectious Diseases. 2008;8, article 170 doi: 10.1186/1471-2334-8-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Oraganization. Global Tuberculosis Control. Surveillance, Planning, Financing. 2007.
- 3.Mathema B, Kurepina NE, Bifani PJ, Kreiswirth BN. Molecular epidemiology of tuberculosis: current insights. Clinical Microbiology Reviews. 2006;19(4):658–685. doi: 10.1128/CMR.00061-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dou HY, Tseng FC, Lu JJ, et al. Associations of Mycobacterium tuberculosis genotypes with different ethnic and migratory populations in Taiwan. Infection, Genetics and Evolution. 2008;8(3):323–330. doi: 10.1016/j.meegid.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 5.Gagneux S, DeRiemer K, Van T, et al. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(8):2869–2873. doi: 10.1073/pnas.0511240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glynn JR, Whiteley J, Bifani PJ, Kremer K, van Soolingen D. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerging Infectious Diseases. 2002;8(8):843–849. doi: 10.3201/eid0808.020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirsh AE, Tsolaki AG, DeRiemer K, Feldman MW, Small PM. Stable association between strains of Mycobacterium tuberculosis and their human host populations. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(14):4871–4876. doi: 10.1073/pnas.0305627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Supply P, Warren RM, Bañuls AL, et al. Linkage disequilibrium between minisatellite loci supports clonal evolution of Mycobacterium tuberculosis in a high tuberculosis incidence area. Molecular Microbiology. 2003;47(2):529–538. doi: 10.1046/j.1365-2958.2003.03315.x. [DOI] [PubMed] [Google Scholar]
- 9.Supply P, Mazars E, Lesjean S, Vincent V, Gicquel B, Locht C. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Molecular Microbiology. 2000;36(3):762–771. doi: 10.1046/j.1365-2958.2000.01905.x. [DOI] [PubMed] [Google Scholar]
- 10.Agostini HT, Yanagihara R, Davis V, Ryschkewitsch CF, Stoner GL. Asian genotypes of JC virus in Native Americans and in a Pacific Island population: markers of viral evolution and human migration. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(26):14542–14546. doi: 10.1073/pnas.94.26.14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falush D, Wirth T, Linz B, et al. Traces of human migrations in Helicobacter pylori populations. Science. 2003;299(5612):1582–1585. doi: 10.1126/science.1080857. [DOI] [PubMed] [Google Scholar]
- 12.Monot M, Honoré N, Garnier T, et al. On the origin of leprosy. Science. 2005;308(5724):1040–1042. doi: 10.1126/science/1109759. [DOI] [PubMed] [Google Scholar]
- 13.Musser JM, Kroll JS, Granoff DM, et al. Global genetic structure and molecular epidemiology of encapsulated Haemophilus influenzae. Reviews of Infectious Diseases. 1990;12(1):75–111. doi: 10.1093/clinids/12.1.75. [DOI] [PubMed] [Google Scholar]
- 14.Wirth T, Wang X, Linz B, et al. Distinguishing human ethnic groups by means of sequences from Helicobacter pylori: lessons from Ladakh. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(14):4746–4751. doi: 10.1073/pnas.0306629101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu YH, Chen CW, Sun HS, Jou R, Lee JJ, Su IJ. Association of NRAMP 1 gene polymorphism with susceptibility to tuberculosis in Taiwanese aboriginals. Journal of the Formosan Medical Association. 2006;105(5):363–369. doi: 10.1016/S0929-6646(09)60131-5. [DOI] [PubMed] [Google Scholar]
- 16.Jou R, Chiang CY, Huang WL. Distribution of the Beijing family genotypes of Mycobacterium tuberculosis in Taiwan. Journal of Clinical Microbiology. 2005;43(1):95–100. doi: 10.1128/JCM.43.1.95-100.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glynn JR, Kremer K, Borgdorff MW, Rodriguez MP, van Soolingen D. Beijing/W genotype Mycobacterium tuberculosis and drug resistance: European concerted action on new generation genetic markers and techniques for the epidemiology and control of tuberculosis. Emerging Infectious Diseases. 2006;12(5):736–743. doi: 10.3201/eid1205.050400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bifani PJ, Mathema B, Kurepina NE, Kreiswirth BN. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends in Microbiology. 2002;10(1):45–52. doi: 10.1016/s0966-842x(01)02277-6. [DOI] [PubMed] [Google Scholar]
- 19.Brudey K, Driscoll JR, Rigouts L, et al. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiology. 2006;6, article 23 doi: 10.1186/1471-2180-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farnia P, Masjedi MR, Mirsaeidi M, et al. Prevalence of Haarlem I and Beijing types of Mycobacterium tuberculosis strains in Iranian and Afghan MDR-TB patients. Journal of Infection. 2006;53(5):331–336. doi: 10.1016/j.jinf.2005.12.020. [DOI] [PubMed] [Google Scholar]
- 21.Kremer K, van Soolingen D, Frothingham R, et al. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: Interlaboratory study of discriminatory power and reproducibility. Journal of Clinical Microbiology. 1999;37(8):2607–2618. doi: 10.1128/jcm.37.8.2607-2618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.A Brief History of Taiwan—A Sparrow Transformed into a Phoenix. http://www.gio.gov.tw/Taiwan-Website/5-gp/history/
- 23.Filliol I, Driscoll JR, van Soolingen D, et al. Snapshot of moving and expanding clones of Mycobacterium tuberculosis and their global distribution assessed by spoligotyping in an international study. Journal of Clinical Microbiology. 2003;41(5):1963–1970. doi: 10.1128/JCM.41.5.1963-1970.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwamoto T, Yoshida S, Suzuki K, Wada T. Population structure analysis of the Mycobacterium tuberculosis Beijing family indicates an association between certain sublineages and multidrug resistance. Antimicrobial Agents and Chemotherapy. 2008;52(10):3805–3809. doi: 10.1128/AAC.00579-08. [DOI] [PMC free article] [PubMed] [Google Scholar]