SUMMARY
mTOR Complex 1 (mTORC1; mammalian target of rapamycin (mTOR) in complex with raptor) is a key regulator of protein synthesis and cell growth in response to nutrient amino acids. Here we report that inositol polyphosphate multikinase (IPMK), which possesses both inositol phosphate kinase and lipid kinase activities, regulates amino acid signaling to mTORC1. This regulation is independent of IPMK's catalytic function, instead reflecting its binding with mTOR and raptor, which maintains the mTOR-raptor association. Thus, IPMK appears to be a physiologic mTOR cofactor, serving as a determinant of mTORC1 stability and amino acid-induced mTOR signaling. Substances that block IPMK-mTORC1 binding may afford therapeutic benefit in nutrient amino acid-regulated conditions such as obesity and diabetes.
INTRODUCTION
Inositol phosphates (IPs) such as diphosphoinositol pentakisphosphate (IP7) regulate diverse processes including endocytosis (Saiardi et al., 2002), chemotactic signaling (Luo at al., 2003), and maintenance of telomere length (Saiardi et al., 2005). In the cloning of IP6 kinases (IP6Ks) responsible for generating IP7, we identified inositol polyphosphate multikinase (IPMK) (Saiardi et al., 1999). In contrast to other members of the IP6K family, IPMK does not form IP7 but instead synthesizes Ins(1,4,5,6)P4 and subsequently Ins(1,3,4,5,6)P5 by adding a phosphate at the three or six positions (Saiardi et al., 1999; Odom et al., 2000). IPMK also displays lipid kinase activity, phosphorylating phosphatidylinositol-4,5-bisphosphate (PIP2) to generate phosphatidyl inositol-3,4,5-trisphosphate (PIP3) (Resnick et al., 2005). IPMK was identified in yeast as an essential gene influencing responses to arginine and hence was labeled ArgIII or Arg82, and is also known as Ipk2 or yeast IPMK (Béchet et al., 1970; Saiardi et al., 1999; Odom et al., 2000). Accordingly, in seeking mammalian functions for IPMK, we have focused upon amino acid regulation of cell growth via the mTOR signaling pathway.
mTOR is a protein kinase which mediates the influence of growth factors as well as nutrient amino acids upon protein synthesis and growth (Wullschleger et al., 2006). mTOR exists in two distinct complexes that include several components: regulatory associated protein of mTOR (raptor) for mTORC1 (Loewith et al., 2002; Kim et al., 2002; Hara et al., 2002) and rapamycin-insensitive companion of mTOR (rictor) for mTOR Complex 2 (mTORC2) (Jacinto et al., 2004; Sarbassov et al., 2004; Wullschleger et al., 2006). The Rag GTPases, raptor that is involved in translocating mTOR in a Rag GTPase-mediated manner (Sancak et al., 2008), and human vacuolar protein sorting 34 (Gulati et al., 2008) have been implicated in amino acid-mTOR signaling, but how amino acids influence and activate mTORC1 has not been well delineated.
Here, we report that mammalian IPMK mediates the influence of amino acid stimulation upon mTOR signaling. The action of IPMK is independent of its catalytic activity. Instead, IPMK stabilizes mTOR-raptor binding in the mTORC1 complex through the amino terminal sequence of IPMK, a uniquely mammalian mTOR-binding site.
RESULTS
IPMK Depletion Prevents Amino Acid-Induced mTOR Signaling
To circumvent the embryonic lethality of conventional IPMK null mice (Frederick et al., 2005), we generated mouse embryonic fibroblasts (MEFs) in which IPMK is inducibly depleted (Figure 1A). Under normal growth conditions, both wild-type and IPMK-depleted MEFs show a similar level of mTORC1 activity as monitored by phosphorylation of ribosomal protein S6 kinase (S6K) (Figure 1A). Withdrawal for 1 hour of serum and amino acids inactivates mTORC1 (Figure 1A; Hara et al., 1998), while serum deprivation alone for 1 hour does not affect S6K phosphorylation in either wild-type or IPMK-depleted MEFs (Figure S1A).
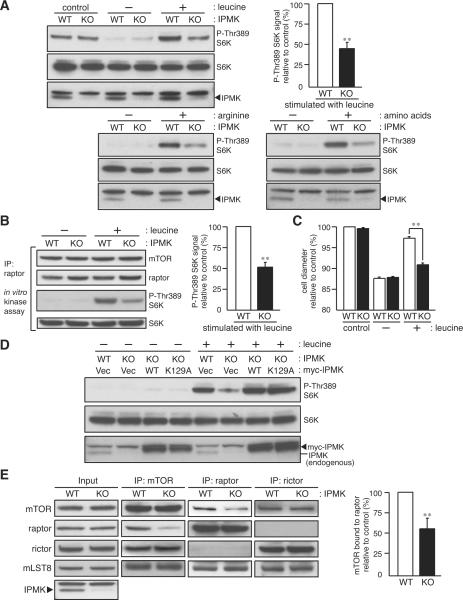
Figure 1. IPMK depletion diminishes amino acid signaling and mTOR-raptor interactions.
(A) Wild-type (WT) and IPMK-depleted (KO) MEFs were grown in 10% serum, nutrient-rich medium (control), deprived of serum and amino acids (leucine, arginine, or total amino acids) for 1 hour. Cells were stimulated by leucine (400 μM), arginine (400 μM), or total amino acids mixture (1×) for 10 min. (B) In vitro mTORC1 activity assay from leucine-treated MEFs. (C) MEFs were deprived of leucine for 24 hours and stimulated with leucine (400 μM) for 12 hours. Cell size was analyzed from at least 8,000 cells per trial (n = 3). Bars represent mean ± SEM. (D) IPMK-depleted MEFs stably expressing wild-type or the catalytically inactive, mutant IPMK (K129A) were analyzed as in (A). (E) mTOR, raptor, and rictor immunoprecipitates were isolated in the presence of DSP, and the amount of indicated mTORC subunits were analyzed. Bars in (A), (B), and (E) represent mean ± SD (n = 4). **p < 0.01.
Wild-type MEFs display marked stimulation of mTORC1 by arginine and leucine, the two most potent nutrient regulators of mTOR, as well as by total amino acids (Figure 1A). Depletion of IPMK reduces mTORC1 signaling in response to all of these amino acid stimuli by about 60% (Figures 1A and S1B). Activity of mTOR kinase, assayed in vitro, is also markedly reduced in IPMK-depleted MEFs (Figure 1B). Moreover, IPMK depletion prevents the stimulation by leucine of cell size (Figure 1C). The diminished signaling does not reflect influences of IPMK upon amino acid uptake, as intracellular import of arginine, leucine, and a methionine/cysteine mixture is unaltered in IPMK-depleted MEFs (Figure S1C). In addition, the activity of mitogen-activated protein kinase, a positive regulator of mTORC1 (Wullschleger et al., 2006) is unaltered in IPMK-depleted MEFs (Figure S1D).
To further examine the specificity of IPMK's role in regulating nutrient effects on mTORC1, we evaluated influences of oxidative stress elicited by H O or glucose deprivation. Both treatments markedly inhibit mTORC1 signaling to the same extent in wild-type and IPMK-depleted MEFs, and glucose restoration reverses mTORC1 activity (Figures S1E and S1F).
IPMK Regulation of mTOR is Independent of Catalytic Activity
We wondered whether catalytic activity of IPMK is required for regulation of mTORC1. Previously, we showed that mutation of IPMK's inositol-binding site abolishes its catalytic activity (Resnick et al., 2005). In IPMK-depleted MEFs, expression of mutant IPMK-K129A restores amino acid-induced mTORC1 signaling just as well as wild-type IPMK (Figure 1D), establishing that its regulation of mTOR is independent of catalytic activity. We were concerned that IPMK-K129A might retain trace amounts of catalytic activity so that overexpression of IPMK-K129A would provide sufficient IPMK products to rescue mTOR signaling defects. Accordingly, we examined the influence of IPMK-K129A overexpression on IP generation. IPMK-depletion, as expected, abolishes IP5 and IP6 formation with their generation rescued by overexpression of wild-type IPMK (Figures S1G, S1H, and S1I). Overexpression of IPMK-K129A in the IPMK-depleted MEFs fails to restore wild-type levels of IP5 and IP6 (Figure S1J), consistent with the notion that IPMK's catalytic activity is not required for amino acid-induced mTOR signaling. Additionally, we examined a form of IPMK mutated within the catalytic domain, which also restores mTORC1 signaling (Figure S2).
IPMK Non-catalytically Stabilizes mTORC1
What non-catalytic functions of IPMK might mediate mTOR activation? We examined a potential role for IPMK in the stabilization of mTORC1. To analyze the in vivo status of mTOR complexes, we isolated mTOR immunoprecipitates prepared from cell lysates treated with dithiobis(succinimidyl propionate) (DSP), an established crosslinker widely-used for capturing mTORC subunits in intact cells (Kim et al., 2002; Sarbassov et al., 2004). Depletion of IPMK markedly reduces mTOR-raptor binding without influencing the interaction of mTOR with rictor or mLST8, a subunit required for stabilizing mTORC2 (Guertin et al., 2006) (Figure 1E). Moreover, the association between mTOR and the Rag GTPase that binds raptor and mediates amino acid signaling to mTORC1, is significantly weakened in IPMK-depleted MEFs, while Rag-raptor binding is unaltered (Figure S1L). Thus, IPMK appears to act non-catalytically to selectively stabilize the association between mTOR and raptor in mTORC1 and thereby mediate amino acid-induced mTOR signaling.
IPMK is an mTOR Cofactor
We wondered whether IPMK regulates the stability of mTORC1 by participating in its protein-protein interaction events. To assess this possibility, we overexpressed GST-tagged human IPMK (GST-hIPMK) in HEK293T cells and detect endogenous mTOR in GST-hIPMK pulldown (Figure 2A). Recombinant IPMK and recombinant mTOR interact with each other in transfected HEK293T cells (Figure 2B). Co-immunoprecipitation of IPMK with either raptor or rictor also reveals that IPMK is a cofactor for both mTORC1 and mTORC2 (Figure 2C).
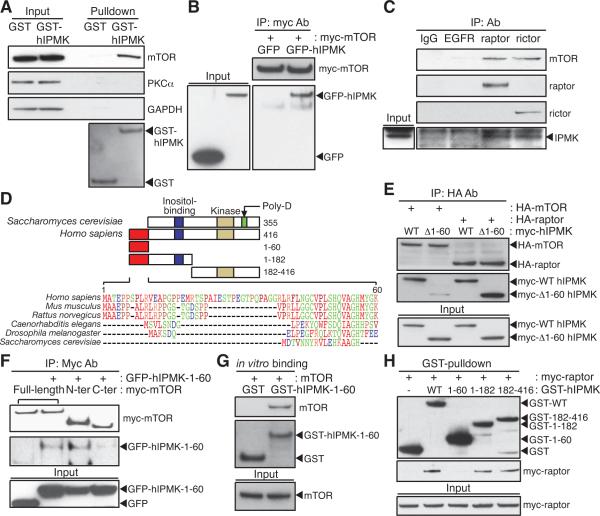
Figure 2. IPMK is an mTOR-associated protein.
(A) GST or GST-hIPMK was expressed in HEK293T cells, followed by pulldown and immunoblotting for endogenous mTOR. (B) GFP or GFP-hIPMK was co-transfected in HEK293T cells with myc-mTOR, and immunoprecipitates were analyzed by immunoblotting. (C) Lysates from IPMK-depleted MEFs stably expressing wild-type mouse IPMK were immunoprecipitated with indicated antibodies. (D) The unique sequence found in mammalian IPMK is aligned and indicated with a red box. Yeast IPMK-specific, polyaspartates sequence (Poly-D; green) and two conserved domains for inositol binding (blue) and kinase activity (yellow) are shown. IPMK fragments used for binding studies are shown in the schematic with the number of the amino acid sequence. (E) HA-mTOR or HA-raptor was co-transfected in HEK293T cells with wild-type or 1–60 deleted human IPMK (Δ1–60) constructs, followed by immunoprecipitation with HA-antibodies and immunoblotting. (F) Myc-wild-type mTOR, 1–1482 (N-ter) fragment, or 1328–2549 (C-ter) fragment was co-transfected in HEK293T cells with GFP or GFP-hIPMK-1–60, and myc-immunoprecipitates were analyzed by immunoblotting. (G) GST or GST-hIPMK-1–60 was incubated with mTOR purified from HEK293T cells. (H) GST or GST-hIPMK was co-transfected in HEK293T cells with myc-raptor, and myc-immunoprecipitates were analyzed by immunoblotting.
Sabatini and associates showed that amino acid deprivation elicits tight binding between mTOR and raptor, which is reversed by amino acid addition (Kim et al., 2002). Likewise, in our experiments, leucine deprivation enhances binding between mTOR, raptor, and IPMK, which is reversed by leucine treatment (Figure S3A). Under crosslinking conditions, leucine deprivation does not alter IPMK-mTORC1 binding, implying that leucine regulates the affinity rather than the stoichiometry of the binding complex (Figure S3B). To examine functional influences of IPMK on mTORC2 signaling, we monitored insulin-stimulated Akt Ser473 phosphorylation (Sarbassov et al., 2005), which is reduced in IPMK-depleted MEFs, as monitored with intact cells and in vitro (Figures S3C and S3D). The insulin-induced activation of Akt/mTORC2 by IPMK appears mechanistically distinct from amino acid-mTORC1 signaling, as it involves IPMK's catalytic activity (Figure S3E).
Functional interactions of IPMK and mTORC1 imply co-localization. Accordingly, we examined the immunofluorescent localizations of GFP-tagged hIPMK and endogenous mTOR in HEK293T cells (Figure S3F). Previous reports showed preferential nuclear staining of IPMK with modest cytosolic levels (Nalaskowski et al., 2002; Fujii and York, 2005; Resnick et al., 2005). In our present experiments, GFP-hIPMK signals occur with endogenous mTOR in a diffusely granular disposition throughout the cytoplasm with weak nuclear localization, supporting functional links of the two proteins. Variable results on IPMK's subcellular distribution are probably related to differences in overexpression and cell type.
The Unique Amino Terminus of IPMK Mediates IPMK-mTOR Binding
In yeast, a role for IPMK as a chaperone for the Arg80/Mcm1 transcription complex is critically dependent upon a polyaspartate domain (Dubois et al., 2000; El Alami et al., 2003), which is absent in mammalian IPMK (Figure 2D). In contrast, mammalian IPMK possesses a unique amino terminal extension of 31–60 amino acids that is absent in flies, worms, and yeast. Deletion of these 60 amino acids in human IPMK markedly reduces IPMK-mTOR binding (Figure 2E), while the 1–60 fragment of hIPMK (hIPMK-1–60) itself binds to mTOR, with especially robust binding to the amino terminal portion of mTOR (Figure 2F). Purified hIPMK-1–60 interacts in vitro with immunopurified mTOR, establishing direct binding between hIPMK-1–60 and mTOR (Figure 2G). IPMK also binds raptor, but the IPMK-raptor interaction occurs through the C-terminal portions of IPMK, not hIPMK-1–60 (Figures 2E and 2H).
Human IPMK-1–60 Peptide Disrupts mTORC1 Stability and Signaling
Because hIPMK-1–60 interacts with the amino terminus of mTOR, a known raptor-binding site (Kim et al., 2002), we explored whether hIPMK-1–60 can act in a dominant-negative fashion. If IPMK is critical for raptor-mTOR binding, might hIPMK-1–60 interfere with this binding? In GFP-transfected control HEK293T cells, mTOR-raptor binding is not notably affected by leucine availability (Figure 3A). Overexpression of GFP-hIPMK-1–60 reduces the mTOR-raptor interaction by 60% in the presence but not the absence of leucine (Figure 3A). By contrast, hIPMK-1–60 does not alter the association between mTOR and rictor (Figure 3A).
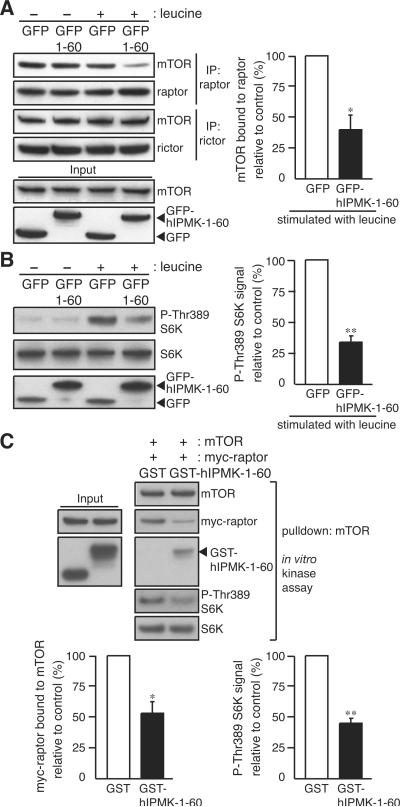
Figure 3. Human IPMK-1–60 peptide interferes with mTOR-raptor binding and amino acid signaling.
(A) HEK293T cells transfected with GFP or GFP-hIPMK-1–60 were deprived of serum and leucine for 1 hour, and stimulated with leucine for 10 min. mTORC1 and mTORC2 were isolated in the presence of DSP, and the amount of mTOR was analyzed. (B) S6K phosphorylation and protein expression were analyzed by immunoblotting. (C) The amount of myc-raptor bound in mTOR pulldown and in vitro mTORC1 activity were analyzed. mTOR immunopurified from leucine-treated HEK293T cells and myc-raptor (100 nM) were co-incubated with GST or GST-hIPMK-1–60 (200 nM). Bars represent mean ± SD (n = 3 in A and C, n = 4 in B). *p < 0.05; **p < 0.01.
We were concerned that hIPMK-1–60 might influence the mTOR-raptor interaction secondary to effects on inositol phosphate metabolism. However, overexpression of GFP-hIPMK-1–60 does not affect IP formation (Figure S1M).
The diminished mTOR-raptor binding elicited by hIPMK-1–60 has functional consequences. Thus, GFP-hIPMK-1–60 reduces mTORC1 signaling by more than 60% (Figure 3B). Dominant-negative influences of hIPMK-1–60 peptide are supported by in vitro experiments wherein the peptide prevents the formation of the mTOR-raptor complex as assessed with or without crosslinking (Figures 3C and S4A) and markedly reduces mTORC1 activity toward S6K (Figure 3C). The hIPMK-1–60 peptide does not influence mTORC2 activity whether assessed in intact cells or in vitro (Figures S4B and S4C). The hIPMK-1–182 and 182–416 fragments, which are responsible for mTOR and raptor binding, also interfere with raptor binding to mTOR and inhibit mTORC1 signaling (Figure S4D). The dominant-negative actions of hIPMK-1–60 establish that IPMK-raptor binding is involved in mTORC1 signaling.
The Amino Terminus of IPMK Stabilizes mTORC1 and Enhances its Signaling
To further explore the importance of the amino terminus of IPMK for mTOR signaling, we examined the effect of wild-type IPMK on mTORC1 stability. Overexpression of wild-type mouse IPMK in IPMK-depleted MEFs doubles mTOR-raptor binding (Figure 4A) and amino acid-induced mTORC1 signaling by more than two fold (Figures 1D and 4B). Mutant mouse IPMK lacking amino acids 1–31, corresponding to hIPMK-1–60 (mIPMK-Δ1–31; see Figure 2D), fails to rescue mTOR-raptor binding, mTORC1 signaling, and cell size defects in IPMK-depleted MEFs (Figures 4A, 4B, and S4E).
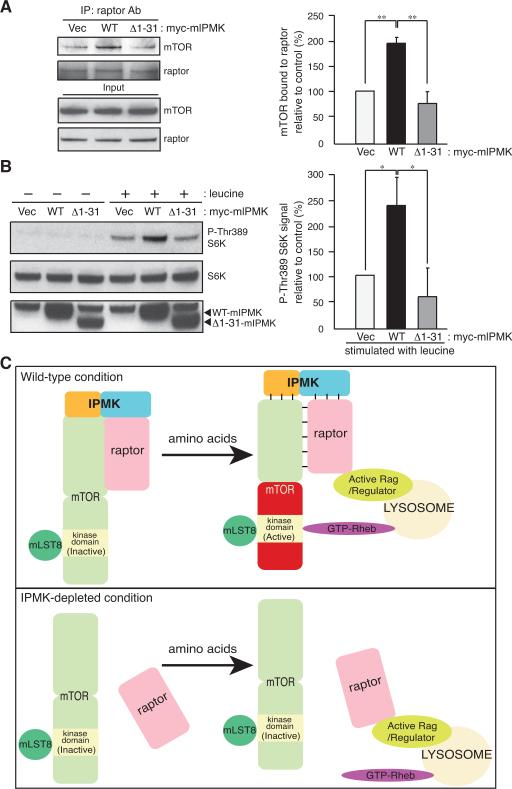
Figure 4. The amino-terminus of IPMK is involved in mTOR-raptor binding and amino acid signaling.
(A) In the presence of DSP, raptor immunoprecipitates were isolated from IPMK-depleted MEFs stably expressing vector (Vec), wild-type (WT), or the deletion mutant IPMK (Δ1–31) followed by immunoblotting. (B) S6K phosphorylation and protein expression were analyzed by immunoblotting. Bars represent mean ± SD (n = 3). *p < 0.05; **p < 0.01. (C) A model depicting regulation of mTORC1 signaling by IPMK in response to amino acids. In the absence of amino acids IPMK binds with high affinity to mTORC1, the N-terminal portion of IPMK interacting with mTOR and the C-terminus with raptor. Amino acid stimulation transforms the complex into a low affinity state, whereupon the complex translocates to the lysosomal surface. The active Rag/Regulator complex associates with raptor, and active GTP-Rheb binds to mTOR thereby enhancing mTOR activity (Sancak et al., 2010). In the absence of IPMK the mTOR-raptor complex dissociates, at least partially. Amino acid treatment still enhances raptor binding to the active Rag/Regulator complex, but GTP-Rheb fails to bind/activate mTOR.
We wondered whether deleting the amino terminal region of mouse IPMK might cause major alterations in IPMK protein folding or stability that would non-specifically impair its activities in rescuing mTOR signaling. Accordingly, we monitored inositol phosphate metabolism in the IPMK-depleted MEFs overexpressing mIPMK-Δ1–31. We detect comparable amounts of IP5 and IP6 (Figure S1K), which suggest that amino terminal deletion in mouse IPMK does not adversely affect the structural and functional integrity of IPMK. Thus, the amino terminus of IPMK is involved in stabilizing IPMK-mTOR binding and enhancing downstream mTORC1 signaling.
DISCUSSION
Our study establishes IPMK as a physiologic determinant of amino acid-induced mTORC1 signaling. Depletion of IPMK reduces amino acid-stimulated mTOR activation by about 60%, comparable to the effects of depletion of major components of the mTORC1 such as raptor (Kim et al., 2002). A mutant IPMK that is devoid of catalytic activity stimulates mTOR signaling as effectively as wild-type IPMK. Thus, regulation of mTOR by IPMK is independent of IPMK's catalytic activity.
We elucidated mechanisms whereby mammalian IPMK regulates mTORC1 signaling. In MEFs, IPMK depletion selectively disrupts mTOR-raptor, but not mTOR-rictor or mTOR-mLST8 interactions. IPMK's stabilization of mTORC1 is dependent upon the amino terminus of IPMK, which is present only in the mammalian form of the enzyme. Overexpression of hIPMK-1–60 selectively disrupts the mTOR-raptor interaction in the presence of leucine and thereby dominant-negatively impairs amino acid-induced mTORC1 signaling. Mutant mouse IPMK lacking amino acids 1–31 fails to maintain mTOR-raptor binding and activation of mTORC1 in response to amino acids. Thus, our findings show that mammalian IPMK is a physiologic mTOR cofactor involved in maintaining the binding of mTOR with raptor, thereby influencing the activation of mTORC1 by nutrient amino acids (Figure 4C). As hIPMK-1–60 peptide does not alter mTORC1 stability under the leucine-deprived condition and IPMK depletion does not fully abolish the mTOR-raptor binding, we presume that other factors act together with IPMK.
Sabatini and associates proposed that mLST8, which occurs in both mTORC1 and mTORC2, maintains the integrity of mTORC2 (Guertin et al., 2006). In MEFs null for mLST8, interactions of mTOR-rictor, but not mTOR-raptor, are lost, leading to depressed mTORC2 signaling. IPMK also associates with mTORC2, indicating that it can participate in the mTORC2 complex, and the decreased mTORC2 signaling toward Akt Ser473 phosphorylation in IPMK-depleted MEFs implies a functional role for IPMK.
The regulation of mTOR signaling by IPMK implies an important role in cellular protein synthesis that is critical for life. Thus genetic deletion of mTOR (Gangloff et al., 2004; Murakami et al., 2004; Guertin et al., 2006) or raptor (Guertin et al., 2006) leads to embryonic lethality at embryonic day 5.5–6.5 (e5.5–6.6) due to growth and proliferation defects. IPMK knockout mice also die early in development at around e9.5 due to severe growth and morphological defects, resembling the phenotype of mTOR or raptor knockout mice (Frederick et al., 2005). This similarity is consistent with a role of IPMK in regulating mTORC1 function.
Obese patients frequently display increased plasma levels of amino acids (Um et al., 2006; Dann et al., 2007), which parallels reduced insulin sensitivity (Krebs, 2005). Nutrient amino acid overload leads to hyperactivated mTORC1 signaling and insulin resistance (Tremblay et al., 2005). Due to its role in protein synthesis and growth, mTOR has been a major target for the development of anti-cancer drugs. Interestingly, obesity is a risk factor for many cancers (Calle and Kaaks, 2004), and anti-diabetic drugs such as metformin, which activates AMP-kinase and thereby inhibits mTORC1 signaling, inhibit cancer cell growth and metabolism (Dowling et al., 2007). Accordingly, drugs that impair IPMK-mediated regulation of the mTORC1 stability and function may be useful in treating obesity, diabetes, and cancer.
Experimental Procedures
Generation of inducible floxed IPMK mice
Floxed IPMK mice were generated at Ozgene. A loxP site was inserted between exons 5 and 6. Floxed IPMK mice were mated with knockin C57BL/6 mice (Jackson Laboratory) carrying the tamoxifen-inducible, Cre-ERT2 driven by a PGK promoter. Details are available as Supplemental Experimental Procedures.
MEFs
MEFs were immortalized by transfecting a SV40 large T-antigen plasmid. Depletion of IPMK in MEFs was achieved by adding 4-hydroxytamoxifen (1 μM) for 48 hours. Ethanol-treated MEFs were used for control.
Deprivation and stimulation of leucine, arginine, and total amino acids
HEK293T cells or MEFs were rinsed with PBS, incubated in leucine-deficient DMEM, arginine-deficient DMEM, or amino acids-deficient, glucose (4500 mg/L)-containing Krebs buffer (20 mM HEPES [pH 7.4], 118 mM NaCl, 4.6 mM KCl, 1 mM MgCl4, 12 mM NaHCO3, 0.5 mM CaCl2, 0.2% (w/v) bovine serum albumin) for 1 hour.
Cells were stimulated by leucine (400 μM), arginine, (400 μM), and amino acids mixture (1×). Lysis, immunoprecipitations, immunoblotting, and materials are available as Supplemental Experimental Procedures.
Cross-linking assay and mTOR kinase assay
Assays were performed as previously described (Sancak et al., 2007; Sancak et al., 2008).
Statistical Analysis
Data were analyzed by paired Student's t-test.
Supplementary Material
Acknowledgments
We thank D. M. Sabatini for reagents. This work was funded by an USPHS grant DA-00266 and Research Scientist Award DA-00074 to S.H.S., Jane Coffin Childs Memorial fellowship (S.K.), R01DK084336 and Bingham Trust Pilot Award (S.F.K.), and Damon Runyon Cancer Research fellowship (D.M.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Béchet J, Grenson M, Wiame JM. Mutations affecting the repressibility of arginine biosynthetic enzymes in Sacchromyces cerevisiae. Eur. J. Biochem. 1970;12:31–39. doi: 10.1111/j.1432-1033.1970.tb00817.x. [DOI] [PubMed] [Google Scholar]
- Calle EE, Kaaks R. Overweight, obesity and cancer: epidemiological evidence and proposed mechanisms. Nat. Rev. Cancer. 2004;4:579–591. doi: 10.1038/nrc1408. [DOI] [PubMed] [Google Scholar]
- Dann SG, Selvaraj A, Thomas G. mTOR Complex1-S6K1 signaling: at the crossroads of obesity, diabetes and cancer. Trends Mol. Med. 2007;13:252–259. doi: 10.1016/j.molmed.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Dowling RJ, Zakikhani M, Fantus IG, Pollak M, Sonenberg N. Metformin inhibits mammalian target of rapamycin-dependent translation initiation in breast cancer cells. Cancer Res. 2007;67:10804–10812. doi: 10.1158/0008-5472.CAN-07-2310. [DOI] [PubMed] [Google Scholar]
- Dubois E, Dewaste V, Erneux C, Messenguy F. Inositol polyphosphate kinase activity of Arg82/ArgRIII is not required for the regulation of the arginine metabolism in yeast. FEBS. Lett. 2000;486:300–304. doi: 10.1016/s0014-5793(00)02318-8. [DOI] [PubMed] [Google Scholar]
- El Alami M, Messenguy F, Scherens B, Dubois E. Arg82p is a bifunctional protein whose inositol polyphosphate kinase activity is essential for nitrogen and PHO gene expression but not for Mcm1p chaperoning in yeast. Mol. Microbiol. 2003;49:457–468. doi: 10.1046/j.1365-2958.2003.03562.x. [DOI] [PubMed] [Google Scholar]
- Frederick JP, Mattiske D, Wofford JA, Megosh LC, Drake LY, Chiou ST, Hogan BL, York JD. An essential role for an inositol polyphosphate multikinase, Ipk2, in mouse embryogenesis and second messenger production. Proc. Natl. Acad. Sci. USA. 2005;102:8454–8459. doi: 10.1073/pnas.0503706102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii M, York JD. A role for rat inositol polyphosphate kinases rIPK2 and rIPK1 in inositol pentakisphosphate and inositol hexakisphosphate production in rat-1 cells. J. Biol. Chem. 2005;280:1156–1164. doi: 10.1074/jbc.M412006200. [DOI] [PubMed] [Google Scholar]
- Gangloff YG, Mueller M, Dann SG, Svoboda P, Sticker M, Spetz JF, Um SH, Brown EJ, Cereghini S, Thomas G, Kozma SC. Disruption of the mouse mTOR gene leads to early postimplantation lethality and prohibits embryonic stem cell development. Mol. Cell. Biol. 2004;24:9508–9516. doi: 10.1128/MCB.24.21.9508-9516.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, Fitzgerald KJ, Sabatini DM. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev. Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Gulati P, Gaspers LD, Dann SG, Joaquin M, Nobukuni T, Natt F, Kozma SC, Thomas AP, Thomas G. Amino acids activate mTOR complex 1 via Ca2+/CaM signaling to hVps34. Cell Metab. 2008;7:456–465. doi: 10.1016/j.cmet.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, Yonezawa K, Weng QP, Kozlowski MT, Belham C, Avruch J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J. Biol. Chem. 1998;273:14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–189. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- Jacinto E, Loewith R, Schmidt A, Lin S, Rüegg MA, Hall A, Hall MN. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 2004;6:1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- Krebs M. Amino acid-dependent modulation of glucose metabolism in humans. Eur. J. Clin. Invest. 2005;35:351–354. doi: 10.1111/j.1365-2362.2005.01506.x. [DOI] [PubMed] [Google Scholar]
- Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, Hall MN. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- Luo HR, Huang YE, Chen JC, Saiardi A, Iijima M, Ye K, Huang Y, Nagata E, Devreotes P, Snyder SH. Inositol pyrophosphates mediate chemotaxis in Dictyostelium via pleckstrin homology domain-PtdIns(3,4,5)P3 interactions. Cell. 2003;114:559–572. doi: 10.1016/s0092-8674(03)00640-8. [DOI] [PubMed] [Google Scholar]
- Murakami M, Ichisaka T, Maeda M, Oshiro N, Hara K, Edenhofer F, Kiyama H, Yonezawa K, Yamanaka S. mTOR is essential for growth and proliferation in early mouse embryos and embryonic stem cells. Mol. Cell. Biol. 2004;24:6710–6718. doi: 10.1128/MCB.24.15.6710-6718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalaskowski MM, Deschermeier C, Fanick W, Mayr GW. The human homologue of yeast ArgRIII protein is an inositol phosphate multikinase with predominantly nuclear localization. Biochem. J. 2002;366:549–556. doi: 10.1042/BJ20020327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom AR, Stahlberg A, Wente SR, York JD. A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science. 2000;287:2026–2029. doi: 10.1126/science.287.5460.2026. [DOI] [PubMed] [Google Scholar]
- Resnick AC, Snowman AM, Kang BN, Hurt KJ, Snyder SH, Saiardi A. Inositol polyphosphate multikinase is a nuclear PI3-kinase with transcriptional regulatory activity. Proc. Natl. Acad. Sci. USA. 2005;102:12783–12788. doi: 10.1073/pnas.0506184102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiardi A, Erdjument-Bromage H, Snowman AM, Tempst P, Snyder SH. Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr. Biol. 1999;9:1323–1326. doi: 10.1016/s0960-9822(00)80055-x. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Nagata E, Luo HR, Snowman AM, Snyder SH. Identification and characterization of a novel inositol hexakisphosphate kinase. J. Biol. Chem. 2001;276:39179–39185. doi: 10.1074/jbc.M106842200. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Sciambi C, McCaffery JM, Wendland B, Snyder SH. Inositol pyrophosphates regulate endocytic trafficking. Proc. Natl. Acad. Sci. USA. 2002;99:14206–14211. doi: 10.1073/pnas.212527899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiardi A, Resnick AC, Snowman AM, Wendland B, Snyder SH. Inositol pyrophosphates regulate cell death and telomere length through phosphoinositide 3-kinase-related protein kinases. Proc. Natl. Acad. Sci. USA. 2005;102:1911–1914. doi: 10.1073/pnas.0409322102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, Carr SA, Sabatini DM. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol. Cell. 2007;25:903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- Tremblay F, Krebs M, Dombrowski L, Brehm A, Bernroider E, Roth E, Nowotny P, Waldhäusl W, Marette A, Roden M. Overactivation of S6 kinase 1 as a cause of human insulin resistance during increased amino acid availability. Diabetes. 2005;54:2674–2684. doi: 10.2337/diabetes.54.9.2674. [DOI] [PubMed] [Google Scholar]
- Um SH, D'Alessio D, Thomas G. Nutrient overload, insulin resistance, and ribosomal protein S6 kinase 1, S6K1. Cell Metab. 2006;3:393–402. doi: 10.1016/j.cmet.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.