Abstract
One of the major environmental problems today is hydrocarbon contamination resulting from the activities related to the petrochemical industry. Accidental releases of petroleum products are of particular concern in the environment. Hydrocarbon components have been known to belong to the family of carcinogens and neurotoxic organic pollutants. Currently accepted disposal methods of incineration or burial insecure landfills can become prohibitively expensive when amounts of contaminants are large. Mechanical and chemical methods generally used to remove hydrocarbons from contaminated sites have limited effectiveness and can be expensive. Bioremediation is the promising technology for the treatment of these contaminated sites since it is cost-effective and will lead to complete mineralization. Bioremediation functions basically on biodegradation, which may refer to complete mineralization of organic contaminants into carbon dioxide, water, inorganic compounds, and cell protein or transformation of complex organic contaminants to other simpler organic compounds by biological agents like microorganisms. Many indigenous microorganisms in water and soil are capable of degrading hydrocarbon contaminants. This paper presents an updated overview of petroleum hydrocarbon degradation by microorganisms under different ecosystems.
1. Introduction
Petroleum-based products are the major source of energy for industry and daily life. Leaks and accidental spills occur regularly during the exploration, production, refining, transport, and storage of petroleum and petroleum products. The amount of natural crude oil seepage was estimated to be 600,000 metric tons per year with a range of uncertainty of 200,000 metric tons per year [1]. Release of hydrocarbons into the environment whether accidentally or due to human activities is a main cause of water and soil pollution [2]. Soil contamination with hydrocarbons causes extensive damage of local system since accumulation of pollutants in animals and plant tissue may cause death or mutations [3]. The technology commonly used for the soil remediation includes mechanical, burying, evaporation, dispersion, and washing. However, these technologies are expensive and can lead to incomplete decomposition of contaminants.
The process of bioremediation, defined as the use of microorganisms to detoxify or remove pollutants owing to their diverse metabolic capabilities is an evolving method for the removal and degradation of many environmental pollutants including the products of petroleum industry [4]. In addition, bioremediation technology is believed to be noninvasive and relatively cost-effective [5]. Biodegradation by natural populations of microorganisms represents one of the primary mechanisms by which petroleum and other hydrocarbon pollutants can be removed from the environment [6] and is cheaper than other remediation technologies [7].
The success of oil spill bioremediation depends on one's ability to establish and maintain conditions that favor enhanced oil biodegradation rates in the contaminated environment. Numerous scientific review articles have covered various factors that influence the rate of oil biodegradation [7–12]. One important requirement is the presence of microorganisms with the appropriate metabolic capabilities. If these microorganisms are present, then optimal rates of growth and hydrocarbon biodegradation can be sustained by ensuring that adequate concentrations of nutrients and oxygen are present and that the pH is between 6 and 9. The physical and chemical characteristics of the oil and oil surface area are also important determinants of bioremediation success. There are the two main approaches to oil spill bioremediation: (a) bioaugmentation, in which known oil-degrading bacteria are added to supplement the existing microbial population, and (b) biostimulation, in which the growth of indigenous oil degraders is stimulated by the addition of nutrients or other growth-limiting cosubstrates.
The success of bioremediation efforts in the cleanup of the oil tanker Exxon Valdez oil spill of 1989 [13] in Prince William Sound and the Gulf of Alaska created tremendous interest in the potential of biodegradation and bioremediation technology. Most existing studies have concentrated on evaluating the factors affecting oil bioremediation or testing favored products and methods through laboratory studies [14]. Only limited numbers of pilot scale and field trials have provided the most convincing demonstrations of this technology which have been reported in the peer-reviewed literature [15–18]. The scope of current understanding of oil bioremediation is also limited because the emphasis of most of these field studies and reviews has been given on the evaluation of bioremediation technology for dealing with large-scale oil spills on marine shorelines.
This paper provides an updated information on microbial degradation of petroleum hydrocarbon contaminants towards the better understanding in bioremediation challenges.
2. Microbial Degradation of Petroleum Hydrocarbons
Biodegradation of petroleum hydrocarbons is a complex process that depends on the nature and on the amount of the hydrocarbons present. Petroleum hydrocarbons can be divided into four classes: the saturates, the aromatics, the asphaltenes (phenols, fatty acids, ketones, esters, and porphyrins), and the resins (pyridines, quinolines, carbazoles, sulfoxides, and amides) [19]. Different factors influencing hydrocarbon degradation have been reported by Cooney et al. [20]. One of the important factors that limit biodegradation of oil pollutants in the environment is their limited availability to microorganisms. Petroleum hydrocarbon compounds bind to soil components, and they are difficult to be removed or degraded [21]. Hydrocarbons differ in their susceptibility to microbial attack. The susceptibility of hydrocarbons to microbial degradation can be generally ranked as follows: linear alkanes > branched alkanes > small aromatics > cyclic alkanes [6, 22]. Some compounds, such as the high molecular weight polycyclic aromatic hydrocarbons (PAHs), may not be degraded at all [23].
Microbial degradation is the major and ultimate natural mechanism by which one can cleanup the petroleum hydrocarbon pollutants from the environment [24–26]. The recognition of biodegraded petroleum-derived aromatic hydrocarbons in marine sediments was reported by Jones et al. [27]. They studied the extensive biodegradation of alkyl aromatics in marine sediments which occurred prior to detectable biodegradation of n-alkane profile of the crude oil and the microorganisms, namely, Arthrobacter, Burkholderia, Mycobacterium, Pseudomonas, Sphingomonas, and Rhodococcus were found to be involved for alkylaromatic degradation. Microbial degradation of petroleum hydrocarbons in a polluted tropical stream in Lagos, Nigeria was reported by Adebusoye et al. [28]. Nine bacterial strains, namely, Pseudomonas fluorescens, P. aeruginosa, Bacillus subtilis, Bacillus sp., Alcaligenes sp., Acinetobacter lwoffi, Flavobacterium sp., Micrococcus roseus, and Corynebacterium sp. were isolated from the polluted stream which could degrade crude oil.
Hydrocarbons in the environment are biodegraded primarily by bacteria, yeast, and fungi. The reported efficiency of biodegradation ranged from 6% [29] to 82% [30] for soil fungi, 0.13% [29] to 50% [30] for soil bacteria, and 0.003% [31] to 100% [32] for marine bacteria. Many scientists reported that mixed populations with overall broad enzymatic capacities are required to degrade complex mixtures of hydrocarbons such as crude oil in soil [33], fresh water [34], and marine environments [35, 36].
Bacteria are the most active agents in petroleum degradation, and they work as primary degraders of spilled oil in environment [37, 38]. Several bacteria are even known to feed exclusively on hydrocarbons [39]. Floodgate [36] listed 25 genera of hydrocarbon degrading bacteria and 25 genera of hydrocarbon degrading fungi which were isolated from marine environment. A similar compilation by Bartha and Bossert [33] included 22 genera of bacteria and 31 genera of fungi. In earlier days, the extent to which bacteria, yeast, and filamentous fungi participate in the biodegradation of petroleum hydrocarbons was the subject of limited study, but appeared to be a function of the ecosystem and local environmental conditions [7]. Crude petroleum oil from petroleum contaminated soil from North East India was reported by Das and Mukherjee [40]. Acinetobacter sp. was found to be capable of utilizing n-alkanes of chain length C10–C40 as a sole source of carbon [41]. Bacterial genera, namely, Gordonia, Brevibacterium, Aeromicrobium, Dietzia, Burkholderia, and Mycobacterium isolated from petroleum contaminated soil proved to be the potential organisms for hydrocarbon degradation [42]. The degradation of poly-aromatic hydrocarbons by Sphingomonas was reported by Daugulis and McCracken [43].
Fungal genera, namely, Amorphoteca, Neosartorya, Talaromyces, and Graphium and yeast genera, namely, Candida, Yarrowia, and Pichia were isolated from petroleum-contaminated soil and proved to be the potential organisms for hydrocarbon degradation [42]. Singh [44] also reported a group of terrestrial fungi, namely, Aspergillus, Cephalosporium, and Pencillium which were also found to be the potential degrader of crude oil hydrocarbons. The yeast species, namely, Candida lipolytica, Rhodotorula mucilaginosa, Geotrichum sp, and Trichosporon mucoides isolated from contaminated water were noted to degrade petroleum compounds [45].
Though algae and protozoa are the important members of the microbial community in both aquatic and terrestrial ecosystems, reports are scanty regarding their involvement in hydrocarbon biodegradation. Walker et al. [51] isolated an alga, Prototheca zopfi which was capable of utilizing crude oil and a mixed hydrocarbon substrate and exhibited extensive degradation of n-alkanes and isoalkanes as well as aromatic hydrocarbons. Cerniglia et al. [52] observed that nine cyanobacteria, five green algae, one red alga, one brown alga, and two diatoms could oxidize naphthalene. Protozoa, by contrast, had not been shown to utilize hydrocarbons.
3. Factors Influencing Petroleum Hydrocarbon Degradation
A number of limiting factors have been recognized to affect the biodegradation of petroleum hydrocarbons, many of which have been discussed by Brusseau [53]. The composition and inherent biodegradability of the petroleum hydrocarbon pollutant is the first and foremost important consideration when the suitability of a remediation approach is to be assessed. Among physical factors, temperature plays an important role in biodegradation of hydrocarbons by directly affecting the chemistry of the pollutants as well as affecting the physiology and diversity of the microbial flora. Atlas [54] found that at low temperatures, the viscosity of the oil increased, while the volatility of the toxic low molecular weight hydrocarbons were reduced, delaying the onset of biodegradation.
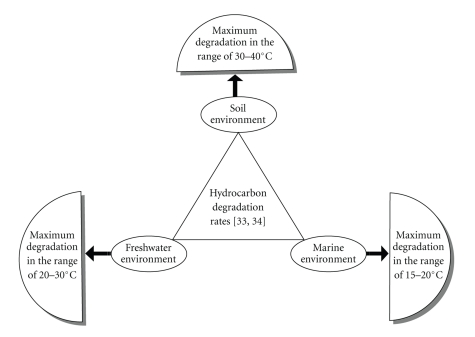
Temperature also affects the solubility of hydrocarbons [62]. Although hydrocarbon biodegradation can occur over a wide range of temperatures, the rate of biodegradation generally decreases with the decreasing temperature. Figure 1 shows that highest degradation rates that generally occur in the range 30–40°C in soil environments, 20–30°C in some freshwater environments and 15–20°C in marine environments [33, 34]. Venosa and Zhu [63] reported that ambient temperature of the environment affected both the properties of spilled oil and the activity of the microorganisms. Significant biodegradation of hydrocarbons have been reported in psychrophilic environments in temperate regions [64, 65].
Figure 1.
Hydrocarbon degradation rates in soil, fresh water, and marine environments.
Nutrients are very important ingredients for successful biodegradation of hydrocarbon pollutants especially nitrogen, phosphorus, and in some cases iron [34]. Some of these nutrients could become limiting factor thus affecting the biodegradation processes. Atlas [35] reported that when a major oil spill occurred in marine and freshwater environments, the supply of carbon was significantly increased and the availability of nitrogen and phosphorus generally became the limiting factor for oil degradation. In marine environments, it was found to be more pronounced due to low levels of nitrogen and phosphorous in seawater [36]. Freshwater wetlands are typically considered to be nutrient deficient due to heavy demands of nutrients by the plants [66]. Therefore, additions of nutrients were necessary to enhance the biodegradation of oil pollutant [67, 68]. On the other hand, excessive nutrient concentrations can also inhibit the biodegradation activity [69]. Several authors have reported the negative effects of high NPK levels on the biodegradation of hydrocarbons [70, 71] especially on aromatics [72]. The effectiveness of fertilizers for the crude oil bioremediation in subarctic intertidal sediments was studied by Pelletier et al. [64]. Use of poultry manure as organic fertilizer in contaminated soil was also reported [73], and biodegradation was found to be enhanced in the presence of poultry manure alone. Maki et al. [74] reported that photo-oxidation increased the biodegradability of petroleum hydrocarbon by increasing its bioavailability and thus enhancing microbial activities.
4. Mechanism of Petroleum Hydrocarbon Degradation
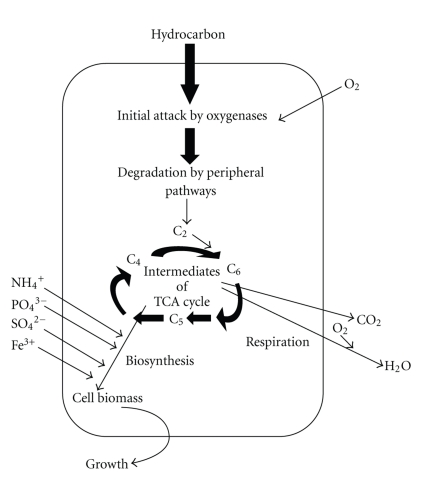
The most rapid and complete degradation of the majority of organic pollutants is brought about under aerobic conditions. Figure 2 shows the main principle of aerobic degradation of hydrocarbons [75]. The initial intracellular attack of organic pollutants is an oxidative process and the activation as well as incorporation of oxygen is the enzymatic key reaction catalyzed by oxygenases and peroxidases. Peripheral degradation pathways convert organic pollutants step by step into intermediates of the central intermediary metabolism, for example, the tricarboxylic acid cycle. Biosynthesis of cell biomass occurs from the central precursor metabolites, for example, acetyl-CoA, succinate, pyruvate. Sugars required for various biosyntheses and growth are synthesized by gluconeogenesis.
Figure 2.
Main principle of aerobic degradation of hydrocarbons by microorganisms.
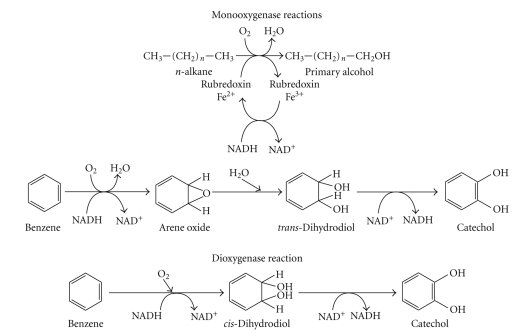
The degradation of petroleum hydrocarbons can be mediated by specific enzyme system. Figure 3 shows the initial attack on xenobiotics by oxygenases [75]. Other mechanisms involved are (1) attachment of microbial cells to the substrates and (2) production of biosurfactants [76]. The uptake mechanism linked to the attachment of cell to oil droplet is still unknown but production of biosurfactants has been well studied.
Figure 3.
Enzymatic reactions involved in the processes of hydrocarbons degradation.
5. Enzymes Participating in Degradation of Hydrocarbons
Cytochrome P450 alkane hydroxylases constitute a super family of ubiquitous Heme-thiolate Monooxygenases which play an important role in the microbial degradation of oil, chlorinated hydrocarbons, fuel additives, and many other compounds [77]. Depending on the chain length, enzyme systems are required to introduce oxygen in the substrate to initiate biodegradation (Table 1). Higher eukaryotes generally contain several different P450 families that consist of large number of individual P450 forms that may contribute as an ensemble of isoforms to the metabolic conversion of given substrate. In microorganisms such P450 multiplicity can only be found in few species [78]. Cytochrome P450 enzyme systems was found to be involved in biodegradation of petroleum hydrocarbons (Table 1). The capability of several yeast species to use n-alkanes and other aliphatic hydrocarbons as a sole source of carbon and energy is mediated by the existence of multiple microsomal Cytochrome P450 forms. These cytochrome P450 enzymes had been isolated from yeast species such as Candida maltosa, Candida tropicalis, and Candida apicola [79]. The diversity of alkaneoxygenase systems in prokaryotes and eukaryotes that are actively participating in the degradation of alkanes under aerobic conditions like Cytochrome P450 enzymes, integral membrane di-iron alkane hydroxylases (e.g., alkB), soluble di-iron methane monooxygenases, and membrane-bound copper containing methane monooxygenases have been discussed by Van Beilen and Funhoff [80].
Table 1.
Enzymes involved in biodegradation of petroleum hydrocarbons.
| Enzymes | Substrates | Microorganisms | References |
|---|---|---|---|
| Soluble Methane Monooxygenases |
C1–C8 alkanes alkenes and cycloalkanes | Methylococcus | |
| Methylosinus | |||
| Methylocystis | McDonald et al. [46] | ||
| Methylomonas | |||
| Methylocella | |||
| Particulate Methane Monooxygenases |
C1–C5 (halogenated) alkanes and cycloalkanes | Methylobacter | |
| Methylococcus, | McDonald et al. [46] | ||
| Methylocystis | |||
| AlkB related Alkane Hydroxylases |
C5–C16 alkanes, fatty acids, alkyl benzenes, cycloalkanes and so forth | Pseudomonas | |
| Burkholderia | Jan et al. [47] | ||
| Rhodococcus, | |||
| Mycobacterium | |||
| Eukaryotic P450 | C10–C16 alkanes, fatty acids | Candida maltosa | |
| Candida tropicalis | Iida et al. [48] | ||
| Yarrowia lipolytica | |||
| Bacterial P450 oxygenase system |
C5–C16 alkanes, cycloalkanes | Acinetobacter | |
| Caulobacter | Van Beilen et al. [49] | ||
| Mycobacterium | |||
| Dioxygenases | C10–C30 alkanes | Acinetobacter sp. | Maeng et al. [50] |
6. Uptake of Hydrocarbons by Biosurfactants
Biosurfactants are heterogeneous group of surface active chemical compounds produced by a wide variety of microorganisms [57, 58, 60, 81–83]. Surfactants enhance solubilization and removal of contaminants [84, 85]. Biodegradation is also enhanced by surfactants due to increased bioavailability of pollutants [86]. Bioremediation of oil sludge using biosurfactants has been reported by Cameotra and Singh [87]. Microbial consortium consisting of two isolates of Pseudomonas aeruginosa and one isolate Rhodococcus erythropolis from soil contaminated with oily sludge was used in this study. The consortium was able to degrade 90% of hydrocarbons in 6 weeks in liquid culture. The ability of the consortium to degrade sludge hydrocarbons was tested in two separate field trials. In addition, the effect of two additives (a nutrient mixture and a crude biosurfactant preparation on the efficiency of the process was also assessed. The biosurfactant used was produced by a consortium member and was identified as being a mixture of 11 rhamnolipid congeners. The consortium degraded 91% of the hydrocarbon content of soil contaminated with 1% (v/v) crude oil sludge in 5 weeks. Separate use of any one additive along with the consortium brought about a 91–95% depletion of the hydrocarbon content in 4 weeks, with the crude biosurfactant preparation being a more effective enhancer of degradation. However, more than 98% hydrocarbon depletion was obtained when both additives were added together with the consortium. The data substantiated the use of a crude biosurfactant for hydrocarbon remediation.
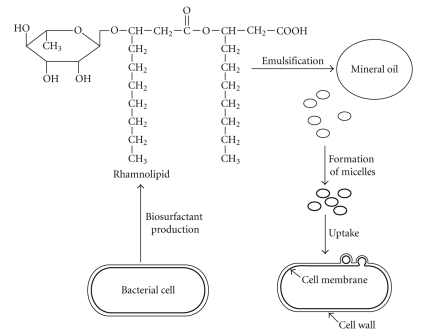
Pseudomonads are the best known bacteria capable of utilizing hydrocarbons as carbon and energy sources and producing biosurfactants [37, 87–89]. Among Pseudomonads, P. aeruginosa is widely studied for the production of glycolipid type biosurfactants. However, glycolipid type biosurfactants are also reported from some other species like P. putida and P. chlororaphis. Biosurfactants increase the oil surface area and that amount of oil is actually available for bacteria to utilize it [90]. Table 2 summarizes the recent reports on biosurfactant production by different microorganisms. Biosurfactants can act as emulsifying agents by decreasing the surface tension and forming micelles. The microdroplets encapsulated in the hydrophobic microbial cell surface are taken inside and degraded. Figure 4 demonstrates the involvement of biosurfactant (rhamnolipids) produced by Pseudomonas sp. and the mechanism of formation of micelles in the uptake of hydrocarbons [75].
Table 2.
Biosurfactants produced by microorganisms.
| Biosurfactants | Microorganisms |
|---|---|
| Sophorolipids | Candida bombicola (Daverey and Pakshirajan, [55]) |
| Rhamnolipids | Pseudomonas aeruginosa (Kumar et al. [56]) |
| Lipomannan | Candida tropicalis (Muthuswamy et al. [57]) |
| Rhamnolipids | Pseudomonas fluorescens (Mahmound et al. [58]) |
| Surfactin | Bacillus subtilis (Youssef et al. [59]) |
| Glycolipid | Aeromonas sp. (Ilori et al. [60]) |
| Glycolipid | Bacillus sp. (Tabatabaee et al. [61]) |
Figure 4.
Involvement of biosurfactant (rhamnolipid) produced by Pseudomonas sp in the uptake of hydrocarbons.
7. Biodegradation of Petroleum Hydrocarbons by Immobilized Cells
Immobilized cells have been used and studied for the bioremediation of numerous toxic chemicals. Immobilization not only simplifies separation and recovery of immobilized cells but also makes the application reusable which reduces the overall cost. Wilsey and Bradely [91] used free suspension and immobilized Pseudomonas sp. to degrade petrol in an aqueous system. The study indicated that immobilization resulted in a combination of increased contact between cell and hydrocarbon droplets and enhanced level of rhamnolipids production. Rhamnolipids caused greater dispersion of water-insoluble n-alkanes in the aqueous phase due to their amphipathic properties and the molecules consist of hydrophilic and hydrophobic moieties reduced the interfacial tension of oil-water systems. This resulted in higher interaction of cells with solubilized hydrocarbon droplets much smaller than the cells and rapid uptake of hydrocarbon in to the cells. Diaz et al. [92] reported that immobilization of bacterial cells enhanced the biodegradation rate of crude oil compared to free living cells in a wide range of culture salinity. Immobilization can be done in batch mode as well as continuous mode. Packed bed reactors are commonly used in continuous mode to degrade hydrocarbons. Cunningham et al. [93] used polyvinyl alcohol (PVA) cryogelation as an entrapment matrix and microorganisms indigenous to the site. They constructed laboratory biopiles to compare immobilised bioaugmentation with liquid culture bioaugmentation and biostimulation. Immobilised systems were found to be the most successful in terms of percentage removal of diesel after 32 days.
Rahman et al. [94] conducted an experiment to study the capacity of immobilized bacteria in alginate beads to degrade hydrocarbons. The results showed that there was no decline in the biodegradation activity of the microbial consortium on the repeated use. It was concluded that immobilization of cells are a promising application in the bioremediation of hydrocarbon contaminated site.
8. Commercially Available Bioremediation Agents
Microbiological cultures, enzyme additives, or nutrient additives that significantly increase the rate of biodegradation to mitigate the effects of the discharge were defied as bioremediation agents by U.S.EPA [95]. Bioremediation agents are classified as bioaugmentation agents and biostimulation agents based on the two main approaches to oil spill bioremediation. Numerous bioremediation products have been proposed and promoted by their vendors, especially during early 1990s, when bioremediation was popularized as “the ultimate solution” to oil spills [96].
The U.S. EPA compiled a list of 15 bioremediation agents [95, 97] as a part of the National Oil and Hazardous Substances Pollution Contingency Plan (NCP) Product Schedule, which was required by the Clean Water Act, the Oil Pollution Act of 1990, and the National Contingency Plan (NCP) as shown in Table 3. But the list was modified, and the number of bioremediation agents was reduced to nine.
Table 3.
Bioremediation agents in NCP product schedule (Adapted from USEPA, 2002).
| Name or Trademark | Product Type | Manufacture |
|---|---|---|
| BET BIOPETRO | MC | BioEnviro Tech, Tomball, TX |
| BILGEPRO | NA | International Environmental Products, LLC, Conshohocken, PA. |
| INIPOL EAP 22 | NA | Societe, CECA S.A., France |
| LAND AND SEA | NA | Land and Sea Restoration LLC, San Antonio, TX |
| RESTORATION MICRO-BLAZE | MC | Verde Environmental, Inc., Houston, TX |
| OIL SPILL EATER II | NA/EA | Oil Spill Eater International, Corporation, Dallas, TX |
| OPPENHEIMER FORMULA | MC | Oppenheimer Biotechnology, Inc., Austin, TX |
| PRISTINE SEA II | MC | Marine Systems, Baton Rouge, LA |
| STEP ONE | MC | B & S Research, Inc., Embarrass, MN |
| SYSTEM E.T. 20. | MC | Quantum Environmental Technologies, Inc(QET), La Jolla, CA |
| VB591TMWATER, VB997TMSOIL, AND BINUTRIX | NA | BioNutraTech, Inc., Houston,TX |
| WMI-2000 | MC | WMI International, Inc |
Abbreviations of product type:
MC: Microbial Culture
EA: Enzyme Additive
NA: Nutrient Additive.
Studies showed that bioremediation products may be effective in the laboratory but significantly less so in the field [14, 17, 18, 98]. This is because laboratory studies cannot always simulate complicated real world conditions such as spatial heterogeneity, biological interactions, climatic effects, and nutrient mass transport limitations. Therefore, field studies and applications are the ultimate tests or the most convincing demonstration of the effectiveness of bioremediation products.
Compared to microbial products, very few nutrient additives have been developed and marketed specifically as commercial bioremediation agents for oil spill cleanup. It is probably because common fertilizers are inexpensive, readily available, and have been shown effective if used properly. However, due to the limitations of common fertilizers (e.g., being rapidly washed out due to tide and wave action), several organic nutrient products, such as oleophilic nutrient products, have recently been evaluated and marketed as bioremediation agents. Four agents, namely, Inipol EAP22, Oil Spill Eater II (OSE II), BIOREN 1, and BIOREN 2, listed on the NCP Product Schedule have also been put into this category.
Inipol EAP22 (Societe, CECA S.A., France) is listed on the NCP Product Schedule as a nutrient additive and probably the most well-known bioremediation agent for oil spill cleanup due to its use in Prince William Sound, Alaska. This nutrient product is a microemulsion-containing urea as a nitrogen source, sodium laureth phosphate as a phosphorus source, 2-butoxy-1-ethanol as a surfactant, and oleic acid to give the material its hydrophobicity. The claimed advantages of Inipol EAP22 include (1) preventing the formation of water-in-oil emulsions by reducing the oil viscosity and interfacial tension; (2) providing controlled release of nitrogen and phosphorus for oil biodegradation; (3) exhibiting no toxicity to flora and fauna and good biodegradability [99].
Oil Spill Eater II (Oil Spill Eater International, Corp.) is another nutrient product listed on the NCP Schedule [97]. This product is listed as a nutrient/enzyme additive and consists of “nitrogen, phosphorus, readily available carbon, and vitamins for quick colonization of naturally occurring bacteria”. A field demonstration was carried out at a bioventing site in a Marine Corps Air Ground Combat Center (MCAGCC) in California to investigate the efficacy of OSEII for enhancing hydrocarbon biodegradation in a fuel-contaminated vadose zone [106].
Researchers from European EUREKA BIOREN program conducted a field trial in an estuary environment to evaluate the effectiveness of two bioremediation products (BIOREN 1 and 2) [114, 115]. The two nutrient products were derived from fish meals in a granular form with urea and super phosphate as nitrogen and phosphorus sources and proteinaceous material as the carbon source. The major difference between the two formulations was that BIOREN 1 contained a biosurfactant. The results showed that the presence of biosurfactant in BIOREN 1 was the most active ingredient which contributed to the increase in oil degradation rates whereas BIOREN 2 (without biosurfactant) was not effective in that respect. The biosurfactant could have contributed to greater bioavailability of hydrocarbons to microbial attack.
9. Phytoremediation
Phytoremediation is an emerging technology that uses plants to manage a wide variety of environmental pollution problems, including the cleanup of soils and groundwater contaminated with hydrocarbons and other hazardous substances. The different mechanisms, namely, hydraulic control, phytovolatilization, rhizoremediation, and phytotransformation. could be utilized for the remediation of a wide variety of contaminants.
Phytoremediation can be cost-effective (a) for large sites with shallow residual levels of contamination by organic, nutrient, or metal pollutants, where contamination does not pose an imminent danger and only “polishing treatment” is required; (b) where vegetation is used as a final cap and closure of the site [116].
Advantages of using phytoremediation include cost-effectiveness, aesthetic advantages, and long-term applicability (Table 4). Furthermore, the use of phytoremediation as a secondary or polishing in situ treatment step minimizes land disturbance and eliminates transportation and liability costs associated with offsite treatment and disposal.
Table 4.
Advantages and disadvantages of phytoremediation over traditional technologies.
| Advantages | Disadvantages |
|---|---|
| Relatively low cost | Longer remediation times |
| Easily implemented and maintained | Climate dependent |
| Several mechanisms for removal | Effects to food web might be unknown |
| Environmentally friendly | Ultimate contaminant fates might be unknown |
| Aesthetically pleasing | Results are variable |
| Reduces landfilled wastes | |
| Harvestable plant material |
Research and application of phytoremediation for the treatment of petroleum hydrocarbon contamination over the past fifteen years have provided much useful information that can be used to design effective remediation systems and drive further improvement and innovation. Phytoremediation could be applied for the remediation of numerous contaminated sites. However, not much is known about contaminant fate and transformation pathways, including the identity of metabolites (Table 4). Little data exists on contaminant removal rates and efficiencies directly attributable to plants under field conditions.
The potential use of phytoremediation at a site contaminated with hydrocarbons was investigated. The Alabama Department of Environmental Management granted a site, which involved about 1500 cubic yards of soil of which 70% of the baseline samples contained over 100 ppm of total petroleum hydrocarbon (TPH). After 1 year of vegetative cover, approximately 83% of the samples were found to contain less than 10-ppm TPH. Removal of total petroleum hydrocarbon (TPH) at several field sites contaminated with crude oil, diesel fuel, or petroleum refinery wastes, at initial TPH concentrations of 1,700 to 16,000 mg/kg were also investigated [117, 118]. Plant growth was found to vary depending upon the species. Presence of some species led to greater TPH disappearance than with other species or in unvegetated soil. Among tropical plants tested for use in Pacific Islands, three coastal trees, kou (Cordia subcordata), milo (Thespesia populnea), and kiawe (Prosopis pallida) and the native shrub beach naupaka (Scaevola serica) tolerated field conditions and facilitated cleanup of soils contaminated with diesel fuel [119]. Grasses were often planted with trees at sites with organic contaminants as the primary remediation method. Tremendous amount of fine roots in the surface soil was found to be effective at binding and transforming hydrophobic contaminants such as TPH, BTEX, and PAHs. Grasses were often planted between rows of trees to provide soil stabilization and protection against wind-blown dust that could move contaminants offsite. Legumes such as alfalfa (Medicago sativa), alsike clover (Trifolium hybridum), and peas (Pisum sp.) could be used to restore nitrogen to poor soils. Fescue (Vulpia myuros), rye (Elymus sp.), clover (Trifolium sp.), and reed canary grass (Phalaris arundinacea) were used successfully at several sites, especially contaminated with petrochemical wastes. Once harvested, the grasses could be disposed off as compost or burned.
Microbial degradation in the rhizosphere might be the most significant mechanism for removal of diesel range organics in vegetated contaminated soils [120]. This occurs because contaminants such as PAHs are highly hydrophobic, and their sorption to soil decreases their bioavailability for plant uptake and phytotransformation.
10. Genetically Modified Bacteria
Applications for genetically engineered microorganisms (GEMs) in bioremediation have received a great deal of attention to improve the degradation of hazardous wastes under laboratory conditions. There are reports on the degradation of environmental pollutants by different bacteria. Table 5 shows some examples of the relevant use of genetic engineering technology to improve bioremediation of hydrocarbon contaminants using bacteria. The genetically engineered bacteria showed higher degradative capacity. However, ecological and environmental concerns and regulatory constraints are major obstacles for testing GEM in the field. These problems must be solved before GEM can provide an effective clean-up process at lower cost.
Table 5.
Genetic engineering for biodegradation of contaminants.
| Microorganisms | Modification | Contaminants | Reference |
|---|---|---|---|
| Pseudomonas. putida | pathway | 4-ethylbenzoate | Ramos et al. [100] |
| P. putida KT2442 | pathway | toluene/benzoate | Panke and Sanchezromero [101] |
| Pseudomonas sp.FRI | pathway | chloro-, methylbenzoates | Rojo et al. [102] |
|
Comamonas. testosteroni VP44 |
substrate specificity | o-, p-monochlorobiphenyls | Hrywna et al. [103] |
| Pseudomonas sp. LB400 | substrate specificity | PCB | Erickson and Mondello [104] |
|
P. pseudoalcaligenes KF707-D2 |
substrate specificity | TCE, toluene, benzene | Suyama et al. [105] |
The use of genetically engineered bacteria was applied to bioremediation process monitoring, strain monitoring, stress response, end-point analysis, and toxicity assessment. Examples of these applications are listed in Table 6. The range of tested contaminants included chlorinated compounds, aromatic hydrocarbons, and nonpolar toxicants. The combination of microbiological and ecological knowledge, biochemical mechanisms, and field engineering designs are essential elements for successful in situ bioremediation using genetically modified bacteria.
Table 6.
Application of genetically modified bacteria for assessing the biodegradation process efficiency.
| Microorganisms | Application | Contaminants | Reference |
|---|---|---|---|
| A. eutrophus H850Lr | process monitoring | PCB | Van Dyke et al. [107] |
| P. putida TVA8 | process monitoring | TCE, BTEX | Applegate et al. [108] |
| P. fluorescens HK44 | process monitoring | naphthalene, anthracene | Sayler and Ripp [109] |
| B. cepacia BRI6001L | strain monitoring | 2,4-D | Masson et al. [110] |
| P. fluorescens 10586s/pUCD607 | stress response | BTEX | Sousa et al. [111] |
| Pseudomonas strain Shk1 | toxicity assessment | 2, 4-dinitrophenol hydroquinone | Kelly et al. [112] |
| A. eutrophus 2050 | end point analysis | non polar narcotics | Layton et al. [113] |
11. Conclusion
Cleaning up of petroleum hydrocarbons in the subsurface environment is a real world problem. A better understanding of the mechanism of biodegradation has a high ecological significance that depends on the indigenous microorganisms to transform or mineralize the organic contaminants. Microbial degradation process aids the elimination of spilled oil from the environment after critical removal of large amounts of the oil by various physical and chemical methods. This is possible because microorganisms have enzyme systems to degrade and utilize different hydrocarbons as a source of carbon and energy.
The use of genetically modified (GM) bacteria represents a research frontier with broad implications. The potential benefits of using genetically modified bacteria are significant. But the need for GM bacteria may be questionable for many cases, considering that indigenous species often perform adequately but we do not tap the full potential of wild species due to our limited understanding of various phytoremediation mechanisms, including the regulation of enzyme systems that degrade pollutants.
Therefore, based on the present review, it may be concluded that microbial degradation can be considered as a key component in the cleanup strategy for petroleum hydrocarbon remediation.
References
- 1.Kvenvolden KA, Cooper CK. Natural seepage of crude oil into the marine environment. Geo-Marine Letters. 2003;23(3-4):140–146. [Google Scholar]
- 2.Holliger C, Gaspard S, Glod G, et al. Contaminated environments in the subsurface and bioremediation: organic contaminants. FEMS Microbiology Reviews. 1997;20(3-4):517–523. doi: 10.1111/j.1574-6976.1997.tb00334.x. [DOI] [PubMed] [Google Scholar]
- 3.Alvarez PJJ, Vogel TM. Substrate interactions of benzene, toluene, and para-xylene during microbial degradation by pure cultures and mixed culture aquifer slurries. Applied and Environmental Microbiology. 1991;57(10):2981–2985. doi: 10.1128/aem.57.10.2981-2985.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Medina-Bellver JI, Marín P, Delgado A, et al. Evidence for in situ crude oil biodegradation after the Prestige oil spill. Environmental Microbiology. 2005;7(6):773–779. doi: 10.1111/j.1462-2920.2005.00742.x. [DOI] [PubMed] [Google Scholar]
- 5.April TM, Foght JM, Currah RS. Hydrocarbon-degrading filamentous fungi isolated from flare pit soils in northern and western Canada. Canadian Journal of Microbiology. 2000;46(1):38–49. [PubMed] [Google Scholar]
- 6.Ulrici W. Rehm HJ, Reed G, editors. Contaminant soil areas, different countries and contaminant monitoring of contaminants. Environmental Process II. Soil Decontamination Biotechnology. 2000;11:5–42. [Google Scholar]
- 7.Leahy JG, Colwell RR. Microbial degradation of hydrocarbons in the environment. Microbiological Reviews. 1990;54(3):305–315. doi: 10.1128/mr.54.3.305-315.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zobell CE. Action of microorganisms on hydrocarbons. Bacteriological Reviews. 1946;10:1–49. [PubMed] [Google Scholar]
- 9.Atlas RM. Microbial degradation of petroleum hydrocarbons: an environmental perspective. Microbiological Reviews. 1981;45(1):180–209. doi: 10.1128/mr.45.1.180-209.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Atlas RM, editor. Petroleum Microbiology. New York, NY, USA: Macmillion; 1984. [Google Scholar]
- 11.Atlas RM, Bartha R. Hydrocabon biodegradation and oil spill bioremediation. Advances in Microbial Ecology. 1992;12:287–338. [Google Scholar]
- 12.Foght JM, Westlake DWS. Biodegradation of hydrocarbons in freshwater. In: Vandermeulen JH, Hrudey SR, editors. Oil in Freshwater: Chemistry, Biology, Countermeasure Technology. New York, NY, USA: Pergamon Press; 1987. pp. 217–230. [Google Scholar]
- 13.Atlas RM, Bartha R. Microbial Ecology. 4th edition. San Francisco, Calif, USA: Benjamin/Cummings; 1998. Fundamentals and applications; pp. 523–530. [Google Scholar]
- 14.Mearns AJ. Cleaning oiled shores: putting bioremediation to the test. Spill Science and Technology Bulletin. 1997;4(4):209–217. [Google Scholar]
- 15.Prince RC. Petroleum spill bioremediation in marine environments. Critical Reviews in Microbiology. 1993;19(4):217–242. doi: 10.3109/10408419309113530. [DOI] [PubMed] [Google Scholar]
- 16.Swannell RPJ, Lee K, Mcdonagh M. Field evaluations of marine oil spill bioremediation. Microbiological Reviews. 1996;60(2):342–365. doi: 10.1128/mr.60.2.342-365.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Venosa AD, Suidan MT, Wrenn BA, et al. Bioremediation of an experimental oil spill on the shoreline of Delaware Bay. Environmental Science and Technology. 1996;30(5):1764–1775. [Google Scholar]
- 18.Venosa AD, King DW, Sorial GA. The baffled flask test for dispersant effectiveness: a round Robin evaluation of reproducibility and repeatability. Spill Science and Technology Bulletin. 2002;7(5-6):299–308. [Google Scholar]
- 19.Colwell RR, Walker JD, Cooney JJ. Ecological aspects of microbial degradation of petroleum in the marine environment. Critical Reviews in Microbiology. 1977;5(4):423–445. doi: 10.3109/10408417709102813. [DOI] [PubMed] [Google Scholar]
- 20.Cooney JJ, Silver SA, Beck EA. Factors influencing hydrocarbon degradation in three freshwater lakes. Microbial Ecology. 1985;11(2):127–137. doi: 10.1007/BF02010485. [DOI] [PubMed] [Google Scholar]
- 21.Barathi S, Vasudevan N. Utilization of petroleum hydrocarbons by Pseudomonas fluorescens isolated from a petroleum-contaminated soil. Environment International. 2001;26(5-6):413–416. doi: 10.1016/s0160-4120(01)00021-6. [DOI] [PubMed] [Google Scholar]
- 22.Perry JJ. Microbial metabolism of cyclic alkanes. In: Atlas RM, editor. Petroleum Microbiology. New York, NY, USA: Macmillan; 1984. pp. 61–98. [Google Scholar]
- 23.Atlas R, Bragg J. Bioremediation of marine oil spills: when and when not—the Exxon Valdez experience. Microbial Biotechnology. 2009;2(2):213–221. doi: 10.1111/j.1751-7915.2008.00079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Atlas RM. Encyclopedia of Microbiology. Baltimore, Md, USA: Academic Press; 1992. Petroleum microbiology; pp. 363–369. [Google Scholar]
- 25.Amund OO, Nwokoye N. Hydrocarbon potentials of yeast isolates from a polluted Lagoon. Journal of Scientific Research and Development. 1993;1:65–68. [Google Scholar]
- 26.Lal B, Khanna S. Degradation of crude oil by Acinetobacter calcoaceticus and Alcaligenes odorans. Journal of Applied Bacteriology. 1996;81(4):355–362. doi: 10.1111/j.1365-2672.1996.tb03519.x. [DOI] [PubMed] [Google Scholar]
- 27.Jones DM, Douglas AG, Parkes RJ, Taylor J, Giger W, Schaffner C. The recognition of biodegraded petroleum-derived aromatic hydrocarbons in recent marine sediments. Marine Pollution Bulletin. 1983;14(3):103–108. [Google Scholar]
- 28.Adebusoye SA, Ilori MO, Amund OO, Teniola OD, Olatope SO. Microbial degradation of petroleum hydrocarbons in a polluted tropical stream. World Journal of Microbiology and Biotechnology. 2007;23(8):1149–1159. [Google Scholar]
- 29.Jones J, Knight M, Byron JA. Effect of gross population by kerosene hydrocarbons on the microflora of a moorland soil. Nature. 1970;227:p. 1166. doi: 10.1038/2271166a0. [DOI] [PubMed] [Google Scholar]
- 30.Pinholt Y, Struwe S, Kjoller A. Microbial changes during oil decomposition in soil. Holarctic Ecology. 1979;2:195–200. [Google Scholar]
- 31.Hollaway SL, Faw GM, Sizemore RK. The bacterial community composition of an active oil field in the Northwestern Gulf of Mexico. Marine Pollution Bulletin. 1980;11(6):153–156. [Google Scholar]
- 32.Mulkins Phillips GJ, Stewart JE. Distribution of hydrocarbon utilizing bacteria in Northwestern Atlantic waters and coastal sediments. Canadian Journal of Microbiology. 1974;20(7):955–962. doi: 10.1139/m74-147. [DOI] [PubMed] [Google Scholar]
- 33.Bartha R, Bossert I. The treatment and disposal of petroleum wastes. In: Atlas RM, editor. Petroleum Microbiology. New York, NY, USA: Macmillan; 1984. pp. 553–578. [Google Scholar]
- 34.Cooney JJ. The fate of petroleum pollutants in fresh water ecosystems. In: Atlas RM, editor. Petroleum Microbiology. New York, NY, USA: Macmillan; 1984. pp. 399–434. [Google Scholar]
- 35.Atlas RM. Effects of hydrocarbons on micro-organisms and biodegradation in Arctic ecosystems. In: Engelhardt FR, editor. Petroleum Effects in the Arctic Environment. London, UK: Elsevier; 1985. pp. 63–99. [Google Scholar]
- 36.Floodgate G. The fate of petroleum in marine ecosystems. In: Atlas RM, editor. Petroleum Microbiology. New York, NY, USA: Macmillion; 1984. pp. 355–398. [Google Scholar]
- 37.Rahman KSM, Rahman TJ, Kourkoutas Y, Petsas I, Marchant R, Banat IM. Enhanced bioremediation of n-alkane in petroleum sludge using bacterial consortium amended with rhamnolipid and micronutrients. Bioresource Technology. 2003;90(2):159–168. doi: 10.1016/s0960-8524(03)00114-7. [DOI] [PubMed] [Google Scholar]
- 38.Brooijmans RJW, Pastink MI, Siezen RJ. Hydrocarbon-degrading bacteria: the oil-spill clean-up crew. Microbial Biotechnology. 2009;2(6):587–594. doi: 10.1111/j.1751-7915.2009.00151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yakimov MM, Timmis KN, Golyshin PN. Obligate oil-degrading marine bacteria. Current Opinion in Biotechnology. 2007;18(3):257–266. doi: 10.1016/j.copbio.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 40.Das K, Mukherjee AK. Crude petroleum-oil biodegradation efficiency of Bacillus subtilis and Pseudomonas aeruginosa strains isolated from a petroleum-oil contaminated soil from North-East India. Bioresource Technology. 2007;98(7):1339–1345. doi: 10.1016/j.biortech.2006.05.032. [DOI] [PubMed] [Google Scholar]
- 41.Throne-Holst M, Wentzel A, Ellingsen TE, Kotlar H-K, Zotchev SB. Identification of novel genes involved in long-chain n-alkane degradation by Acinetobacter sp. strain DSM 17874. Applied and Environmental Microbiology. 2007;73(10):3327–3332. doi: 10.1128/AEM.00064-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chaillan F, Le Flèche A, Bury E, et al. Identification and biodegradation potential of tropical aerobic hydrocarbon-degrading microorganisms. Research in Microbiology. 2004;155(7):587–595. doi: 10.1016/j.resmic.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 43.Daugulis AJ, McCracken CM. Microbial degradation of high and low molecular weight polyaromatic hydrocarbons in a two-phase partitioning bioreactor by two strains of Sphingomonas sp. Biotechnology Letters. 2003;25(17):1441–1444. doi: 10.1023/a:1025007729355. [DOI] [PubMed] [Google Scholar]
- 44.Singh H. Mycoremediation: Fungal Bioremediation. New York, NY, USA: Wiley-Interscience; 2006. [Google Scholar]
- 45.Bogusławska-Was E, Dąbrowski W. The seasonal variability of yeasts and yeast-like organisms in water and bottom sediment of the Szczecin Lagoon. International Journal of Hygiene and Environmental Health. 2001;203(5-6):451–458. doi: 10.1078/1438-4639-00056. [DOI] [PubMed] [Google Scholar]
- 46.McDonald IR, Miguez CB, Rogge G, et al. Diversity of soluble methane monooxygenase-containing methanotrophs isolated from polluted environments. FEMS Microbiology Letters. 2006;255(2):225–232. doi: 10.1111/j.1574-6968.2005.00090.x. [DOI] [PubMed] [Google Scholar]
- 47.Jan B, Beilen V, Neuenschwunder M, et al. Rubredoxins involved in alkane degradation. The Journal of Bacteriology. 2003;184(6):1722–1732. doi: 10.1128/JB.184.6.1722-1732.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iida T, Sumita T, Ohta A, Takagi M. The cytochrome P450ALK multigene family of an n-alkane-assimilating yeast, Yarrowia lipolytica: cloning and characterization of genes coding for new CYP52 family members. Yeast. 2000;16(12):1077–1087. doi: 10.1002/1097-0061(20000915)16:12<1077::AID-YEA601>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 49.Van Beilen JB, Funhoff EG, Van Loon A, et al. Cytochrome P450 alkane hydroxylases of the CYP153 family are common in alkane-degrading eubacteria lacking integral membrane alkane hydroxylases. Applied and Environmental Microbiology. 2006;72(1):59–65. doi: 10.1128/AEM.72.1.59-65.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maeng JHO, Sakai Y, Tani Y, Kato N. Isolation and characterization of a novel oxygenase that catalyzes the first step of n-alkane oxidation in Acinetobacter sp. strain M-1. Journal of Bacteriology. 1996;178(13):3695–3700. doi: 10.1128/jb.178.13.3695-3700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walker JD, Colwell RR, Vaituzis Z, Meyer SA. Petroleum degrading achlorophyllous alga Prototheca zopfii. Nature. 1975;254(5499):423–424. [Google Scholar]
- 52.Cerniglia CE, Gibson DT, Van Baalen C. Oxidation of naphthalene by cyanobacteria and microalgae. Journal of General Microbiology. 1980;116(2):495–500. [Google Scholar]
- 53.Brusseau ML. The impact of physical, chemical and biological factors on biodegradation. In: Serra R, editor. In: Proceedings of the International Conference on Biotechnology for Soil Remediation: Scientific Bases and Practical Applications; 1998; Milan, Italy. C.I.P.A. S.R.L.; pp. 81–98. [Google Scholar]
- 54.Atlas RM. Effects of temperature and crude oil composition on petroleum biodegradation. Journal of Applied Microbiology. 1975;30(3):396–403. doi: 10.1128/am.30.3.396-403.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Daverey A, Pakshirajan K. Production of sophorolipids by the yeast Candida bombicola using simple and low cost fermentative media. Food Research International. 2009;42(4):499–504. [Google Scholar]
- 56.Kumar M, León V, De Sisto Materano A, Ilzins OA, Luis L. Biosurfactant production and hydrocarbon-degradation by halotolerant and thermotolerant Pseudomonas sp. World Journal of Microbiology and Biotechnology. 2008;24(7):1047–1057. [Google Scholar]
- 57.Muthusamy K, Gopalakrishnan S, Ravi TK, Sivachidambaram P. Biosurfactants: properties, commercial production and application. Current Science. 2008;94(6):736–747. [Google Scholar]
- 58.Mahmound A, Aziza Y, Abdeltif A, Rachida M. Biosurfactant production by Bacillus strain injected in the petroleum reservoirs. Journal of Industrial Microbiology & Biotechnology. 2008;35:1303–1306. [Google Scholar]
- 59.Youssef N, Simpson DR, Duncan KE, et al. In situ biosurfactant production by Bacillus strains injected into a limestone petroleum reservoir. Applied and Environmental Microbiology. 2007;73(4):1239–1247. doi: 10.1128/AEM.02264-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ilori MO, Amobi CJ, Odocha AC. Factors affecting biosurfactant production by oil degrading Aeromonas spp. isolated from a tropical environment. Chemosphere. 2005;61(7):985–992. doi: 10.1016/j.chemosphere.2005.03.066. [DOI] [PubMed] [Google Scholar]
- 61.Tabatabaee A, Assadi MM, Noohi AA, Sajadian VA. Isolation of biosurfactant producing bacteria from oil reservoirs. Iranian Journal of Environmental Health Science & Engineering. 2005;2(1):6–12. [Google Scholar]
- 62.Foght JM, Westlake DWS, Johnson WM, Ridgway HF. Environmental gasoline-utilizing isolates and clinical isolates of Pseudomonas aeruginosa are taxonomically indistinguishable by chemotaxonomic and molecular techniques. Microbiology. 1996;142(9):2333–2340. doi: 10.1099/00221287-142-9-2333. [DOI] [PubMed] [Google Scholar]
- 63.Venosa AD, Zhu X. Biodegradation of crude oil contaminating marine shorelines and freshwater wetlands. Spill Science and Technology Bulletin. 2003;8(2):163–178. [Google Scholar]
- 64.Pelletier E, Delille D, Delille B. Crude oil bioremediation in sub-Antarctic intertidal sediments: chemistry and toxicity of oiled residues. Marine Environmental Research. 2004;57(4):311–327. doi: 10.1016/j.marenvres.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 65.Delille D, Coulon F, Pelletier E. Effects of temperature warming during a bioremediation study of natural and nutrient-amended hydrocarbon-contaminated sub-Antarctic soils. Cold Regions Science and Technology. 2004;40(1-2):61–70. [Google Scholar]
- 66.Mitsch WJ, Gosselink JG. Wetlands. 2nd edition. New York, NY, USA: John Wiley & Sons; 1993. [Google Scholar]
- 67.Choi S-C, Kwon KK, Sohn JH, Kim S-J. Evaluation of fertilizer additions to stimulate oil biodegradation in sand seashore mesocosms. Journal of Microbiology and Biotechnology. 2002;12(3):431–436. [Google Scholar]
- 68.Kim S-J, Choi DH, Sim DS, Oh Y-S. Evaluation of bioremediation effectiveness on crude oil-contaminated sand. Chemosphere. 2005;59(6):845–852. doi: 10.1016/j.chemosphere.2004.10.058. [DOI] [PubMed] [Google Scholar]
- 69.Chaillan F, Chaîneau CH, Point V, Saliot A, Oudot J. Factors inhibiting bioremediation of soil contaminated with weathered oils and drill cuttings. Environmental Pollution. 2006;144(1):255–265. doi: 10.1016/j.envpol.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 70.Oudot J, Merlin FX, Pinvidic P. Weathering rates of oil components in a bioremediation experiment in estuarine sediments. Marine Environmental Research. 1998;45(2):113–125. [Google Scholar]
- 71.Chaîneau CH, Rougeux G, Yéprémian C, Oudot J. Effects of nutrient concentration on the biodegradation of crude oil and associated microbial populations in the soil. Soil Biology and Biochemistry. 2005;37(8):1490–1497. [Google Scholar]
- 72.Carmichael LM, Pfaender FK. The effect of inorganic and organic supplements on the microbial degradation of phenanthrene and pyrene in soils. Biodegradation. 1997;8(1):1–13. doi: 10.1023/a:1008258720649. [DOI] [PubMed] [Google Scholar]
- 73.Okolo JC, Amadi EN, Odu CTI. Effects of soil treatments containing poultry manure on crude oil degradation in a sandy loam soil. Applied Ecology and Environmental Research. 2005;3(1):47–53. [Google Scholar]
- 74.Maki H, Sasaki T, Haramaya S. Photooxidation of biodegradable crude oil and toxicity of the photooxidized products. Chemosphere. 2005;44:1145–1151. doi: 10.1016/s0045-6535(00)00292-7. [DOI] [PubMed] [Google Scholar]
- 75.Fritsche W, Hofrichter M. Aerobic degradation by microorganisms. In: Klein J, editor. Environmental Processes- Soil Decontamination. Weinheim, Germany: Wiley-VCH; 2000. pp. 146–155. [Google Scholar]
- 76.Hommel RK. Formation and phylogenetic role of biosurfactants. Journal of Applied Microbiology. 1990;89(1):158–119. [Google Scholar]
- 77.Van Beilen JB, Funhoff EG. Alkane hydroxylases involved in microbial alkane degradation. Applied Microbiology and Biotechnology. 2007;74(1):13–21. doi: 10.1007/s00253-006-0748-0. [DOI] [PubMed] [Google Scholar]
- 78.Zimmer T, Ohkuma M, Ohta A, Takagi M, Schunck W-H. The CYP52 multigene family of Candida maltosa encodes functionally diverse n-alkane-inducible cytochromes p450. Biochemical and Biophysical Research Communications. 1996;224(3):784–789. doi: 10.1006/bbrc.1996.1100. [DOI] [PubMed] [Google Scholar]
- 79.Scheuer U, Zimmer T, Becher D, Schauer F, Schunck W-H. Oxygenation cascade in conversion of n-alkanes to α,ω-dioic acids catalyzed by cytochrome P450 52A3. Journal of Biological Chemistry. 1998;273(49):32528–32534. doi: 10.1074/jbc.273.49.32528. [DOI] [PubMed] [Google Scholar]
- 80.Van Beilen JB, Funhoff EG. Expanding the alkane oxygenase toolbox: new enzymes and applications. Current Opinion in Biotechnology. 2005;16(3):308–314. doi: 10.1016/j.copbio.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 81.Ilori MO, Adebusoye SA, Ojo AC. Isolation and characterization of hydrocarbon-degrading and biosurfactant-producing yeast strains obtained from a polluted lagoon water. World Journal of Microbiology and Biotechnology. 2008;24(11):2539–2545. [Google Scholar]
- 82.Kiran GS, Hema TA, Gandhimathi R, et al. Optimization and production of a biosurfactant from the sponge-associated marine fungus Aspergillus ustus MSF3. Colloids and Surfaces B. 2009;73(2):250–256. doi: 10.1016/j.colsurfb.2009.05.025. [DOI] [PubMed] [Google Scholar]
- 83.Obayori OS, Ilori MO, Adebusoye SA, Oyetibo GO, Omotayo AE, Amund OO. Degradation of hydrocarbons and biosurfactant production by Pseudomonas sp. strain LP1. World Journal of Microbiology and Biotechnology. 2009;25(9):1615–1623. [Google Scholar]
- 84.Brusseau ML, Miller RM, Zhang Y, Wang X, Bai GY. Biosurfactant and cosolvent enhanced remediation of contaminated media. ACS Symposium Series. 1995;594:82–94. [Google Scholar]
- 85.Bai G, Brusseau ML, Miller RM. Biosurfactant-enhanced removal of residual hydrocarbon from soil. Journal of Contaminant Hydrology. 1997;25(1-2):157–170. [Google Scholar]
- 86.Barkay T, Navon-Venezia S, Ron EZ, Rosenberg E. Enhancement of solubilization and biodegradation of polyaromatic hydrocarbons by the bioemulsifier alasan. Applied and Environmental Microbiology. 1999;65(6):2697–2702. doi: 10.1128/aem.65.6.2697-2702.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cameotra SS, Singh P. Bioremediation of oil sludge using crude biosurfactants. International Biodeterioration and Biodegradation. 2008;62(3):274–280. [Google Scholar]
- 88.Beal R, Betts WB. Role of rhamnolipid biosurfactants in the uptake and mineralization of hexadecane in Pseudomonas aeruginosa. Journal of Applied Microbiology. 2000;89(1):158–168. doi: 10.1046/j.1365-2672.2000.01104.x. [DOI] [PubMed] [Google Scholar]
- 89.Pornsunthorntawee O, Wongpanit P, Chavadej S, Abe M, Rujiravanit R. Structural and physicochemical characterization of crude biosurfactant produced by Pseudomonas aeruginosa SP4 isolated from petroleum—contaminated soil. Bioresource Technology. 2008;99(6):1589–1595. doi: 10.1016/j.biortech.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 90.Nikolopoulou M, Kalogerakis N. Biostimulation strategies for fresh and chronically polluted marine environments with petroleum hydrocarbons. Journal of Chemical Technology and Biotechnology. 2009;84(6):802–807. [Google Scholar]
- 91.Wilson NG, Bradley G. The effect of immobilization on rhamnolipid production by Pseudomonas fluorescens. Journal of Applied Bacteriology. 1996;81(5):525–530. [Google Scholar]
- 92.Díaz MP, Boyd KG, Grigson SJW, Burgess JG. Biodegradation of crude oil across a wide range of salinities by an extremely halotolerant bacterial consortium MPD-M, immobilized onto polypropylene fibers. Biotechnology and Bioengineering. 2002;79(2):145–153. doi: 10.1002/bit.10318. [DOI] [PubMed] [Google Scholar]
- 93.Cunningham CJ, Ivshina IB, Lozinsky VI, Kuyukina MS, Philp JC. Bioremediation of diesel-contaminated soil by microorganisms immobilised in polyvinyl alcohol. International Biodeterioration and Biodegradation. 2004;54(2-3):167–174. [Google Scholar]
- 94.Rahman RNZA, Ghazali FM, Salleh AB, Basri M. Biodegradation of hydrocarbon contamination by immobilized bacterial cells. Journal of Microbiology. 2006;44(3):354–359. [PubMed] [Google Scholar]
- 95.Nichols WJ. The U.S. Environmental Protect Agency: National Oil and Hazardous Substances Pollution Contingency Plan, Subpart J Product Schedule (40 CFR 300.900). In: Proceedings of the International Oil Spill Conference; 2001; Washington, DC, USA. American Petroleum Institute; pp. 1479–1483. [Google Scholar]
- 96.Hoff RZ. Bioremediation: an overview of its development and use for oil spill cleanup. Marine Pollution Bulletin. 1993;26(9):476–481. [Google Scholar]
- 97.U.S. EPA. Spill NCP Product Schedule. 2002, http://www.epa.gov/oilspill.
- 98.Lee K, Tremblay GH, Gauthier J, Cobanli SE, Griffin M. Bioaugmentation and biostimulation: a paradox between laboratory and field results. In: Proceedings of the International Oil Spill Conference; 1997; Washington, DC, USA. American Petroleum Institute; pp. 697–705. [Google Scholar]
- 99.Ladousse A, Tramier B. Results of 12 years of research in spilled oil bioremediation: inipol EAP 22. In: Proceedings of the International Oil Spill Conference; 1991; Washington, DC, USA. American Petroleum Institute; pp. 577–581. [Google Scholar]
- 100.Ramos JL, Wasserfallen A, Rose K, Timmis KN. Redesigning metabolic routes: manipulation of TOL plasmid pathway for catabolism of alkylbenzoates. Science. 1987;235(4788):593–596. doi: 10.1126/science.3468623. [DOI] [PubMed] [Google Scholar]
- 101.Panke S, Sánchez-Romero JM, De Lorenzo V. Engineering of quasi-natural Pseudomonas putida strains toluene metabolism through an ortho-cleavage degradation pathway. Applied and Environmental Microbiology. 1998;64(2):748–751. doi: 10.1128/aem.64.2.748-751.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rojo F, Pieper DH, Engesser K-H, Knackmuss H-J, Timmis KN. Assemblage of ortho cleavage route for simultaneous degradation of chloro- and methylaromatics. Science. 1987;238(4832):1395–1398. doi: 10.1126/science.3479842. [DOI] [PubMed] [Google Scholar]
- 103.Hrywna Y, Tsoi TV, Maltseva OV, Quensen JF, III, Tiedje JM. Construction and characterization of two recombinant bacteria that grow on ortho- and para-substituted chlorobiphenyls. Applied and Environmental Microbiology. 1999;65(5):2163–2169. doi: 10.1128/aem.65.5.2163-2169.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Erickson BD, Mondello FJ. Enhanced biodegradation of polychlorinated biphenyls after site-directed mutagenesis of a biphenyl dioxygenase gene. Applied and Environmental Microbiology. 1993;59(11):3858–3862. doi: 10.1128/aem.59.11.3858-3862.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Suyama A, Iwakiri R, Kimura N, Nishi A, Nakamura K, Furukawa K. Engineering hybrid pseudomonads capable of utilizing a wide range of aromatic hydrocarbons and of efficient degradation of trichloroethylene. Journal of Bacteriology. 1996;178(14):4039–4046. doi: 10.1128/jb.178.14.4039-4046.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zwick TC, Foote EA, Pollack AJ, et al. In-Situ and On-Site Bioremediation. Columbus, Ohio, USA: Battelle Press; 1997. Effects of nutrient addition during bioventing of fuel contaminated soils in an arid environment; pp. 403–409. [Google Scholar]
- 107.Van Dyke MI, Lee H, Trevors JT. Survival of luxAB-marked Alcaligenes eutrophus H850 in PCB-contaminated soil and sediment. Journal of Chemical Technology and Biotechnology. 1996;65(2):115–122. doi: 10.1002/(SICI)1097-4660(199602)65:2<115::AID-JCTB391>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 108.Applegate BM, Kehrmeyer SR, Sayler GS. A chromosomally based tod-luxCDABE whole-cell reporter for benzene, toluene, ethybenzene, and xylene (BTEX) sensing. Applied and Environmental Microbiology. 1998;64(7):2730–2735. doi: 10.1128/aem.64.7.2730-2735.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sayler GS, Ripp S. Field applications of genetically engineered microorganisms for bioremediation processes. Current Opinion in Biotechnology. 2000;11(3):286–289. doi: 10.1016/s0958-1669(00)00097-5. [DOI] [PubMed] [Google Scholar]
- 110.Masson L, Tabashnik BE, Mazza A, et al. Mutagenic analysis of a conserved region of domain III in the Cry1ac toxin of Bacillus thuringiensis. Applied and Environmental Microbiology. 2002;68(1):194–200. doi: 10.1128/AEM.68.1.194-200.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sousa C, De Lorenzo V, Cebolla A. Modulation of gene expression through chromosomal positioning in Escherichia coli. Microbiology. 1997;143(6):2071–2078. doi: 10.1099/00221287-143-6-2071. [DOI] [PubMed] [Google Scholar]
- 112.Kelly CJ, Lajoie CA, Layton AC, Sayler GS. Bioluminescent reporter bacterium for toxicity monitoring in biological wastewater treatment systems. Water Environment Research. 1999;71(1):31–35. [Google Scholar]
- 113.Layton AC, Gregory B, Schultz TW, Sayler GS. Validation of genetically engineered bioluminescent surfactant resistant bacteria as toxicity assessment tools. Ecotoxicology and Environmental Safety. 1999;43(2):222–228. doi: 10.1006/eesa.1999.1792. [DOI] [PubMed] [Google Scholar]
- 114.Le Floch S, Merlin F-X, Guillerme M, Dalmazzone C, Le Corre P. A field experimentation on bioremediation: bioren. Environmental Technology. 1999;20(8):897–907. [Google Scholar]
- 115.Le Floch S, Merlin FX, Guillerme M, et al. In-Situ and On-Site Bioremediation. Columbus, Ohio, USA: Battelle Press; 1997. Bioren: recent experiment on oil polluted shoreline in temperate climate; pp. 411–417. [Google Scholar]
- 116.Schnoor JL, Licht LA, McCutcheon SC, Wolfe NL, Carreira LH. Phytoremediation of organic and nutrient contaminants. Environmental Science and Technology. 1995;29(7):318A–323A. doi: 10.1021/es00007a747. [DOI] [PubMed] [Google Scholar]
- 117.Hecht D, Badiane G. Phytoremediation. New Internationalist. June 1998 [Google Scholar]
- 118.Nedunuri KV, Govindaraju RS, Banks MK, Schwab AP, Chen Z. Evaluation of phytoremediation for field-scale degradation of total petroleum hydrocarbons. Journal of Environmental Engineering. 2000;126(6):483–490. [Google Scholar]
- 119.U.S. Army Corps of Engineers. Agriculturally Based Bioremediation of Petroleum-Contaminated Soils and Shallow Groundwater in Pacific Island Ecosystems. 2003. [Google Scholar]
- 120.Miya RK, Firestone MK. Enhanced phenanthrene biodegradation in soil by slender oat root exudates and root debris. Journal of Environmental Quality. 2001;30(6):1911–1918. doi: 10.2134/jeq2001.1911. [DOI] [PubMed] [Google Scholar]