Abstract
Novel design solid state bioreactor, FERMSOSTAT, had been evaluated in cellulase production studies using local isolate Aspergillus niger USM AI 1 grown on sugarcane bagasse and palm kernel cake at 1 : 1 (w/w) ratio. Under optimised SSF conditions of 0.5 kg substrate; 70% (w/w) moisture content; 30°C; aeration at 4 L/h · g fermented substrate for 5 min and mixing at 0.5 rpm for 5 min, about 3.4 U/g of Filter paper activity (FPase) was obtained. At the same time, comparative studies of the enzymes production under the same SSF conditions indicated that FPase produced by A. niger USM AI 1 was about 35.3% higher compared to Trichoderma reesei. This shows that the performance of this newly designed SSF bioreactor is acceptable and potentially used as prototype for larger-scale bioreactor design.
1. Introduction
Malaysia spent a lot of money yearly on various types of enzymes for local industries use and research purposes. This was because no attempts have been made to produce the enzymes for commercial using local resources. Solid state fermentation (SSF), an alternative for submerged fermentation for enzyme production, was found to be more favorable, which can be performed under limited financial and labour requirements. Agroindustrial residuals have potentially been used as substrate in SSF not only for enzymes production but also other secondary metabolites. In this aspect, countries like Malaysia with abundant agro-industrial residual especially oil palm industry, will be at great advantage.
In addition, many research activities focused on SSF, which had lead to a wide range of applications not only at laboratory scale [1–5] but also at pilot and industrial scale [6–9]. In fermentation, bioreactor provides control environment for growth and activity of the microorganism that carried out biological reactions [10]. On the other hand, selection of an appropriate type of bioreactor is also crucial since bioreactor is the “heart” of the fermentation process. Furthermore, there are many types of SSF bioreactors and all their performances are different [11].
This study was to produce cellulase enzyme using Aspergillus niger USM AI 1 grown on sugar cane bagasse and palm kernel cake (PKC). The major factors that affect the SSF process for enzyme production have been investigated. These included amount of substrate, moisture content, aeration rate and time, mixing rate, and mixing intensity. At the same time, comparative studies with Trichoderma reesei have been carried out.
2. Materials and Methods
2.1. Microorganism
Aspergillus niger USM AI 1 was used throughout this study for the production of cellulase. It was obtained from Industrial Biotechnology Research Laboratory (IBRL) culture collection, USM Penang. It is a local isolate, which was obtained from the Northern Region of Peninsular Malaysia in year 2002. Trichoderma reseei, which is an established culture for cellulase production was used as a benchmark for the production of cellulase by A. niger USM AI 1. In order to compare the enzymes production, the determined optimum SSF conditions were used by T. reseei on the production of cellulase.
2.2. Solid State Fermentation Condition
2.2.1. Fungus Cultivation for Spore Production and Inoculum Preparation
Fungus spore was obtained by growing the culture on Potato Dextrose Agar (AES, France) under room temperature in medium bottles. The spore was harvested after 4-5 days of cultivation with sterile distilled water containing 0.1% (w/v) Tween 80 [12]. The spore suspension was passed through a 0.5 mm sieve to eliminate mycelia and the spore concentration was estimated by direct microscopic counting using hemocytometer [13]. The spore suspension at 1 × 108 spore/ml and volume of 20% (v/w) was mixed with the prior autoclaved growth medium. This medium was used as inoculum for SSF process.
2.2.2. Substrates Preparation and Sterilization
A mixture of sugarcane bagasse (≤2 mm) and palm kernel cake (≤0.5 mm) at 1:1 ratio was used as substrates for FPase production. The substrates were thoroughly mixed by hand prior to transfer into a newly developed fermenter, namely, FERMSOSTAT. The substrate was sterilized in situ by hot air sterilization at 130°C for 3 hours [14]. After the sterilization process, the fermenter and substrates were allowed to cool down to ambient temperature for overnight before inoculation of the substrates were carried out.
2.2.3. Medium Composition
The growth medium used in SSF process consisted of mineral salts and trace elements. The composition of the mineral salts was composed of (g/L); NH4NO3, 5.0; KH2PO4, 5.0; Corn steep liquor, 2.0; NaCl, 1.0; MgSO4.7H2O, 1.0 and trace elements (mg/L); FeSO4.7H2O, 5.0; MnSO4.4H2O, 1.60; ZnSO4.7H2O, 3.45; CoCl2, 2.0. The pH was adjusted to 7.0 prior to autoclave [15]. The pH of the growth medium was not controlled during the course of fermentation.
2.2.4. Inoculation Process
Air/water pressure type of spraying nozzles was used to spray the spore suspension and growth medium over the solid substrate. Continuous mixing of substrate by impeller was carried out during the inoculation. The air supplied was discontinued after all the spore suspension was sprayed on the substrates. However, mixing was continued for another 15 min in order to allow the absorption of the growth medium by the solid substrate.
2.2.5. Sampling Process
The growth profile of A. niger USM AI 1 and enzymes production was carried out for 6 to 7 days. About 10 g of substrate was sampled out from each of the sampling port in every 24 h. The substrate was mixed for 5 min prior to sampling process. The sample was used to determine the enzyme production and glucosamine content.
2.2.6. Enzymes Extraction
The enzyme was extracted by mixing the fermented substrates with distilled water containing 0.1% (w/v) Tween 80. The mixture was left for 2 hours in room temperature. The solid residue was separated from the enzymic solution by filtering through Whatman No. 1 filter paper [16].
2.2.7. Determination of FPase Activity
The cellulase activity is measured as FPase Activity Units. A 1 × 6 cm strip of Whatman number 1 filter paper was added to a total volume of 1.5 ml of culture filtrate in 50 mM citrate buffer (pH 4.5). The samples were incubated for 1 h at 60°C. The hydrolysis was terminated by addition of 3 ml of DNS solution, followed by 5 min boiling. After cooling, 20 ml of distilled water were added and the absorbance was read at 540 nm using glucose as standard [17].
One unit of enzyme activity was defined as the amount of enzymes, which catalyzes the liberation of 1 μmol of reducing sugar per minute under specified assay condition. Enzyme activity was expressed as unit (U) per g of fermented substrate. Enzymes productivity was expressed as unit (U) per mg of glucosamine content of the fungal growth. All assays were carried out in triplicate and the results were presented as mean of the triplicate experiments.
2.2.8. Determination of Fungal Growth
The growth of A. niger USM AI 1 was examined determining the glucosamine content of the fungus as described by Swift [18]. The glucosamine content was measured spectrophotometrically at 530 nm using glucosamine as standard.
2.2.9. Optimization of FPase Production
Factors that affect the growth of A. niger USM AI 1 and the production of FPase was determined, which included amount of substrates, moisture content, incubation temperature, aeration rate and aeration time, mixing rate, and mixing intensity. The optimum condition obtained from each experiment was used unless otherwise stated.
3. Results and Discussions
3.1. Effect of Amount Substrate
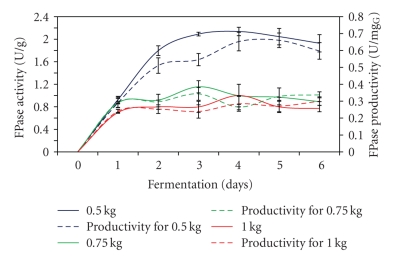
The highest FPase of 2.14 U/g was obtained after 4 days of fermentation process by using 0.5 kg of substrate with about 0.66 U/mgG of FPase productivity (Figure 1). As shown in the figure, about 50% decrease in FPase activity was detected when 0.75 kg and 1.0 kg of substrate were used in the fermentation process. While Roussos et al. [19] reported about 56% reduction in FPase production when the amount substrate used in Zymotis was increased from 4 kg to 12 kg. On the other hand, according to Tao et al. [20], 11.0 U/g and 4.0 U/g of FPase activity were detected when 2 cm and 10 cm of substrate bed depths were used in SSF tray system, respectively. Significant reduction in enzymes production was detected with an increasing use of amount of substrate in the fermentation process. During sterilization, the depth of the substrate for 0.5 kg was 4 cm compared to 6 cm when 1.0 kg of substrate was used. The chances of each particle that attained 130C during the 3 h sterilization for the 6 cm depth (1 kg) substrate was relatively less than the 4 cm depth (0.5 kg) substrate. This may be due to the limitation of heat transfer especially at the center of the bed. This might be owing to the fact that sterilization cum substrate pretreatment makes the substrate more amenable to microbial attack [21].
Figure 1.
Effect of different amount of substrates on FPase production.
3.2. Effect of Moisture Content
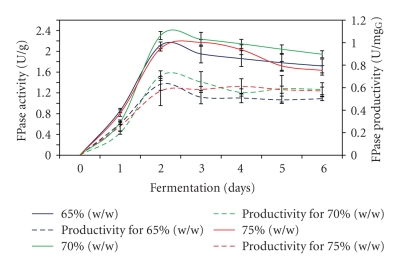
Moisture content was a crucial factor in any SSF process because this variable had influenced growth and biosynthesis of microbe as well as secretion of different metabolites such as enzymes. The result demonstrated that FPase reaching maximum level after 2 days of fermentation process using 65% and 70% (w/w) of moisture content compared with 3 days when 75% (w/w) of moisture content was used (Figure 2). Under the optimum moisture content of 70% (w/w), the highest FPase activity of 2.3 U/g and FPase productivity of 0.71 U/mgG glucosamine were gained. On the other hand, Krishna [22] and Chandra et al. [23] obtained the highest FPase activities of 2.8 U/g and 2.9 U/g under optimum moisture content of 70% and 50% (w/w), respectively. The result obtained showed no marginal differences in FPase activity at different levels of moisture content used in the fermentation process.
Figure 2.
Effect of different moisture contents on FPase production.
3.3. Effect of Incubation Temperature
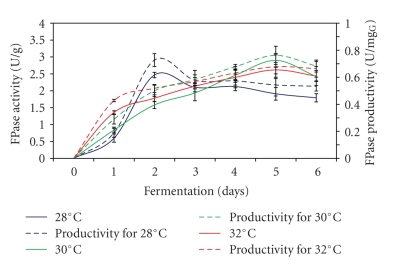
The effect of different incubation temperature on FPase production and productivity is shown in Figure 3. Similar pattern of enzyme production and productivity profiles were exhibited for all the different incubation temperatures used. FPase production achieved maximum level after 5 days of fermentation process at 30°C and 32°C compared to 2 days incubated at 28°C. Under optimum incubation temperature (32°C), the highest FPase activity of 2.9 U/g was obtained with the enzyme productivity of 0.76 U/mgG glucosamine. The result obtained was in agreement with Alam et al. [24], who observed the highest FPase activity (8.2 U/g) when carried out SSF at 32°C using Trichoderma harzianum T2008 grown on empty fruit bunches for 4 days in Erlenmeyer flask. As shown in the figure, the different incubation temperatures used in the fermentation process had showed no great influence on FPase production. This possibly due to the narrow range of incubation temperatures used.
Figure 3.
Effect of different incubation temperatures on FPase production.
3.4. Effect of Aeration Rate
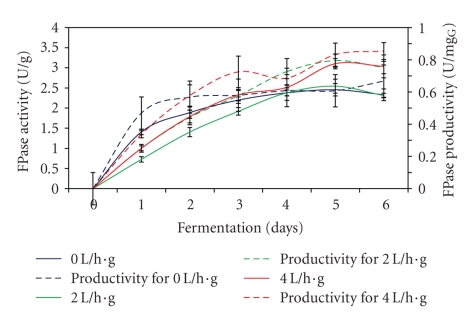
The FPase production reaching maximum level after 5 days regardless of the different aeration used during the fermentation process (Figure 4). Aeration at 4 L/h·g fermented substrate was needed for optimum FPase production with 3.0 U/g of activity and 0.83 U/mgG glucosamine of productivity. Meanwhile, no aeration and aeration at 2 L/h·g fermented substrate shown no or slight differences in the enzyme production. On the other hand, only 0.39 U/g higher in FPase production was observed when aeration used in the fermentation process was increased from 0.06 L/h·g to 0.12 L/h·g wheat straw [25]. Furthermore, Alam et al., [24] obtained FPase activity of 10.1 U/g when carried out SSF process at very low airflow rate of 0.04 L/h·g empty fruit bunches. The optimum aeration in SSF processes depends on the nature of the microorganism used, the oxygen requirements for product synthesis, the amount of heat to be removed from the mass, the degree to which carbon dioxide and other volatile metabolites must be eliminated, the thickness and the available air spaces of the substrate [21]. It can be concluded that aeration has strong influence on FPase production, in which by increasing aeration from 2 L/h·g to 4 L/h·g fermented substrate, the FPase production was increased about 22%.
Figure 4.
Effect of different aeration rates on FPase production.
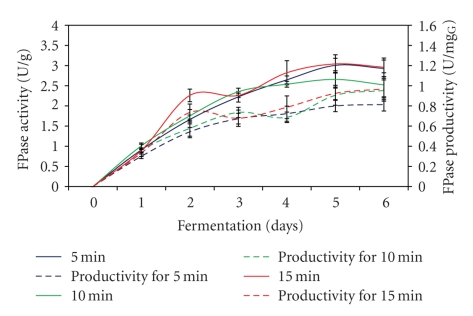
3.5. Effect of Aeration Time
The effect of different aeration times on FPase production and productivity is shown in Figure 5. As demonstrated in the figure, FPase enzyme required 5 fermentation days to reach maximum level regardless of the different aeration times used during the fermentation process. In addition, no difference in FPase production was observed when 5 and 15 min of aeration time were used compared to 10 min which showed marginal differences. Fifteen minutes of aeration time showed optimum FPase production of 3.04 U/g followed by 3.01 U/g and 2.66 U/g when 5 min and 10 min of aeration time were used during the fermentation process, respectively. As also indicated in the figure, optimum aeration time (15 min) also resulted in the highest FPase productivity of 0.97 U/mgG glucosamine. While Mo et al. [26] obtained the highest FPase activity of 17.7 U/g under optimum airflow rate of 7 L/min for 15 min, the production dropped to 16.1 U/g when higher airflow rate (10 L/min) was used. In addition, the result obtained in present work was not similar to Milagres et al. [27]. The authors reported that higher aeration rate used in the fermentation process resulted in a decline in enzyme production. This is mostly due to the fact that rapidly decreased moisture level with higher air supply would occur.
Figure 5.
Effect of different aeration time on FPase production.
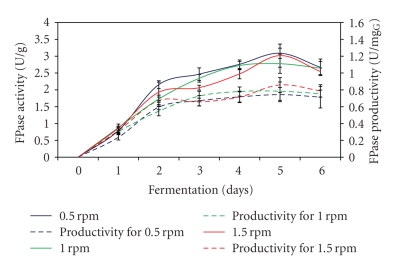
3.6. Effect of Mixing Rate
The result indicated that for all different mixing rates used in the fermentation process, the enzyme production increased rapidly and reached maximum level after 5 days of the fermentation process (Figure 6). As also shown in the figure, mixing at 0.5 rpm for 5 min gives the highest FPase activity of 3.09 U/g and productivity of 0.84 U/mgG glucosamine. On the other hand, Kalogeris et al. [28] obtained FPase activity of about 4.3 U/g when the SSF process was carried out with mixing at 10 rpm for 1 min at 3 hours interval. No strong effect of mixing rate on FPase production was observed in this study. This was because FPase production achieved maximum level after 5 days regardless of the different mixing rates used in the fermentation process. This probably is due to narrow range (0.5 rpm interval) of mixing rate used during the fermentation process.
Figure 6.
Effect of different mixing rates on FPase production.
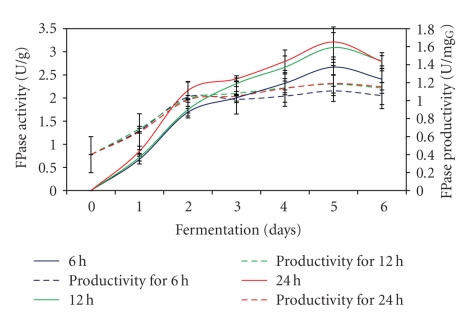
3.7. Effect of Mixing Intensity
The FPase production and productivity reaching maximum level after 5 days of the fermentation process regardless of the difference in the mixing intensity used (Figure 7). At the same time, the FPase production increased with decreasing the mixing intensity used in the fermentation process. The highest FPase activity and productivity of 3.21 U/g and 0.83 U/mgG glucosamine were detected when mixing intensity of every 24 h interval was used during the fermentation process. However, the enzyme production dropped about 17% to 2.67 U/g when 6 h interval of mixing intensity was applied during the fermentation process. Different results of agitation on microorganism growth and productivity in SSF have been reported by different authors but intermittent agitation often gives positive result [14, 29–33]. Therefore, it is necessary to study the effect of agitation for a particular SSF process. The result obtained in this study indicated that higher mixing intensity resulted in a decrease in enzymes production. This may be due to disruption of fungal attachment to the solid substrates and damage to fungal mycelial due to shear forces [34].
Figure 7.
Effect of different mixing intensities on FPase production.
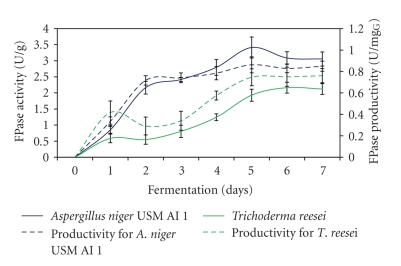
3.8. Comparison of Cellulases Production with Trichoderma reesei
FPase produced by A. niger USM AI 1 reaching maximum level after 5 days of fermentation process but after 6 days when the enzyme was produced by T. reesei (Figure 8). However, these findings were not similar to Chandra et al. [23] and Tao et al. [20], who obtained maximum level of FPase after 3 days of the fermentation process using Aspergillus sp. and Trichoderma sp., respectively. Present work shows that marginal differences in FPase produced by both fungal were observed, in which the highest FPase activity of 3.4 U/g and 2.2 U/g was produced by A. niger USM AI 1 and T. reesei, respectively. On the other hand, highest FPase productivity of 0.89 U/mgG and 0.78 U/mgG glucosamine were obtained by A. niger USM AI 1 and T. reesei after 7 days of the fermentation process, respectively. In can be concluded that enzyme produced by A. niger USM AI 1 required shorter fermentation days to achieve maximum level compared to enzymes produced by T. reesei. This is probably due to slow growth of T. reesei compared to A. niger USM AI 1.
Figure 8.
Production of FPase enzyme by A. niger USM AI 1 and T. reesei under optimized fermentation conditions.
4. Conclusions
In conclusion, under optimum SSF conditions of 0.5 kg substrate; 70% (w/w) moisture content; 30°C; aeration at 4 L/h·g fermented substrate for 5 min and mixing at 0.5 rpm for 5 min, about 3.4 U/g of FPase was produced by A. niger USM AI 1. At the same time, under the above same SSF conditions, about 2.2 U/g of FPase activity was obtained from T. reesei. Although the enzymes produced by T. reesei were lower compared to A. niger USM AI 1, it must keep in mind that the SSF conditions used for the enzymes production was optimum for A. niger USM AI 1 but not for T. reesei. Therefore, further optimization of SSF conditions need to be carried out for T. reesei in order to confirm which fungus was superior for FPase production using novel developed SSF bioreactor, FERMSOSTAT.
Acknowledgments
This work was mainly financed by Intensive Research Priority Area (IRPA) grant from Ministry of Science, Technology and The Environment, Malaysia. The author (C. K. Lee) would like to express his greatest appreciation and gratitude to Professor Dr. Darah Ibrahim and Professor Ibrahim Che Omar for their unlimited advice support, invaluable help, support and guidance. Moreover, the author would like to thanks USM for giving him USM fellowship.
References
- 1.Gupte A, Madamwar D. Solid state fermentation of lignocellulosic waste for cellulase and β-glucosidase production by cocultivation of Aspergillus ellipticus and Aspergillus fumigatus. Biotechnology Progress. 1997;13(2):166–169. [Google Scholar]
- 2.Gutierrez-Correa M, Tengerdy RR. Xylanase production by fungal mixed culture solid substrate fermentation on sugar cane bagasse. Biotechnology Letters. 1998;20(1):45–47. [Google Scholar]
- 3.Hang YD, Woodams EE. Production of citric acid from corncobs by Aspergillus niger. Bioresource Technology. 1998;65(3):251–253. [Google Scholar]
- 4.Kotwal SM, Gote MM, Sainkar SR, Khan MI, Khire JM. Production of α-galactosidase by thermophilic fungus Humicola sp. in solid-state fermentation and its application in soyamilk hydrolysis. Process Biochemistry. 1998;33(3):337–343. [Google Scholar]
- 5.Sekar C, Balaraman K. Optimization studies on the production of cyclosporin A by solid state fermentation. Bioprocess Engineering. 1998;18(4):293–296. [Google Scholar]
- 6.Durand A, Chereau D. A new pilot reactor for solid-state fermentation: application to the protein enrichment of sugar beet pulp. Biotechnology and Bioengineering. 1988;31(5):476–486. doi: 10.1002/bit.260310513. [DOI] [PubMed] [Google Scholar]
- 7.Xue M, Liu D, Zhang H, Qi H, Lei Z. A pilot process of solid state fermentation from sugar beet pulp for the production of microbial protein. Journal of Fermentation and Bioengineering. 1992;73(3):203–205. [Google Scholar]
- 8.Durand A, Renaud R, Maratray J, Almanza S, Diez M. INRA-Dijon reactors for solid state fermentation: designs and applications. Journal of Scientific and Industrial Research. 1996;55(5-6):317–332. [Google Scholar]
- 9.Fernández M, Pérez-Correa JR, Solar I, Agosin E. Automation of a solid substrate cultivation pilot reactor. Bioprocess Engineering. 1996;16(1):1–4. [Google Scholar]
- 10.Pandey A. Aspects of fermenter design for solid-state fermentations. Process Biochemistry. 1991;26(6):355–361. [Google Scholar]
- 11.Mitchell DA, Berovic M, Krieger N. Introduction to solid state fermentation bioreactors. In: Mitchell DA, Krieger N, Berovi M, editors. Solid State Bioreactors: Fundamentals of Design and Operation. Berlin, Germany: Springer; 2006. pp. 33–44. [Google Scholar]
- 12.Smits JP, Rinzema A, Tramper J, Van Sonsbeek HM, Knol W. Solid-state fermentation of wheat bran by Trichoderma reesei QM9414: substrate composition changes, C balance, enzyme production, growth and kinetics. Applied Microbiology and Biotechnology. 1996;46(5-6):489–496. [Google Scholar]
- 13.Raimbault M, Alazard D. Culture method to study fungal growth in solid fermentation. European Journal of Applied Microbiology and Biotechnology. 1980;9(3):199–209. [Google Scholar]
- 14.Lonsane BK, Saucedo-Castaneda G, Raimbault M, et al. Scale-up strategies for solid state fermentation systems. Process Biochemistry. 1992;27(5):259–273. [Google Scholar]
- 15.Kheng PP, Darah I, Poppe L, Szakacs G, Ibrahim CO. Xylanase production by local isolate, Trichoderma spp. FETL C3-2 via solid-state fermentation using agricultural wastes as substrates. Malaysian Journal of Microbiology. 2006;2:7–14. [Google Scholar]
- 16.Aikat K, Bhattacharyya BC. Protease extraction in solid state fermentation of wheat bran by a local strain of Rhizopus oryzae and growth studies by the soft gel technique. Process Biochemistry. 2000;35(9):907–914. [Google Scholar]
- 17.Ghose TK. Measurement of cellulase activities. Pure Applied Chemistry. 1987;59:257–268. [Google Scholar]
- 18.Swift MJ. The estimation of mycelial biomass by determination of the hexosamine content of wood tissue decayed by fungi. Soil Biology and Biochemistry. 1973;5(3):321–332. [Google Scholar]
- 19.Roussos S, Raimbault M, Prebois J-P, Lonsane BK. Zymotis, a large scale solid state fermenter design and evaluation. Applied Biochemistry and Biotechnology. 1993;42(1):37–52. [Google Scholar]
- 20.Tao S, Zuohu L, Deming L. A novel design of solid state fermenter and its evaluation for cellulase production by Trichoderma viride SL-1. Biotechnology Techniques. 1996;10(11):889–894. [Google Scholar]
- 21.Lonsane BK, Childyal NP, Budiatman S, Ramakrishna SV. Engineering aspects of solid state fermentation. Enzyme and Microbial Technology. 1985;7(6):258–265. [Google Scholar]
- 22.Krishna C. Production of bacterial cellulases by solid state bioprocessing of banana wastes. Bioresource Technology. 1999;69(3):231–239. [Google Scholar]
- 23.Chandra MS, Viswanath B, Reddy BR. Cellulolytic enzymes on lignocellulosic substrates in solid state fermentation by Aspergillus niger. Indian Journal of Microbiology. 2007;47(4):323–328. doi: 10.1007/s12088-007-0059-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alam MDZ, Mamun AA, Qudsieh IY, Muyibi SA, Salleh HM, Omar NM. Solid state bioconversion of oil palm empty fruit bunches for cellulase enzyme production using a rotary drum bioreactor. Biochemical Engineering Journal. 2009;46(1):61–64. [Google Scholar]
- 25.Kalogeris E, Iniotaki F, Topakas E, Christakopoulos P, Kekos D, Macris BJ. Performance of an intermittent agitation rotating drum type bioreactor for solid-state fermentation of wheat straw. Bioresource Technology. 2003;86(3):207–213. doi: 10.1016/s0960-8524(02)00175-x. [DOI] [PubMed] [Google Scholar]
- 26.Mo H, Zhang X, Li Z. Control of gas phase for enhanced cellulase production by Penicillium decumbens in solid-state culture. Process Biochemistry. 2004;39(10):1293–1297. [Google Scholar]
- 27.Milagres AMF, Santos E, Piovan T, Roberto IC. Production of xylanase by Thermoascus aurantiacus from sugar cane bagasse in an aerated growth fermentor. Process Biochemistry. 2004;39(11):1387–1391. [Google Scholar]
- 28.Kalogeris E, Fountoukides G, Kekos D, Macris BJ. Design of a solid-state bioreactor for thermophilic microorganisms. Bioresource Technology. 1999;67(3):313–315. [Google Scholar]
- 29.Silman RW. Enzyme formation during solid-substrate fermentation in rotating vessels. Biotechnology and Bioengineering. 1980;22(2):411–420. doi: 10.1002/bit.260220213. [DOI] [PubMed] [Google Scholar]
- 30.Mitchell DA, Doelle HW, Greenfield PF. Improvement of growth of Rhizopus oligosporus on a model solid substrate. Biotechnology Letters. 1988;10(7):497–502. [Google Scholar]
- 31.Kargi F, Curme JA. Solid-state fermentation of sweet sorghum to ethanol in a rotary-drum fermentor. Biotechnology and Bioengineering. 1985;27(8):1122–1125. doi: 10.1002/bit.260270806. [DOI] [PubMed] [Google Scholar]
- 32.Kumar PKR, Lonsane BK. Potential of fed-batch culture in solid state fermentation for production of gibberellic acid. Biotechnology Letters. 1987;9(3):179–182. [Google Scholar]
- 33.Kumar PKR, Lonsane BK. Batch and fed batch solid state fermentation: kinetics of cell growth, hydrolytic enzymes production and gibberellic acid production. Process Biochemistry. 1988;23:43–47. [Google Scholar]
- 34.Lonsane BK, Durand A, Renaud R, et al. Product leaching and downstream processing. In: Doelle HW, Mitchell DA, Rolz CW, editors. Solid Substrate Cultivation. London, UK: Elsevier Science; 1992. pp. 147–171. [Google Scholar]