Abstract
Human diffuse large B-cell lymphoma cell line RC-K8 has an altered EP300 locus that encodes a C-terminally truncated histone acetyltransferase (HAT) protein (p300ΔC). We now show that p300ΔC contains 1047 N-terminal amino acids of p300 fused to 25 amino acids encoded by sequences from chromosome 6. Over-expressed p300ΔC localized to nuclear subdomains and interacted with transcription factor REL. p300ΔC did not function as a co-activator for REL-directed transactivation, and blocked the ability of wild-type p300 to enhance transcriptional activation by REL. Knock down of p300ΔC in RC-K8 cells reduced their growth in both liquid culture and soft agar. Truncations of p300 were not found in eight other B-lymphoma cell lines. These results suggest that p300ΔC contributes to the oncogenic state of RC-K8 cells by acting as a defective co-activator.
Keywords: p300, RC-K8, Chromosomal rearrangement, REL, Diffuse large B-cell lymphoma
1. Introduction
p300 is a ubiquitously expressed histone acetyltransferase (HAT) that acts as a co-activator for many different transcription factors. Histone acetylation plays an important role in the transcriptional activation of many genes, and p300 is involved in the regulation of genes controlling cell proliferation, apoptosis, differentiation, the cell cycle, and DNA repair [1,2]. Genome-wide mapping has shown that p300 is present at approximately 6000 different promoter and enhancer regions in human T cells [3]. These p300 target genes include oncogenes (MDM2, FOS, and CDC25A), tumor suppressor genes (TP73, BRCA1, RB1, and CDKN2A), cytokine genes (TGFB1, IL2, IL12B, and TNFA) and several caspase genes (CASP3, CASP8, and CASP9) [3]. Moreover, p300 can function as a co-activator for many transcription factors involved in cell growth and survival, including the NF-κB proteins RelA and REL, p53, estrogen receptor α, BRCA1, and Notch1 [1,2,4-8]. However, p300 is also involved in transcriptional repression of some genes, such as MYC [9].
p300 has been implicated in cancer in several ways. First, wild-type p300 displays certain characteristics of a tumor suppressor. Although a complete knockout of p300 in mice is lethal [10], histiocytic sarcomas develop from p300-/- cell populations in chimeric mice [11]. Expression of wild-type p300 slows the growth of two cancer cell lines with biallelic inactivating mutations in EP300 [12]. Second, mutations in the p300 gene (EP300), most of which cause C-terminal truncations deleting all or part of the HAT domain, have been found in colorectal, gastric, breast, pancreatic, cervical, and ovarian cancers [2,7,13-15]. Additionally, translocations have been identified that result in the C-terminal half of p300 being fused to the monocytic leukemia zinc-finger protein (MOZ) in acute myeloid leukemia patients or to the mixed lineage leukemia protein (MLL) in myelodysplastic syndrome patients [16-18]. Third, reduced p300 expression due to increased expression of EP300-targeting microRNAs has been reported for several pancreatic ductal carcinoma cell lines [19]. Finally, cytoplasmic localization of p300, which is normally a nuclear protein, has been observed in breast cancer cells [20].
We have recently shown that p300 can act as a co-activator for the proto-oncoprotein REL [8], which is an NF-κB family transcription factor misregulated in many B-cell lymphomas [21]. Moreover, only a C terminally-truncated p300 protein (called p300ΔC) is expressed in the diffuse large B-cell lymphoma (DLBCL) cell line RC-K8 [8], which requires REL activity for its proliferation [22]. In this report, we have characterized the genomic alteration that leads to expression of p300ΔC in RC-K8 cells, as well as the structure and function of p300ΔC. Of particular note, we found that knock down of p300ΔC expression in RC-K8 cells caused these cells to proliferate more slowly in liquid culture and to form fewer colonies in soft agar. These results suggest that expression of the truncated p300 protein contributes to the malignant state of the RC-K8 cell line and that similar p300 mutations play an active role in oncogenesis in other tumor cell types.
2. Materials and methods
2.1 Cloning of the p300ΔC cDNA
The p300ΔC cDNA was amplified using the SMART RACE cDNA Amplification Kit according to the manufacturer's protocol (Clontech, Mountain View, CA, USA). One μg of total RNA was isolated from RC-K8 cells and reverse transcribed into cDNA using M-MLV reverse transcriptase (Promega, Madison, WI, USA) and the 3'RACE CDS Primer A (Clontech). As a first step in amplifying the p300ΔC cDNA, the reaction was diluted 1:100, and 2.5 μl was used as template in a touchdown PCR containing a forward primer complementary to exon 15 sequences of EP300, the Universal Primer A Mix (Clontech), and Crimson Taq DNA Polymerase (New England Biolabs, Ipswich, MA, USA). The product of this reaction was then diluted 1:50, and 5 μl was used as template in a nested PCR containing a forward primer complementary to exon 16 sequences of EP300, the Nested Universal Primer A (Clontech), and Crimson Taq DNA Polymerase (New England Biolabs) to amplify p300 cDNA. The final PCR product was subjected to agarose gel electrophoresis, and an approximately 300 bp band was excised and sequenced (Eurofins MWG Operon, Huntsville, AL, USA). To directly amplify the p300ΔC cDNA, 5 μl of the diluted cDNA was used as template in a PCR containing a forward primer complementary to exon 9, a reverse primer complementary to non-p300 sequences in the EP300ΔC cDNA, and Crimson Taq DNA Polymerse (New England Biolabs). This PCR product was then digested with NdeI and HindIII and was used to replace the corresponding wild-type 3’ EP300 cDNA sequences in pCMVβ-p300.
2.2 Plasmids
DNA manipulations were carried out by standard methods [23]. Complete details of all subclones and primers used in this study are described in supplementary information and at www.nfkb.org.
2.3 Cell culture
A293 and BOSC23 human embryonic kidney cells and DF-1 chicken fibroblasts were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Biologos, Montgomery, IL, USA) as previously described [24]. RC-K8 and other human B-lymphoma cells were cultured in DMEM or RPMI supplemented with 10-20% heat-inactivated FBS. The human B-lymphoma cell lines as follows: DLBCL (RC-K8, Pfeiffer, SUDHL6); Hodgkins lymphoma (KMH2, L428, HDMYZ), and Burkitt's lymphoma (Namalwa, Raji, Ramos). KMH2 cells were a gift of Dr. Cyril Benes (Massachusetts General Hospital, Charlestown, MA, USA); all other cell lines were obtained from Dr. Ellen Cahir-McFarland (Channing Labs, Boston, MA, USA).
For transfections, A293, BOSC23, and DF-1 cells were seeded such that they were approximately 60% confluent on the following day when transfections were performed using polyethylenimine (PEI) (Polysciences, Warrington, PA, USA). On the day of transfection, DNA:PEI was incubated at a ratio of 1:3 for A293 and BOSC23 cells or at 1:6 for DF-1 cells in serum-free DMEM (200 μl for a 35-mm plate; 500 μl for a 100-mm plate) for 15 min at room temperature. The DNA/PEI mixture was then added to two (35-mm plate) or ten ml (100-mm plate) of DMEM containing 10% FBS, and the final mixture was then added to the cells. The next day, the transfection media was replaced with fresh DMEM containing 10% FBS. Cells were harvested and lysed 24 h later.
Short hairpin RNAs (shRNA) for EP300 (5'-ACCAGATGCCTCGAATAA-3'; [9]) and control (5’-GCAAGCTGCCCGTGCCCTG-3’; [25]) sequences were designed using the shRNA Sequence Designer (Clontech) and were subcloned into the pSIREN-RetroQ retroviral vector (Clontech). Viral stocks were generated by transfecting BOSC23 cells with 10 μg pSIREN vectors and 5 μg pCL-10a1 packaging vector. Forty-eight hours after transfection, media containing viral particles was harvested. Two ml of viral supernatant was used to infect 106 RC-K8 cells in the presence of 8 μg/ml polybrene. Two days later, infected cells were selected with 2.5 μg/ml puromycin (Sigma, St. Louis, MO, USA) for 2-4 weeks.
2.4 Western blotting and indirect immunofluorescence
Western blotting was performed as described previously [8]. The following antisera were used: rabbit anti-p300 (1:200; anti-N-terminal, sc-584, Santa Cruz Biotechnology, Santa Cruz, CA, USA), mouse anti-MYC (1:1000, sc-40, Santa Cruz Biotechnology), and rabbit anti-β-tubulin (1:500; sc-9104, Santa Cruz Biotechnology).
Indirect immunofluorescence was performed as described previously [24]. DF-1 cells were plated two days after transfection onto glass coverslips. The subcellular localization of p300 and p300ΔC was determined by indirect immunofluorescence using anti-p300 (1:50; sc-584, Santa Cruz Biotechnology) primary antibody and fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG secondary antibody (1:80; Sigma). Nuclei were stained with 4',6-diamidino-2-phenylindole (DAPI). Cells were visualized using a fluorescent microscope (Olympus FLUOVIEW Laser Scanner Microscope BX 50, Center Valley, PA, USA).
2.5 GST pulldown assays
GST pulldown assays were performed as previously described [8]. Five percent of the protein-bound beads from each sample were separated on an SDS-polyacrylamide gel and stained with Coomassie Blue to verify that approximately equal amounts of each GST-fusion protein had been used in the pulldown assays. The remaining beads were incubated with 1 mg of A293 or 3 mg of RC-K8 whole cell extract for 2 h at 4°C. One percent of the amount of extract used for one pulldown (30 μg) was included on the gel as an input lane. The membrane was then subjected to anti-p300 Western blotting.
2.6 Luciferase reporter assays
Luciferase reporter assays were performed using the Luciferase Assay System (Promega) as described previously [8]. A293 cells in 35-mm plates were transfected with 0.5 μg of reporter plasmid pGL2-3x-κB-luciferase and 0.5 μg of normalization plasmid RSV-βgal. Cells were co-transfected with 0.5 μg of pcDNA-REL or vector alone, along with 0.5 μg of pCMVβ-p300, pCMVβ-p300ΔC, or vector alone. In competition assays (Fig. 3B), cells were transfected with 0.5 μg of pCMVβ-p300 and pcDNA-REL, and then increasing amounts of pCMVβ-p300ΔC. Luciferase and β-galactosidase activities were determined, and values were normalized to the relevant vector control (1.0).
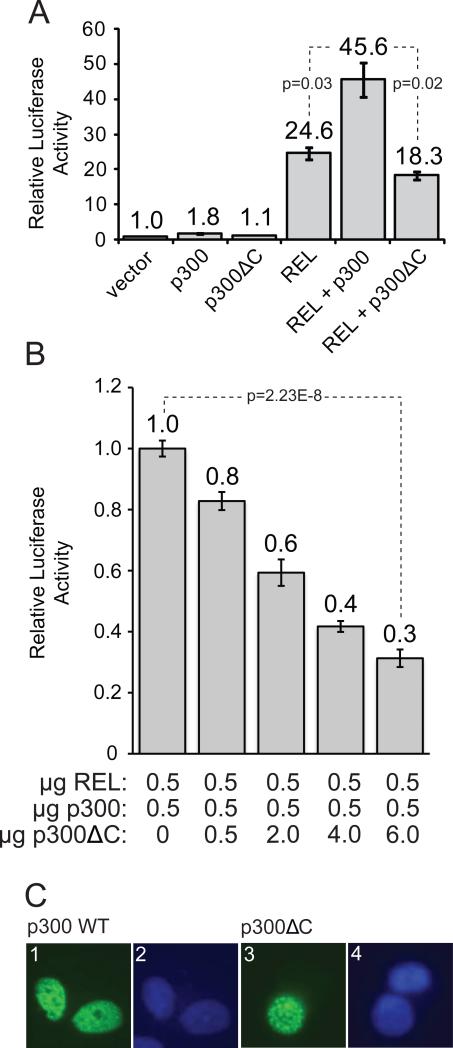
Fig. 3.
p300ΔC localized to the nucleus, but did not enhance REL activity. (A) A293 cells were transfected with 0.5 μg of a 3x-κB-site luciferase reporter plasmid and 0.5 μg of normalization plasmid RSV-βgal. Cells were co-transfected with 0.5 μg of pcDNA-REL or vector alone, along with 0.5 μg of pCMVβ-p300, pCMVβ-p300ΔC, or vector alone. Luciferase and β-galactosidase activites were determined, and values were normalized to control transfections as indicated (1.0). Values are the averages of three experiments performed with triplicate samples. (B) 3x-κB-site luciferase reporter assays were performed as in (A) with constant amounts (0.5 μg) of pcDNA-REL and pCMVβ-p300, but increasing amounts of pCMVβ-p300ΔC, as indicated. (C) DF-1 chicken fibroblasts were transfected with 4 μg of pCMVβ-p300 (panels 1 and 2) or pCMVβ-p300ΔC (panels 3 and 4). Indirect immunofluorescence was performed using a primary anti-p300 antibody and an FITC-conjugated anti-rabbit secondary antibody (panels 1 and 3). Nuclei were visualized by DAPI staining (panels 2 and 4).
2.7 Cell proliferation and soft agar assays
One hundred thousand RC-K8 cells, retrovirally transduced to express p300 shRNA or control shRNA, were plated in 16-mm wells in 0.5 ml DMEM containing 20% FBS. On each of five days after plating, cells from three wells of each cell type were counted using a hemocytometer.
Soft agar colony assays were performed as described previously [26]. Equal numbers of RC-K8 cells expressing the indicated shRNA (250 and 500 cells) were placed in soft agar containing DMEM with 20% FBS and 0.3% Bacto Agar (Difco, Franklin Lakes, NJ, USA), and plates were incubated at 37°C in a humid incubator with 5% CO2. Macroscopic colonies were counted 14 days after plating.
2.8 Statistical analysis
Statistical analyses were performed using a Student's two-tailed, paired t-test.
3. Results
3.1 Characterization of the EP300 genomic locus and cDNA in RC-K8 cells
We previously showed that one copy of the EP300 gene in RC-K8 B-lymphoma cells has a genomic alteration after exon 17, which results in the expression of a 3’ truncated mRNA and a C terminally-truncated p300 protein (p300ΔC) [8]. To characterize the precise molecular alteration in EP300 in RC-K8 cells, we first performed a series of 3'RACE reactions using an exon 15 primer and an oligo-dT primer, followed by nested reactions. From these reactions, we obtained preliminary cDNA sequence information that enabled us to design specific primers for the 3’ non-EP300 sequences in the p300ΔC mRNA. As shown in Fig. 1A, PCR amplification using a 5’ exon 17 EP300 primer and a 3’ non-EP300 primer in p300ΔC yielded primarily a ~120-bp band from cDNA and a ~220-bp band with genomic DNA from RC-K8 cells. These bands were not amplified using cDNA or genomic DNA from human BJAB cells, which express only wild-type p300 [8].
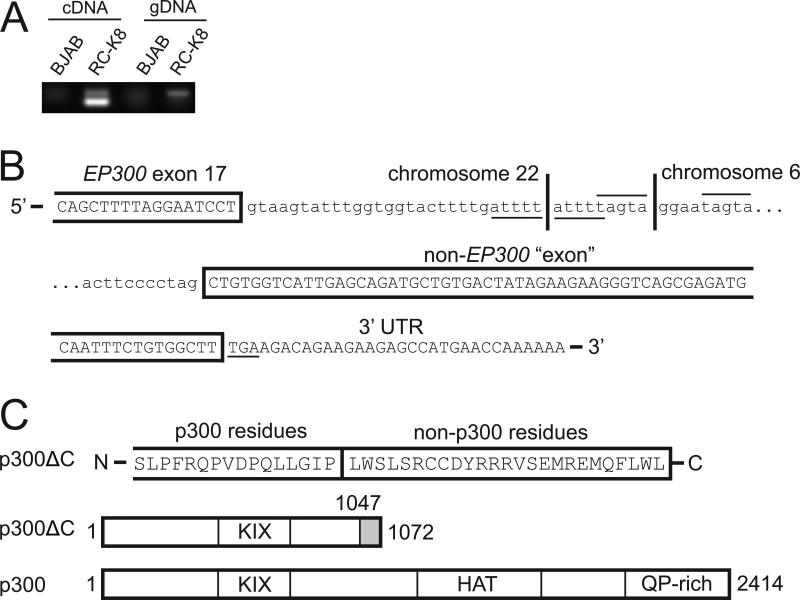
Fig. 1.
Structure of the EP300ΔC locus and p300ΔC protein in the RC-K8 diffuse large B-cell lymphoma cell line. (A) PCR amplification using a 5’ exon 17 EP300 primer and a 3’ non-EP300 primer in p300ΔC yielded primarily a 122-bp band with cDNA and a 223-bp band with genomic DNA from RC-K8 cells, but not BJAB cells. (B) The EP300ΔC genomic locus has 28 bp of EP300 intron after exon 17, followed by 9 bp of unknown sequence, and at least 180 bp of DNA sequence from chromosome 6. The intervening 9-bp sequence contains sequences identical to those found on either side of the breakpoint fusion, as indicated by lines above and below these repeated sequences. In the EP300ΔC cDNA, 110 bases of genomic DNA are removed by splicing, such that exon 17 sequences of EP300 are spliced to a site located 73 bases into the now-adjacent chromosome 6 sequences. The p300ΔC predicted stop codon (TGA) is underlined. Amino acid-coding sequences are boxed and sequences removed by splicing are indicated by lowercase type. (C) p300ΔC is predicted to be a 1072-aa protein that contains aa 1-1047 of p300 (encoded by exons 1-17 of EP300), followed by 25 C-terminal aa encoded by the fused chromosome 6 sequences. p300ΔC lacks the HAT and QP-rich domains of the normal p300 protein. The EP300-junction genomic and mRNA sequences can be accessed at GenBank (accession numbers HM212648 and HM212647, respectively). UTR, untranslated region; KIX, CREB-binding domain; HAT, histone acetyltransferase domain; QP-rich, glutamine/proline-rich domain.
DNA sequencing of these amplified products showed that the altered EP300 genomic locus has 28 bp of EP300 intron after exon 17, followed by 9 bp of unknown sequence, and (at least) 180 bp of DNA sequence from chromosome 6 (Fig. 1B). These non-EP300 sequences are from the reverse strand of the intron after exon 9 of the BCKDHB gene (branched chain keto acid dehydrogenase E1β) on chromosome 6. In the p300ΔC cDNA, 110 bases are removed by splicing such that exon 17 sequences of EP300 are spliced to a site located 73 bp into the now adjacent chromosome 6 sequences (Fig. 1B). Thus, the p300ΔC mRNA encodes a 1072-amino acid (aa) protein that contains aa 1-1047 of p300 (encoded by exons 1-17 of EP300), followed by 25 C-terminal aa derived from non-coding sequences from chromosome 6. Therefore, p300ΔC lacks the HAT and QP-rich domains present in the normal p300 protein (Fig. 1C).
3.2 REL interacts with ectopically expressed p300ΔC
As a first step in characterizing the p300ΔC protein, we created a pcDNA-based expression vector for p300ΔC. This vector directed the expression of a protein in A293 cells that co-migrated with p300ΔC from RC-K8 cells and that was much smaller than the endogenous p300 protein in A293 cells and BJAB cells (Fig. 2A). This result indicates that we isolated the bona fide p300ΔC cDNA from RC-K8 cells.
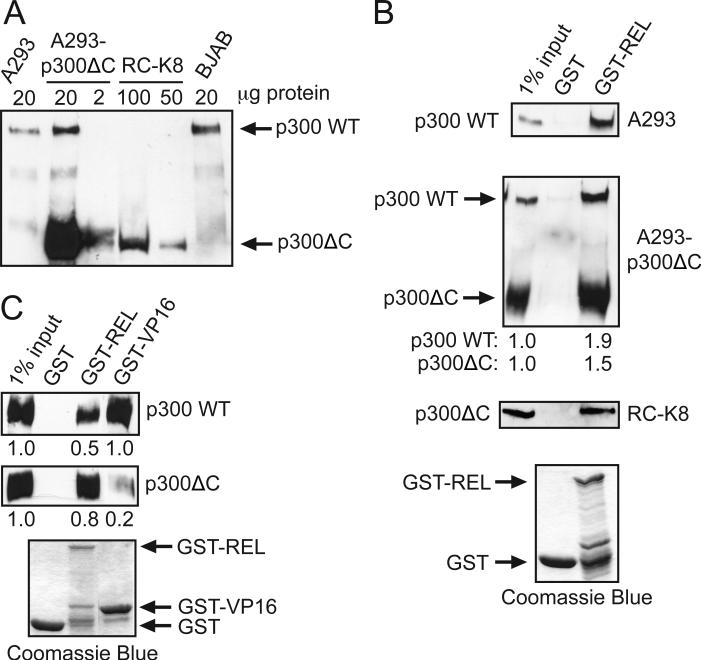
Fig. 2.
REL interacted with ectopically expressed p300ΔC. (A) A293 cell-expressed p300ΔC co-migrates with p300ΔC from RC-K8 cells. Anti-p300 Western blotting was performed on extracts from control A293 cells, p300ΔC-transfected A293 cells, RC-K8 cells and BJAB cells. For each lane, the amount of cell extract protein loaded is indicated. (B) GST-REL interacted with ectopically expressed p300ΔC. GST-REL (aa 324-587) pulldowns were performed on whole cell extracts from A293 cells (top panel), p300ΔC-transfected A293 cells (second panel), and RC-K8 cells (third panel). Bound proteins were subjected to anti-p300 Western blotting (top three panels) to detect endogenous p300 or p300ΔC, as indicated. For each GST pulldown, Coomassie Blue staining was performed on 5% of GST alone or the indicated GST-fusion protein used in the pulldowns (lower panel). In the second panel, the amounts of pulled down p300 and p300ΔC were quantified by scanning of the films; the values below the panel indicate the amount of each protein relative to the intensity of the band for the given protein in the input lane (1.0). (C) p300ΔC interacted weakly with VP16. GST-REL (aa 324-587) and GST-VP16 (aa 452-490) pulldowns were performed on whole cell extracts of A293 cells overexpressing p300 (top panel) and p300ΔC (second panel). Bound proteins were subjected to anti-p300 Western blotting (top two panels). For each GST pulldown, Coomassie Blue staining was performed on 5% of GST alone or the indicated GST-fusion protein used in the pulldowns (lower panel). In the top two panels, bands were quantified as for (B).
Like p300ΔC from RC-K8 cells [8], p300ΔC over-expressed in A293 cells interacted in GST pulldown assays with C-terminal transactivation domain sequences (aa 324-587) from the REL transcription factor (Fig. 2B). To determine whether p300ΔC can interact with other transcription factors, we tested its ability to interact with the herpes simplex virus VP16 transcriptional activator, which was previously shown to interact with wild-type p300 [27]. We performed GST pulldown assays using GST fusion proteins containing the transactivation domain sequences of REL (aa 324-587) and VP16 (aa 452-490) with whole cell extracts from A293 cells over-expressing either wild-type p300 or p300ΔC. While GST-REL interacted to a similar extent with both p300 and p300ΔC, GST-VP16 interacted with wild-type p300 approximately five-fold more efficiently than with p300ΔC (Fig. 2C). These results suggest that p300ΔC varies in its ability to interact with specific transcription factors when compared to wild-type p300.
3.3. Functional characterization of the p300ΔC protein
To determine whether the p300ΔC protein can act as a co-activator, we tested the ability of p300ΔC to enhance the transactivating ability of REL in reporter gene assays in A293 cells. Consistent with previous results [8], wild-type p300 enhanced the ability of REL to activate a κB-site reporter by approximately two-fold (Fig. 3A). In contrast, p300ΔC did not enhance REL-dependent transactivation (Fig. 3A). Moreover, transfection of excess p300ΔC expression plasmid blocked the ability of wild-type p300 to enhance REL-dependent transactivation (Fig. 3B).
We next compared the subcellular localization of p300 and p300ΔC in transfected cells. As shown in Fig. 3C, both wild-type p300 and p300ΔC were predominantly localized in discrete regions of the nucleus (“speckles”) in transfected chicken fibroblast cells. Together, these results indicate that p300ΔC can enter the nucleus and interact with its target protein REL, but cannot function as a co-activator for REL-dependent transactivation.
3.4. Knock down of p300ΔC slows the proliferation and reduces the soft agar colony-forming ability of RC-K8 cells
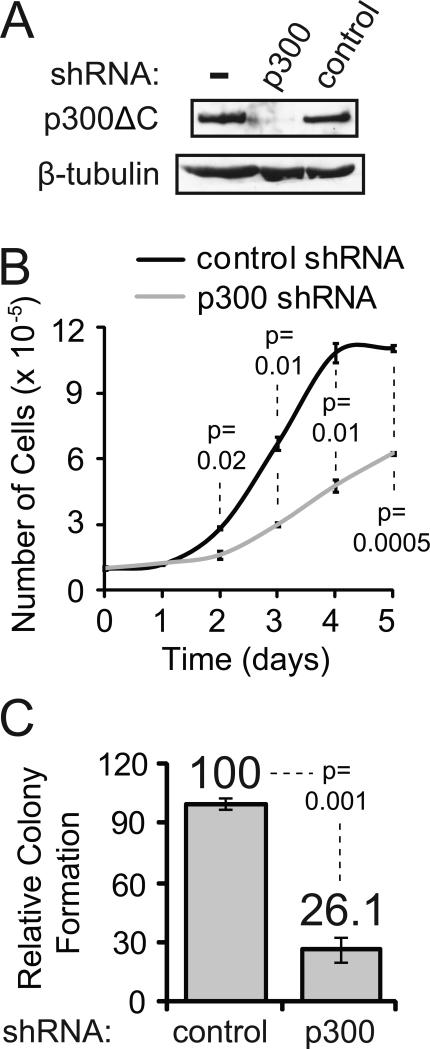
To determine whether p300ΔC plays a role in the proliferation of RC-K8 cells, we sought to knock down its expression in RC-K8 cells. RC-K8 cells were infected with a retroviral vector containing a short hairpin RNA (shRNA) that has previously been shown to knock down expression of wild-type p300 [9]. Western blotting confirmed that p300ΔC expression was greatly reduced in stable pools of RC-K8 cells expressing p300 shRNA as compared to non-transduced RC-K8 cells or RC-K8 cells expressing a control, non-targeting shRNA (Fig. 4A). We next compared the proliferation of RC-K8 cells expressing p300 shRNA to RC-K8 cells expressing control shRNA. As shown in Fig. 4B, p300ΔC knockdown cells proliferated significantly slower than cells expressing the control shRNA. Furthermore, p300ΔC knockdown cells formed approximately four-fold fewer colonies in soft agar as compared to RC-K8 cells expressing the control shRNA (Fig 4C). These results indicate that p300ΔC is essential for the optimal proliferation as well as growth in suspension of RC-K8 cells.
Fig. 4.
Knock down of p300ΔC slowed the proliferation and reduced the soft agar colony-forming ability of RC-K8 cells. (A) Anti-p300 Western blotting was performed on whole cell extracts from RC-K8 cells, RC-K8 cells expressing p300 shRNA, and RC-K8 cells expressing control shRNA (top panel). Anti-β-tubulin Western blotting was performed as a loading control (lower panel). Sixty μg of total protein was loaded in each lane. (B) 1×105 RC-K8 cells expressing p300 or control shRNA were plated into 16-mm wells. Cells were counted from three wells of each cell type every day for five days. (C) Soft agar colony assays were performed using RC-K8 cells expressing p300 or control shRNA. Results are the averages of four experiments performed with triplicate plates containing 250 and 500 cells. Colony numbers are relative to the number of colonies obtained with RC-K8 cells expressing the control shRNA (100).
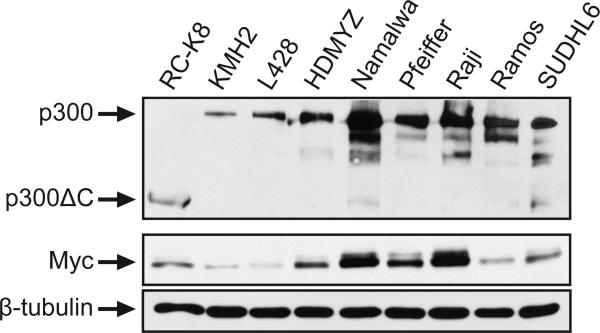
3.5. Alterations in p300 are not seen in eight additional B-lymphoma cell lines
We previously showed that five other human B-lymphoma cell lines (BJAB, Daudi, SUDHL4, IB4, BL41), as well as an avian transformed B-cell line, did not have obvious truncations in p300 [8]. To broaden this survey, we performed anti-p300 Western blotting on eight additional human B-lymphoma cell lines. None of these eight B-lymphoma cell lines appeared to have mutations that caused truncation of p300 (Fig. 5). Because wild-type p300 has been implicated in the repression of MYC transcription [9], we hypothesized that inactivation of WT p300 in RC-K8 cells might lead to increased MYC protein expression. However, we found that there was a range of MYC expression among these cell lines, and there was no obvious increase in MYC levels in RC-K8 cells as compared to the other B-lymphoma cell lines with apparently wild-type p300 (Fig. 5).
Fig. 5.
Expression of p300 and MYC in a panel of human B-lymphoma cell lines. Extracts from the indicated human B-lymphoma cell lines were analyzed by Western blotting for p300, MYC, and β-tubulin (as a loading control). In a given panel, all lanes contain equal amounts of cellular protein, except for the first lane in the top panel, which contains twice the amount of protein in order to detect the low levels of p300ΔC in RC-K8 cells (see also Fig. 2A).
4. Discussion
We have characterized the altered EP300 genomic locus, mRNA and protein (p300ΔC) from the human diffuse large B-cell lymphoma cell line RC-K8. Although truncations and mutations have been described for p300 and the related co-activator CBP in several types of cancer [2,7,13-15], the RC-K8 cell line is, to date, the only example of a p300/CBP C-terminal truncation occurring in a B-cell lymphoma [8].
DNA sequencing of the breakpoint in the altered EP300 locus in RC-K8 cells showed that sequences in the intron after exon 17 of EP300 are fused to sequences normally found on chromosome 6, within an intron of the BCKDHB gene. At the site of the EP300-chromosome 6 fusion in RC-K8 cells, there is a 9-bp sequence of unknown origin (see Fig. 1). The first 5 of these 9 bp are identical to the sequence on the chromosome 22 (EP300) side of the junction, and the last 5 bp are identical to an adjacent sequence on the chromosome 6 side. We noted a similar occurrence at the rearranged REL-NRG junction in RC-K8 cells where 12 of 26 new bases at the chromosomal fusion junction are identical to the proximal downstream NRG sequences [28]. The presence of duplications at breakpoint junctions suggests that these alterations are the result of abnormal breakage-joining such as normally occurs in V(D)J recombination. Of note, the immunoglobulin recombinase genes RAG1 and RAG2 have been reported to be NF-κB target genes [29], suggesting that the high NF-κB activity in RC-K8 cells could promote aberrant recombination.
In RT-PCR reactions of RC-K8 cell mRNA using a p300 primer and a 3’ p300ΔC-specific primer, we amplified a major band of ~120 bp that corresponds to the spliced version of the p300ΔC mRNA, as well as a minor band that co-migrated with the ~220-bp band amplified by PCR of genomic DNA (Fig. 1A). The 220-bp band seen in this RT-PCR probably arose from either contamination of the mRNA with genomic DNA or inefficient splicing of the pre-mRNA for p300ΔC. Of note, the sequence at the downstream splice site in the chromosome 6 non-EP300 sequences that is joined to exon 17 of EP300 is identical to the consensus splice acceptor sequence (AG at the 3’ end of the intron; see Fig. 1B).
The cloning of the p300ΔC cDNA from RC-K8 cells enabled us to carry out a functional characterization of the p300ΔC protein. Not surprisingly, given the deletion of the HAT domain, p300ΔC is defective as a transcriptional co-activator for REL in A293 cells (Figs. 3A,B). It is likely that p300ΔC is generally defective for transcriptional enhancement activity, given that numerous studies have shown that HAT activity is required for p300/CBP co-activator activity [7,30-32]. Furthermore, although both p300 and p300ΔC interact readily with REL, p300ΔC had a reduced ability to interact with transcription factor VP16 as compared to wild-type p300 (Fig. 2C). Therefore, in RC-K8 cells, p300ΔC is likely to have a broad range of effects on the many different cellular transcription factors with which p300 normally interacts to effect target gene expression. Of note, MYC protein expression was not especially reduced in RC-K8 cells (Fig. 5), even though wild-type p300 has been implicated in repression of MYC transcription [9].
Like wild-type p300, p300ΔC localizes to discrete “speckles” in the nucleus of chicken cells (Fig. 3B) and can interact with REL in the nucleus of RC-K8 cells [8]. Several groups have reported that p300 and CBP localize to nuclear subdomains, often called speckles, which correspond to sites of active transcription in interphase nuclei [33,34]. Therefore, even though p300ΔC lacks the HAT domain, it is likely to localize normally in RC-K8 cells. However, the subnuclear localization of p300ΔC and its lack of co-activator activity remain to be confirmed in RC-K8 cells themselves.
RC-K8 cells have several chromosomal alterations ([35]; http://www.dsmz.de/human_and_animal_cell_lines/info.php?dsmz_nr=561) including one within the REL locus on chromosome 2 [36]. Consequently, a chimeric REL protein, termed REL-NRG, is expressed in RC-K8 cells [28,36]. The RC-K8 cell line also contains two other mutations that impact the REL/NF-κB pathway: mutations that inactivate the NF-κB inhibitor IκBα [22] and inactivating mutations in the A20 ubiquitin-modifying protein [37]. RC-K8 cells contain abundant levels of nuclear REL (and REL-NRG) and express high levels of several REL/NF-κB target genes [22]. As we argued previously [8], the combination of REL-activating (IκBα and A20) and REL-inhibiting (REL-NRG and p300ΔC) mutations is likely to keep nuclear REL transcriptional activity within an effective range for oncogenic activity. Indeed, several studies have shown that optimal transformation of chicken lymphoid cells occurs when REL's transcriptional activation ability is at a moderate level [21,26,38-42]. Given that over-expression of REL can be toxic [43], the reduced REL activity afforded by mutations such as truncation of p300 may be better suited for sustained B-cell proliferation and oncogenicity in cases where REL (or other p300-dependent transcription factors) has unrestrained activity. Nevertheless, truncations of p300 were not detected in two B-lymphoma cell lines---KMH2 and L428---that also have inactivating mutations in IκBα and high NF-κB activity [44,45] (Fig. 5). To date, we have detected an obvious truncation of p300 only in the RC-K8 cell line, among 14 B-lymphoma cell lines that we have screened (Fig. 5; and ref. 8).
Consistent with p300ΔC contributing to the oncogenic state in RC-K8 cells, the full-length p300 protein is not expressed in RC-K8 cells (Fig. 2A; [8]), and knock down of p300ΔC reduced the growth of RC-K8 cells in both liquid medium and soft agar (Fig. 4B,C). Although over-expression of p300ΔC blocked the ability of wild-type p300 to enhance REL-dependent transactivation in reporter gene assays, it did so efficiently only when p300ΔC was in substantial excess to wild-type p300 (Fig. 3B). Thus, if p300ΔC contributes to the oncogenic state of RC-K8 cells by reducing normal p300-mediated co-activation, then reduced WT p300 expression may have been selected in these cells to maximize p300ΔC activity. As a defective co-activator bound to REL (or other transcription factors) in the nucleus, p300ΔC would likely reduce transactivation of several target genes. In addition, p300ΔC could block other co-activators from binding to REL (or other transcription factors). Consistent with this model, CBP can readily bind REL transactivation sequences in vitro, but CBP does not efficiently co-precipitate with REL from RC-K8 cells [8]. Alternatively, the N-terminal sequences retained in p300ΔC contribute to the growth of RC-K8 cells in a way that is not related to p300-dependent transcriptional co-activation.
Previous studies have suggested that EP300 is a tumor suppressor gene [11-15,19], primarily based on the deletion of the HAT domain in certain cancers as well as the ability of re-expressed p300 to reduce tumor cell growth in certain cells with p300 mutations. However, most of the p300 alterations in these cancer cell lines are C-terminal truncations accompanied by loss of heterozygosity (of the wild-type EP300 allele), suggesting that retention of the truncated EP300 copy is important for growth. Moreover, mice with a conditional knockout of the p300 gene in B cells do not develop lymphoma [46]. Therefore, we suggest that EP300 is not a tumor suppressor per se, but rather that expression of truncated, defective p300 proteins actively contributes to tumorigenesis in some cases. Future experiments will be aimed at determining how p300ΔC affects gene expression in RC-K8 cells and whether p300 mutations occur in other human B-lymphoma cell lines, especially those that have high levels of REL/NF-κB activity.
Supplementary Material
Acknowledgements
This work was supported by NIH grant CA047763 and ARRA supplement CA047763-21S3. R.C.T. was supported by NHLBI Hematology Training Grant T32 HL007501. We thank Drs. Cyril Benes and Ellen Cahir-McFarland for human lymphoma cell lines. We thank Drs. John Celenza, Cynthia Bradham, Tina Matos, and Tony Capobianco for helpful discussions.
Abbreviations
- aa
amino acid(s)
- bp
base pair
- DLBCL
diffuse large B-cell lymphoma
- FBS
fetal bovine serum
- DMEM
Dulbecco's modified Eagle's medium
- GST
glutathione S-transferase
- HAT
histone acetyltransferase
- NRG
non-REL gene
- PCR
polymerase chain reaction
- PEI
polyethylenimine
- QP
glutamine proline
- RACE
rapid amplification of cDNA ends
- RT-PCR
reverse transcriptase-PCR
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: None declared.
References
- 1.Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 2.Iyer NG, Özdag H, Caldas C. p300/CBP and cancer. Oncogene. 2004;23:4225–4231. doi: 10.1038/sj.onc.1207118. [DOI] [PubMed] [Google Scholar]
- 3.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive gene. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerritsen ME, Williams AJ, Neish AS, Moore S, Shi Y, Collins T. CREB-binding protein/p300 are transcriptional co-activators of p65. Proc. Natl. Acad. Sci. USA. 1997;94:2927–2932. doi: 10.1073/pnas.94.7.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grossman SR. p300/CBP/p53 interaction and regulation of the p53 response. Eur. J. Biochem. 2001;268:2773–2778. doi: 10.1046/j.1432-1327.2001.02226.x. [DOI] [PubMed] [Google Scholar]
- 6.Oswald F, Täuber B, Dobner T, Bourteele S, Kostezka U, Adler G, Liptay S, Schmid BM. p300 acts as a transcriptional coactivator for mammalian Notch-1. Mol. Cell. Biol. 2001;21:7761–7774. doi: 10.1128/MCB.21.22.7761-7774.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalkhoven E. CBP and p300: HATs for different occasions. Biochem. Pharmacol. 2004;68:1145–1155. doi: 10.1016/j.bcp.2004.03.045. [DOI] [PubMed] [Google Scholar]
- 8.Garbati M, Alço G, Gilmore TD. The histone acetyltransferase p300 co-activator is functionally inactivated in the human diffuse large B-cell lymphoma cell line RC-K8. Cancer Lett. 2010;291:237–245. doi: 10.1016/j.canlet.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sankar N, Baluchamy S, Kadeppagari R-K, Singhal G, Weitzman S. p300 provides a corepressor function by cooperating with YY1 and HDAC3 to repress c-Myc. Oncogene. 2008;27:5717–5728. doi: 10.1038/onc.2008.181. [DOI] [PubMed] [Google Scholar]
- 10.Yao TP, Oh SP, Fuchs M, Zhou ND, Ch'ng LE, Newsome D, Bronson RT, Li E, Livingston DM, Eckner R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 11.Rebel VI, Kung AL, Tanner EA, Yang H, Bronson RT, Livingston DM. Distinct roles for CREB-binding protein and p300 in hematopoietic stem cell self-renewal. Proc. Natl. Acad. Sci. USA. 2002;99:14789–14794. doi: 10.1073/pnas.232568499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suganuma T, Kawabata M, Ohshima T, Ikeda M-A. Growth suppression of human carcinoma cells by reintroduction of the p300 co-activator. Proc. Natl. Acad. Sci. USA. 2008;99:13073–13078. doi: 10.1073/pnas.192586699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muraoka M, Konishi M, Kikuchi-Yanoshita R, Tanaka K, Shitara N, Chong JM, Iwama T, Miyaki M. p300 gene alterations in colorectal and gastric carcinomas. Oncogene. 1996;12:1565–1569. [PubMed] [Google Scholar]
- 14.Gayther SA, Batley SJ, Linger L, Bannister A, Thorpe K, Chin S-F, Daigo Y, Russell P, Wilson A, Sowter HM, Delhanty JD, Ponder BA, Kouzarides T, Caldas C. Mutations truncating the EP300 acetylase in human cancers. Nat. Genet. 2000;24:300–303. doi: 10.1038/73536. [DOI] [PubMed] [Google Scholar]
- 15.Bryan EJ, Jokubaitis VJ, Chamberlain NL, Baxter SW, Dawson E, Choong DYH, Campbell IG. Mutation analysis of EP300 in colon, breast and ovarian carcinomas. Int. J. Cancer. 2002;102:137–141. doi: 10.1002/ijc.10682. [DOI] [PubMed] [Google Scholar]
- 16.Laï JL, Jouet JP, Bauters F, Deminatti M. Chronic myelogenous leukemia with translocation (8:22): report of a new case. Cancer Genet. Cytogenet. 1985;17:365–366. doi: 10.1016/0165-4608(85)90121-9. [DOI] [PubMed] [Google Scholar]
- 17.Ida K, Kitabayashi I, Taki T, Taniwaka M, Noro K, Yamamoto M, Ohki M, Hayashi Y. Adenoviral E1A-associated protein p300 is involved in acute myeloid leukemia with t(11;22)(q23;q13) Blood. 1997;90:4699–4704. [PubMed] [Google Scholar]
- 18.Kitabayashi I, Aikawa Y, Yokoyama A, Hosoda F, Nagai M, Kakazu N, Abe T, Ohki M. Fusion of MOZ and p300 histone acetyltransferases in acute monocytic leukemia with a t(8;22)(p11;q13) chromosome translocation. Leukemia. 2001;15:89–94. doi: 10.1038/sj.leu.2401983. [DOI] [PubMed] [Google Scholar]
- 19.Mees ST, Mardin WA, Wendel C, Baeumer N, Willscher E, Senninger N, Schleicher C, Colombo-Benkmann M, Haier J. EP300—a miRNA-regulated metastasis suppressor gene in ductal adenocarcinomas of the pancreas. Int. J. Cancer. 2010;126:114–124. doi: 10.1002/ijc.24695. [DOI] [PubMed] [Google Scholar]
- 20.Fermento ME, Gandini NA, Lang CA, Perez JE, Maturi HV, Curino AC, Facchinetti MM. Intracellular distribution of p300 and its differential recruitment to aggresomes in breast cancer. Exp. Mol. Pathol. 2010;88:256–264. doi: 10.1016/j.yexmp.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 21.Gilmore TD, Kalaitzidis D, Liang M-C, Starczynowski DT. The c-Rel transcription factor and B-cell proliferation: a deal with the devil. Oncogene. 2004;23:2275–2286. doi: 10.1038/sj.onc.1207410. [DOI] [PubMed] [Google Scholar]
- 22.Kalaitzidis D, Davis RE, Rosenwald A, Staudt LM, Gilmore TD. The human B-cell lymphoma cell line RC-K8 has multiple genetic alterations that dysregulate the Rel/NF-κB signal transduction pathway. Oncogene. 2002;21:8759–8768. doi: 10.1038/sj.onc.1206033. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 24.Starczynowski DT, Reynolds JG, Gilmore TD. Mutations of tumor necrosis factor α-responsive serine residues within the C-terminal transactivation domain of human transcription factor REL can enhance its in vitro transforming ability. Oncogene. 2005;24:7355–7368. doi: 10.1038/sj.onc.1208902. [DOI] [PubMed] [Google Scholar]
- 25.Scherr M, Battmer K, Ganser A, Eder M. Modulation of gene expression by lentiviral mediated delivery of small interfering RNA. Cell Cycle. 2003;2:251–257. [PubMed] [Google Scholar]
- 26.Chin M, Herscovitch M, Zhang N, Waxman DJ, Gilmore TD. Overexpression of an activated REL mutant enhances the transformed state of the human B-lymphoma BJAB cell line and alters its gene expression profile. Oncogene. 2009;28:2100–2111. doi: 10.1038/onc.2009.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L, Grossman SR, Kieff E. Epstein-Barr virus nuclear protein 2 interacts with p300, CBP, and PCAF histone acetyltransferases in activation of the LMP1 promoter. Proc. Natl. Acad. Sci. USA. 2000;97:430–435. doi: 10.1073/pnas.97.1.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalaitzidis D, Gilmore TD. Genomic organization and expression of the rearranged REL proto-oncogene in the human B-cell lymphoma cell line RC-K8. Genes Chromosomes Cancer. 2002;34:129–135. doi: 10.1002/gcc.10051. [DOI] [PubMed] [Google Scholar]
- 29.Verkoczy L, Aït-Azzouzene D, Skog P, Märtensson A, Lang J, Duong B, Nemazee D. A role for nuclear factor kappaB/rel transcription factors in the regulation of recombinase activator genes. Immunity. 2005;22:519–531. doi: 10.1016/j.immuni.2005.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bordoli L, Hüsser S, Lüthi U, Netsch M, Osmani H, Eckner R. Functional analysis of the p300 acetyltransferase domain: the PHD finger of p300 but not of CBP is dispensable for enzymatic activity. Nucleic Acids Res. 2001;29:4462–4471. doi: 10.1093/nar/29.21.4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu C, Lu J, Tan J, Li L, Huang B. Human interleukin-5 expression is synergistically regulated by histone acetyltransferase CBP/p300 and transcription factors C/EBP, NF-AT and AP-1. Cytokine. 2004;27:93–100. doi: 10.1016/j.cyto.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 32.Sun H, Lu J, Wei L, Wang X, Xu X, Dong M, Huang B. Histone acetyltransferase activity of p300 enhances the activation of IL-12 p40 promoter. Mol. Immunol. 2004;41:1241–1246. doi: 10.1016/j.molimm.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 33.von Mikecz A, Shang S, Montminy M, Tan EM, Hemmerich P. CREB-binding protein (CBP)/p300 and RNA polymerase II colocalize in transcriptionally active domains in the nucleus. J. Cell Biol. 2000;150:265–273. doi: 10.1083/jcb.150.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lassot I, Estabaud E, Emilani S, Benkirane M, Benerous R, Margottin-Gouguet F. p300 modulates ATF4 stability and transcriptional activity independently of its acetyltransferase domain. J. Biol. Chem. 2005;280:41537–41545. doi: 10.1074/jbc.M505294200. [DOI] [PubMed] [Google Scholar]
- 35.Kubonishi I, Niiya K, Yamashita M, Yano S, Abe T, Ohtsuki T, Mihoshi I. Characterization of a new human lymphoma cell line (RC-K8) with t(11;14) chromosome abnormality. Cancer. 1986;58:1453–1460. doi: 10.1002/1097-0142(19861001)58:7<1453::aid-cncr2820580713>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 36.Lu D, Thompson JD, Gorski GK, Rice NR, Mayer MG, Yunis JJ. Alterations at the rel locus in human lymphoma. Oncogene. 1991;6:1235–1241. [PubMed] [Google Scholar]
- 37.Compagno M, Lim WK, Grunn A, Nandula SV, Brahmachary M, Shen Q, Bertoni F, Ponzoni M, Scandurra M, Califano A, Bhagat G, Chadburn A, Dalla-Favera R, Pasqualucci L. Mutations of multiple genes cause deregulation of NF-κB in diffuse large B-cell lymphoma. Nature. 2009;459:717–721. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kamens J, Richardson P, Mosialos G, Brent R, Gilmore T. Oncogenic transformation by vrel requires an amino-terminal activation domain. Mol. Cell. Biol. 1990;10:2840–2847. doi: 10.1128/mcb.10.6.2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hrdlicková R, Nehyba J, Humphries EH. In vivo evolution of c-rel oncogenic potential. J. Virol. 1994;68:2371–2382. doi: 10.1128/jvi.68.4.2371-2382.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gilmore TD. Multiple mutations contribute to the oncogenicity of the retroviral oncoprotein v-Rel. Oncogene. 1999;18:6925–6937. doi: 10.1038/sj.onc.1203222. [DOI] [PubMed] [Google Scholar]
- 41.Starczynowski DT, Reynolds JG, Gilmore TD. Deletion of either C-terminal transactivation subdomain enhances the in vitro transforming activity of human transcription factor REL in chicken spleen cells. Oncogene. 2003;22:6928–6936. doi: 10.1038/sj.onc.1206801. [DOI] [PubMed] [Google Scholar]
- 42.Fan Y, Gélinas C. An optimal range of transcription potency is necessary for efficient cell transformation by c-Rel to ensure optimal nuclear localization and gene-specific activation. Oncogene. 2007;26:4038–4033. doi: 10.1038/sj.onc.1210164. [DOI] [PubMed] [Google Scholar]
- 43.Bash J, Zong WX, Gélinas C. c-Rel arrests proliferation of HeLa cells and affects critical cell regulators of the G1/S-phase transition. Mol. Cell. Biol. 1997;17:6526–6536. doi: 10.1128/mcb.17.11.6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wood KM, Roff M, Hay RT. Defective IκBα in Hodgkin cell lines with constitutively active NF-κB. Oncogene. 1998;16:2131–2139. doi: 10.1038/sj.onc.1201735. [DOI] [PubMed] [Google Scholar]
- 45.Emmerich F, Meiser M, Hummel M, Demel G, Foss HD, Jundt F, Mathas S, Krappmann D, Scheidereit C, Stein H, Dörkin B. Overexpression of IκBα without inhibition of NF-κB activity and mutations in the IκBα gene in Reed-Sternberg cells. Blood. 1999;94:3129–3134. [PubMed] [Google Scholar]
- 46.Xu W, Fukuyama T, Ney PA, Wang D, Rehg J, Boyd K, van Deursen JMA, Brindle PK. Global transcriptional coactivators CREB-binding protein and p300 are highly essential collectively but not individually in peripheral B cells. Blood. 2006;107:4407–4416. doi: 10.1182/blood-2005-08-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.