Summary
Protein quality control (PQC) degradation systems protect the cell from the toxic accumulation of misfolded proteins. Because any protein can become misfolded, these systems must be able to distinguish abnormal proteins from normal ones, yet be capable of recognizing the wide variety of distinctly shaped misfolded proteins they are likely to encounter. How individual PQC degradation systems accomplish this remains an open question. Here we show that the yeast nuclear PQC ubiquitin ligase San1 directly recognizes its misfolded substrates via intrinsically disordered N- and C-terminal domains. These disordered domains are punctuated with small segments of order and high sequence conservation that serve as substrate-recognition sites San1 uses to target its different substrates. We propose that these substrate-recognition sites, interspersed among flexible, disordered regions, provide San1 an inherent plasticity that allows it to bind its many, differently shaped misfolded substrates.
Introduction
Correct folding of proteins is essential for normal cellular physiology. When folding goes awry, misfolded proteins can acquire toxic aggregation-prone states that lead to cellular dysfunction. This presents an ongoing challenge for the cell because of the continuous stochastic production of misfolded proteins through synthesis errors, environmental stresses, and damage by reactive metabolites. Genetic mutations can also cause structural aberrancies, creating misfolded mutant proteins with a propensity for aggregation and cellular toxicity. Protein aggregation has been linked to >35 human disorders including Alzheimer's, Parkinson's, Huntington's, and amyotrophic lateral sclerosis (Skovronsky et al., 2006). Interestingly, most aggregation disorders are associated with toxic accumulation and aggregation of misfolded proteins in the nucleus (Woulfe, 2007), suggesting the nucleus may be particularly susceptible to the effects of misfolded proteins. However, we know little about how the cell normally prevents nuclear accumulation of misfolded proteins.
One of the main ways the cell manages misfolded proteins is through Protein Quality Control (PQC) degradation. A key aspect of PQC degradation systems is that they must be able to distinguish abnormally folded proteins from normal ones. This presents a problem for the PQC degradation systems in that any protein can become misfolded and will do so in ways distinct from other proteins. Yet, each distinctly misfolded protein must be recognized by a limited set of PQC degradation systems. How PQC degradation systems broadly recognize their variant misfolded substrates has been a longstanding question in PQC degradation.
Studies of PQC degradation in the cytoplasm and ER have shed some light on this question. In the mammalian cytoplasm, the ubiquitin ligase CHIP interacts with Hsp70 chaperones and ubiquitinates Hsp70 client proteins that cannot be properly folded (McDonough and Patterson, 2003). In the yeast cytoplasm, PQC degradation mediated by the ubiquitin ligase Ubr1 uses Hsp70 and Hsp110 chaperones (Heck et al., 2010; Nillegoda et al., 2010), though the role of the chaperones in substrate selection is not yet clear. In the yeast ER, different ubiquitin ligase complexes target abnormal proteins based on where the structural lesion manifests itself in relation to the ER membrane. The ubiquitin ligase Hrd1 ubiquitinates proteins with structural lesions in luminal or membrane domains whereas the ubiquitin ligase Doa10 ubiquitinates proteins with structural lesions in cytoplasmic domains (Vembar and Brodsky, 2008). Hrd1 uses its partner Hrd3, the ER chaperone Kar2, and the lectin Yos9 for luminal substrate recognition (Carvalho et al., 2006; Denic et al., 2006; Gauss et al., 2006a; Gauss et al., 2006b). Hrd1 also uses its transmembrane domain as a specificity determinant to distinctly recognize different membrane substrates (Sato et al., 2009). Although substrate recognition is less clear in the case of Doa10, cytoplasmic chaperones are involved in some aspects of Doa10-mediated PQC degradation (Metzger et al., 2008).
Studies of PQC degradation in the nucleus have also identified ubiquitin ligases that target misfolded proteins. In yeast, the nuclear-localized ubiquitin ligase San1 specifically ubiquitinates abnormal nuclear proteins for proteasome degradation (Gardner et al., 2005). Subsequent studies in mammalian cells have identified potential roles in nuclear PQC degradation for the nuclear ubiquitin ligases PML IV and UHRF-2 (Fu et al., 2005; Iwata et al., 2009; Janer et al., 2006). At this point, the means by which nuclear PQC ligases target their substrates is not known.
To understand substrate selection in nuclear PQC degradation, we focused on how San1 recognizes its substrates. San1 does not contain any known chaperone-binding motifs, although it might bind substrates through other adaptor proteins. Alternatively, San1 might possess one or more domains that directly bind its substrates. Here we report San1 directly targets substrates using highly disordered domains that contain embedded substrate-recognition modules.
Results
San1 directly interacts with substrates
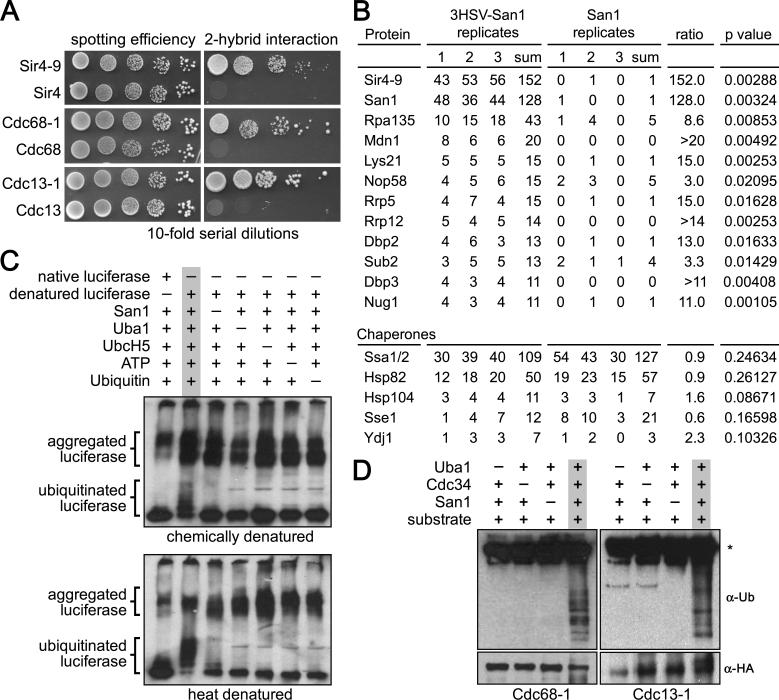
To understand how San1 interacts with substrates, we first developed a 2-hybrid approach. We previously found that San1 specifically targets the mutant temperature-sensitive proteins Sir4-9, Cdc68-1, and Cdc13-1 for degradation whereas normal Sir4, Cdc68, and Cdc13 are not degraded (Gardner et al., 2005). For the 2-hybrid to report San1's PQC function accurately, San1 must interact with the mutant forms of these proteins and not with the normal forms. To test this, we fused to the Gal4 DNA-binding domain (GBD) a ubiquitination-deficient version of San1, which contains the C279S substitution in the RING domain (Gardner et al., 2005). We also fused the Gal4 activation domain (GAD) to either normal or mutant Sir4, Cdc68, and Cdc13. The GAD fusions were coexpressed with GBD-San1C279S in cells containing the HIS3 gene under the control of a Gal4-inducible promoter. This construct requires a successful 2-hybrid interaction for transcription and colony growth on media lacking histidine. As anticipated, we observed an interaction between San1 and mutant Sir4-9, Cdc68-1, and Cdc13-1 but not with normal Sir4, Cdc68, and Cdc13 (Figure 1A).
Figure 1. San1 directly interacts with substrates.
(A) Cells expressing GBD-San1C279S and each GAD fusion were spotted onto media plus or minus histidine to measure spotting efficiency and 2-hybrid interaction. (B) The top 12 proteins with ≥3-fold enrichment in the 3HSV-tagged coIP are shown at the top. Chaperones known to function in PQC degradation are shown at the bottom. Numbers are spectral counts for each protein in each replicate (C) Chemical or heat denatured luciferase was added to a reaction containing the San1 ubiquitination cascade. Western blot was probed with anti-luciferase antibody. Dashes above the lanes indicate which reagent was excluded from the reaction. (D) 2xHA-tagged substrates were immunoprecipitated from E. coli cells expressing the San1 ubiquitination cascade. Western blots were probed with anti-ubiquitin antibodies to assess substrate ubiquitination or anti-HA antibodies to assess substrate immunoprecipitation. Dashes above the lanes indicate which components were not expressed. Asterisk marks the antibody band.
We next devised a co-immunoprecipitation (coIP) tandem mass spectrometry (MS) approach to survey all San1-interacting proteins including substrates and/or partners that might mediate substrate binding. To identify both stable interactors (such as partners) and transient interactors (such as substrates), we used in vivo formaldehyde crosslinking to maintain all interactions throughout the coIP. We previously appended a 3xHSV epitope tag to natively expressed San1 in a sir4-9 strain and found it was functional for Sir4-9 degradation (Gardner et al., 2005). We used this strain to immunoprecipitate San1 from cells expressing the known San1 substrate Sir4-9. We also used a congenic sir4-9 strain with untagged San1 as a negative control. All coIPs were performed in triplicate with independent cultures. We categorized a protein as interacting with San1 if the summed spectral counts for that protein across the tagged replicates exceeded the summed spectral counts across the untagged replicates by ≥3-fold, and if the spectral counts in the tagged replicates were statistically different from the untagged replicates as measured by a t-test (p ≤0.05).
Of the 328 total proteins identified by MS (Table S1), 25 met the enrichment criteria. Those proteins with ≥10 summed spectral counts in the tagged replicates are listed in Figure 1B. The San1 substrate Sir4-9 had the greatest number of spectral counts in the tagged coIP replicates and showed the greatest enrichment over the untagged replicates. We did not observe enrichment for any chaperone in sir4-9 cells (Figure 1B, bottom). We also did not identify any other protein with a function consistent with PQC; 8 of the 10 other enriched proteins are nucleolar and function in ribosome biogenesis. Perhaps San1 regulates a subset of nucleolar proteins or, more likely, these proteins are prone to misfolding under the growth conditions used. Because Sir4-9 had the greatest enrichment in the coIP and no other protein identified fit the role of an intermediary, the interaction between San1 and Sir4-9 is likely direct.
We next determined if San1 could directly ubiquitinate a misfolded substrate using an in vitro ubiquitination assay in which we generated the misfolded substrate through heat or chemical denaturation. We performed the assay using purified San1, ubiquitin cascade components (Uba1, UbcH5a, and ubiquitin), and luciferase that was either native or denatured by preincubation in 4M urea or incubation at 42°C. In the presence of all San1-pathway components, we detected a ladder of ubiquitinated species with denatured luciferase and not with native luciferase (Figure 1C). We did not observe ubiquitination of the aggregated forms of denatured luciferase suggesting that San1 is unable to recognize a misfolded protein once it aggregates.
We also attempted the in vitro ubiquitination assay using purified forms of San1's native yeast substrates. We found, however, that most San1 substrates aggregate during purification from E. coli. To circumvent this problem, we developed an “in coli” ubiquitination assay in which we co-express either San1 yeast substrate Cdc68-1 or Cdc13-1 with San1, Uba1, Cdc34, and ubiquitin in E. coli – a prokaryote that lacks the ubiquitin-proteasome machinery and thus any protein that would function with San1. In this way, San1 has the opportunity to access each substrate as it is made and before it has a chance to aggregate. Using this technique, we found that San1 ubiquitinated Cdc68-1 and Cdc13-1 without the aid of any yeast protein other than the required ubiquitination cascade factors (Figure 1D). Taken together, the results of the 2-hybrid, co-IP, in vitro and in coli assays all support the conclusion that San1 recognizes its misfolded substrates via direct interaction.
San1 is intrinsically disordered
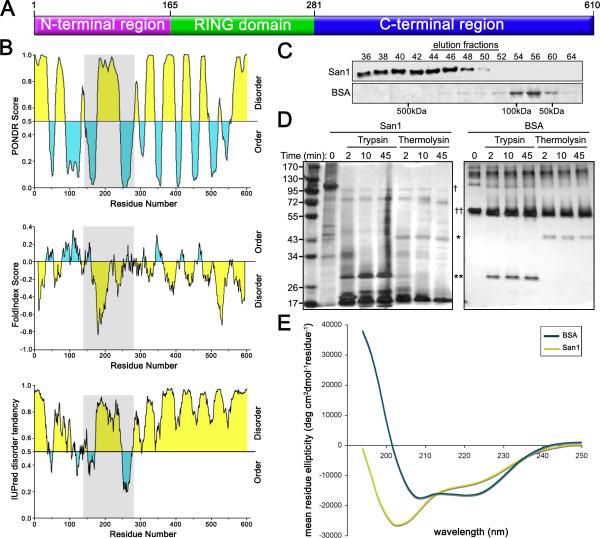
To understand how San1 might bind its substrates, we examined San1's sequence characteristics and domain composition. San1 has an N-terminal region (residues 1-164), a RING domain (residues 165-280), and a C-terminal region (residues 281-610) (Figure 2A). The RING domain confers ubiquitin ligase activity (Dasgupta et al., 2004; Gardner et al., 2005). However, the N- and C-terminal regions do not contain any known domains that could inform us of function. We next applied multiple protein disorder prediction methods to San1's sequence. These methods consistently predicted San1 to be >60% disordered with a similar topology of disorder (Figure 2B). The top panel was generated using PONDR VL-XT which predicts disorder based on sequence attributes typically found in regions that are absent in x-ray and NMR structures (Romero et al., 2001). The middle panel was generated using FoldIndex which plots charge:hydropathy in a moving window across a sequence (Prilusky et al., 2005). High charge and low hydropathy are characteristic of disordered proteins (Uversky et al., 2000). The bottom panel was generated using IUPred which identifies disordered regions that cannot form stabilizing interresidue interactions and predicts disorder by calculating pairwise interresidue interaction energies (Dosztanyi et al., 2005).
Figure 2. San1 is disordered.
(A) A cartoon representing overall topology of San1. (B) Disorder predictions of San1. Top panel is PONDR (http://www.pondr.com/). Middle panel is FoldIndex (http://bip.weizmann.ac.il/fldbin/findex). Bottom panel is IUPred (http://iupred.enzim.hu/index.html). Predicted disordered regions are in yellow and predicted ordered regions are in blue. San1's RING domain is in gray. (C) Purified San1 or BSA was loaded onto a Superdex 200 column and eluted with 50mM NaCl, 15mM Na2PO4, pH7.3. Western blot of San1 was probed with anti-HSV antibodies. BSA gel was stained with Coomassie. (D) Purified San1 or BSA was incubated with trypsin or thermolysin for the indicated times. Proteins were separated by SDS-PAGE and visualized by silver stain. The locations of trypsin (*), thermolysin (**), San1 (†), and BSA (††) are marked. (E) CD spectra of San1 (yellow) and BSA (blue) were recorded at 0.2 mg/ml for San1 and 0.15 mg/ml for BSA in 50mM NaCl and 15mM Na2HPO4, pH7.3 at 25°C.
San1's predicted disorder prompted us to measure if active San1 purified from E. coli is actually disordered using standard biochemical methods to probe disorder: gel filtration, limited proteolysis, and circular dichroism. Because intrinsically disordered proteins have larger hydrodynamic volumes than similarly sized structured proteins, they typically migrate at molecular weights much larger than predicted during gel filtration (Receveur-Brechot et al., 2006). It was previously found that San1 purified from yeast migrates at ~500 kDa rather than its calculated size of ~66 kDa and it was proposed this was due to San1 existing in a multi-protein complex in yeast (Dasgupta et al., 2004). In light of the disorder predictions, we repeated the gel filtration assay using San1 purified from E. coli and found that San1 migrated at ~500 kDa despite the absence of any yeast proteins (Figure 2C), indicating that San1 is either disordered and/or forms multimers with itself. To examine if San1 forms multimers, we coexpressed differently tagged versions of San1 in E. coli and found that neither version copurified with the other (E.K.F. and R.G.G., unpublished data), indicating that San1 does not self-associate.
We next probed San1's structure with limited proteolysis which takes advantage of the fact that protease cleavage sites are usually inaccessible in structured regions but readily accessible in regions of disorder (Fontana et al., 2004). We exposed San1 and the similarly sized bovine serum albumin (BSA) to trypsin or thermolysin for varying lengths of time and analyzed their proteolysis by SDS-PAGE. We found San1 was completely digested at the earliest time points whereas BSA resisted digestion throughout the time course of protease treatment (Figure 2D). We also observed that intact San1 migrated at ~100 kDa, larger than its calculated ~66 kDa mass. Intrinsically disordered proteins often show this behavior due to a low content of hydrophobic residues (Tompa, 2002).
Lastly, we analyzed San1's secondary structure by circular dichroism (CD) which is a technique used to determine the α-helical, β-sheet, and random coil content of proteins (Greenfield, 2006). The CD spectrum of San1 had a minimum close to 200 nm (Figure 2E), which is characteristic of a protein with mostly random coil content and is typical for intrinsically disordered proteins (Receveur-Brechot et al., 2006). By contrast, the CD spectrum of BSA had minima at 208 and 222 nm, which is characteristic of a protein with mostly α-helical content (Figure 2E). The San1 spectrum had a small valley at ~224 nm indicating San1 possesses some α-helical content. This is consistent with a secondary structure prediction using Jpred (Cole et al., 2008) which predicts San1 possesses ~15% α-helical and ~5% β-sheet content (Figure 3C). Altogether, the experimental data confirmed the theoretical predictions that San1 is intrinsically disordered.
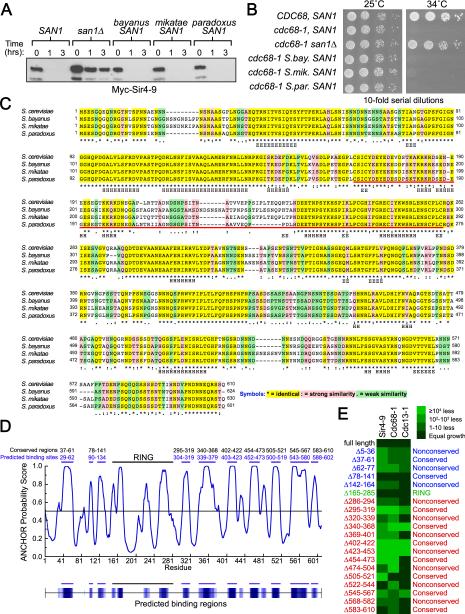
Figure 3. N- & C-terminal regions of San1 are involved in substrate interactions.
(A) Cycloheximide-chase assays of cells expressing Myc-Sir4-9 and San1 homologs were performed to assess complementation. Western blots were probed with anti-Myc antibodies. Time after cycloheximide addition is indicated above each lane. (B) Growth assays of cdc68-1 cells were performed to assess complementation of San1 homologs. Cells were incubated at permissive (25°C) and restrictive temperatures (34°C). (C) ClustalW alignment of S. cerevisiae San1 with its homologs from related Saccharomyces species was performed using Fungal Alignment viewer in the Yeast Genome Database (http://www.yeastgenome.org/cgibin/FUNGI/showAlign). The red bar underlines the RING domain. Secondary structure predicted by Jpred (http://www.compbio.dundee.ac.uk/www-jpred/) is indicated below each residue, with “H” indicating α-helix and “E” indicating β-sheet. (D) ANCHOR prediction of binding sites (http://anchor.enzim.hu/). Blue bars at the top mark the predicted binding sites. Black bars marked conserved regions. (E) Cells expressing the indicated San1 deletion (top) and GAD fusion (left) were spotted media plus or minus histidine to measure spotting efficiency and the 2-hybrid interaction.
San1 binds each substrate using different elements in its disordered N- & C-terminal regions
What might intrinsic disorder mean for San1's function in terms of substrate recognition? Intrinsic disorder can endow a protein with a wide range of conformational states that confer the ability to bind many differently shaped molecules (Dunker et al., 2008). San1's intrinsic disorder in the N- and C-terminal regions could provide San1 the conformational flexibility to bind many differently shaped proteins that have become misfolded in distinct ways. There is precedent for this type of recognition in PQC; intrinsically disordered regions in small heat shock proteins (sHSPs) are involved in the binding of misfolded substrates (Haslbeck et al., 2004; Jaya et al., 2009; Stromer et al., 2004).
From the sHSP precedent, we hypothesized that San1 contains substrate-recognition sites embedded within its disordered N- and C-terminal regions. If so, we reasoned that San1's intrinsic disorder should be maintained in San1 homologs and substrate-recognition sites in the disordered regions should be conserved. We therefore examined San1's homologs from different Saccharomyces species. Each homolog functioned equivalently to S. cerevisiae San1 for Sir4-9 degradation and cdc68-1 temperature sensitivity (Figure 3A & B). Each homolog also shared the same predicted disorder topology as S. cerevisiae San1 (Figure S1), indicating disorder is likely important for San1 function. To identify conserved regions within San1 that might be involved in substrate interactions, we compared S. cerevisiae San1's sequence with the homologs’ sequences using ClustalW (Thompson et al., 1994). We observed discrete identical segments distributed along San1's length that suggest conserved function (Figure 3C, yellow highlighting).
We noticed that the pattern of alternating identical and nonidentical segments in the ClustalW alignment mirrored the pattern of alternating order and disorder predicted in San1's PONDR VL-XT topology. This pattern is intriguing in light of the emerging idea that many intrinsically disordered proteins are punctuated by small ordered regions that serve as binding sites for other proteins (Dunker et al., 2008). PONDR VL-XT predicts binding sites as dips in disorder (Dunker et al., 2008), and such dips are readily observed in San1's PONDR VL-XT profile (Figure 2B, top panel, blue regions). These dips map to the identical segments identified by ClustalW. Using ANCHOR, another predictor of binding sites within disordered regions (Dosztanyi et al., 2009), we obtained a similar result as with PONDR with the predicted binding sites mapping to the identical segments (Figure 3D).
Note that while PONDR and ANCHOR can predict binding sites, they can't distinguish segments that bind substrates from those that bind cofactors. For example, the RING domain contains a disordered segment flanked by two ordered segments predicted to be binding sites (Figures 2B and 3D). The ordered segments contain the Zn2+-coordinating Cys and His residues required for ubiquitin conjugase interaction and thus comprise the binding site for a cofactor, not substrates. Because of this potential ambiguity, we decided to conduct a systematic deletion analysis of San1 and use the 2-hybrid assay to identify actual substrate-binding sites in San1.
Prior to conducting the analysis, we envisioned two models for how San1 engages substrates. The first model is based on the conventional view of protein-protein interactions. In this model, San1 binds all substrates identically using the same defined substrate-binding site, similar to how Hsp70 chaperones bind substrates (Mayer and Bukau, 2005). If this model were correct, particular deletions in San1 would affect each substrate's interactions with San1 equally and identically. The second model combines San1's disorder and binding-site predictions with ideas of how the plasticity of disorder might be used to accommodate the binding of many differently shaped molecules. In this model, San1 binds each substrate differently using a distinct combination of substrate-recognition sites dispersed throughout its disordered regions. If this model were correct, deletions in San1's disordered regions would have varying effects on each individual substrate's interactions.
To identify substrate-binding sites and resolve which model is correct, we made 20 small deletions in San1 that spanned the length of each small identical and nonidentical segment identified by ClustalW. We then assessed how each deletion affected San1's ability to interact with Sir4-9, Cdc68-1, and Cdc13-1 in the 2-hybrid assay. Steady-state protein levels of each GBD-San1 deletion mutant were examined to ensure that any changes in interactions were not due to reduced San1 levels (Figure S2). We present the 2-hybrid interaction data as a heat map where the green color indicates the degree to which each San1 deletion affected the substrate interaction (Figure 3E). As expected, the RING domain deletion (residues 165-285) had little effect on substrate interactions consistent with its primary function in ubiquitin conjugase recruitment. In agreement with the second model, no single interaction profile emerged for San1-substrate interactions; multiple deletions in the N- and C-terminal disordered regions differentially affected each substrate interaction with San1 having a distinct interaction profile with each substrate. The identical, ordered segments were generally more important for substrate interactions particularly those spanning residues 295-319, 340-368, 402-422, 454-473, and 545-567. Their overall importance is consistent with these regions serving as conserved substrate-recognition modules.
Identification of additional substrates of San1
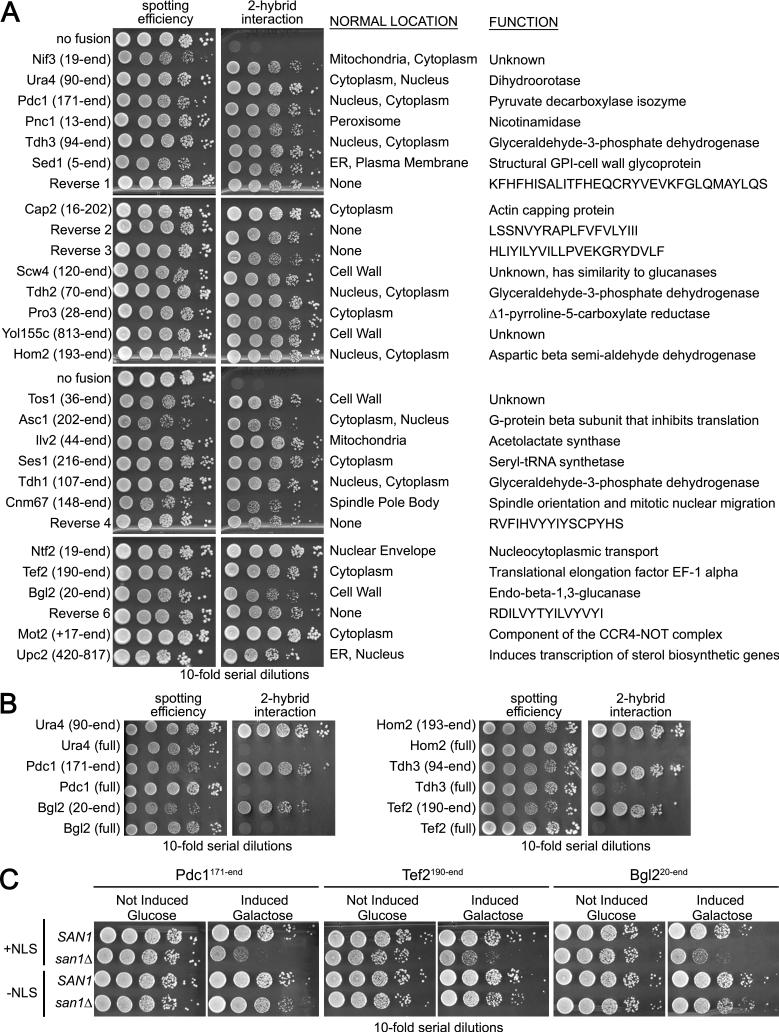
The 2-hybrid deletion analysis provided compelling evidence that San1 interacts in distinct ways with each substrate through substrate-recognition modules located in the disordered regions. This is a small set of substrates, however, and a larger collection of substrates would allow us to test the hypothesis more comprehensively. Because San1 interacts with substrates in the 2-hybrid, we decided to use a 2-hybrid selection to discover additional substrates. We transformed a plasmid library made from yeast cDNA fused to the GAD into cells expressing GBD-San1C279S with the HIS3 gene under control of a Gal4-inducible promoter. From 4×106 transformants, 98 grew into colonies on media lacking histidine. Of these, 32 different cDNAs were represented, 28 of which retested positive for an interaction with San1 (Figure 4A).
Figure 4. 2-hybrid genetic selection uncovers additional San1-interacting proteins.
(A) Cells expressing GBD-San1C279S and each of the listed proteins fused to GAD were spotted onto media plus or minus histidine to measure spotting efficiency and 2-hybrid interaction. The portion of each protein fused to GAD is in parentheses. The sequences of the reverse fusions are on the right. (B) Full-length versions of GAD fusions do not interact with San1. (C) Wild-type SAN1 or mutant san1Δ cells expressing the indicated GAD fusion, with or without an NLS, were spotted in 10-fold serial dilutions onto media with glucose to measure spotting efficiency or galactose to induce expression and toxicity.
None of the San1 interactors has a known function related to nuclear PQC degradation (Figure 4A). Thus, we suspected they aren't partners of San1 but are substrates due to some structural abnormality. We analyzed the actual sequences of the proteins fused to the GAD to see if they could be considered abnormal and found that 22 are truncated, 5 are encoded by cDNAs in their reverse orientation thus generating small “gibberish” sequences, and 1 is full length but with an added 5’ non-coding “gibberish” sequence (Figure 4A). Because most of the proteins identified were truncated, we retested the 2-hybrid interaction with full-length versions of these proteins and observed that San1 did not interact with the full-length versions (Figure 4B), again demonstrating San1's specificity for abnormal proteins in the 2-hybrid assay.
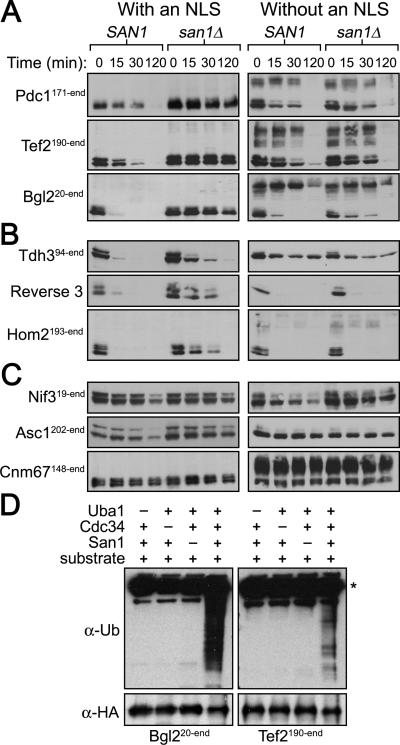
To verify the fusion proteins are San1 substrates and undergo San1-mediated degradation, we transformed the GAD-fusion plasmids into san1Δ cells. In many cases, we obtained few, if anytransformants indicating that some of the GAD fusions might be toxic in San1's absence. This is a predicted phenotype for aggregation-prone misfolded proteins targeted by San1 for degradation. Because the fusions are expressed from the constitutive ADH1 promoter, we placed them behind the galactose-inducible GAL1 promoter in order to regulate their expression. We examined toxicity by comparing the growth of SAN1 and san1Δ cells containing each GAD fusion on media that repressed (glucose) or induced (galactose) expression. Of the 28 proteins, 11 conferred observable growth defects in san1Δ cells (Figures 4C and S3). As expected from San1's nuclear PQC function, the toxicity of all fusions depended upon their nuclear localization. When the NLS was removed from the GAD, the growth defects for all fusions was attenuated (Figures 4C and S3).
We analyzed the degradation of each GAD fusion and found 25 of 28 required San1 to be fully degraded: 14 underwent primarily San1-dependent degradation (Figures 5A and S4A), 11 were subject to degradation that was partially dependent on San1 (Figures 5B and S4A), and 3 were stable when San1 was present (Figure 5C). Concordant with San1's nuclear PQC function, removal of the NLS from the GAD of the fusions caused the GAD-NLS fusions to become stable or degraded in a San1-independent manner (Figures 5A&B and S4B). We also examined San1-dependent ubiquitination of select GAD-fusion substrates using the in coli assay and found they were ubiquitinated in a San1-dependent manner (Figure 5D), indicating San1 directly targets these substrates as well.
Figure 5. Most GAD fusions are San1 substrates.
(A, B, C) Cycloheximide-chase assays of cells expressing the indicated GAD fusion were performed to assess stability in the presence or absence of SAN1. GAD fusion expression was induced by addition of galactose for 3 hours prior to cycloheximide addition. Time after cycloheximide addition is indicated above each lane. Anti-GAD antibodies were used to detect each GAD fusion. (D) 2xHA-tagged GAD substrates were immunoprecipitated from E. coli cells expressing the San1 ubiquitination cascade. Western blots were probed with anti-ubiquitin antibodies to assess substrate ubiquitination or anti-HA antibodies to assess substrate IP. Dashes above the lanes indicate which components were not expressed. Asterisk marks the antibody band.
GAD fusions with Asc1, Cnm67, and Nif3 were stable in SAN1 cells (Figure 5C), raising the possibility that these proteins might function as cofactors in general San1-mediated degradation. However, deletion of ASC1, CNM67, or NIF3 had no effect on the degradation of representative San1 substrates (Figure S4C), indicating these proteins are not general cofactors though they may recruit San1 for the degradation of specific, as yet unknown substrates.
San1 recognizes the 2-hybrid-derived substrates in distinct ways
Using the 2-hybrid assay and the same small San1 deletions described in Figure 3, we assessed which portions of San1's N- and C-terminal regions were involved in interactions with this larger substrate collection. An important point for this collection is that the San1 2-hybrid interaction could be assessed not only on media minus histidine, but on media minus adenine and media minus histidine plus 3-aminotriazole (+3AT), both of which are higher stringency media that require a stronger 2-hybrid interaction to register growth. This was not the case for Sir4-9, Cdc68-1, or Cdc13-1. Growth on higher stringency media is consistent with the 2-hybrid-derived substrates having a greater affinity for San1 than the original substrates.
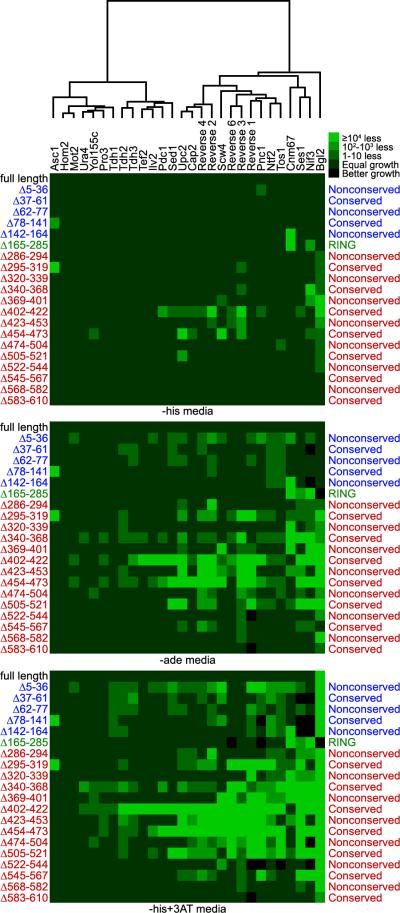
We assayed each interaction in duplicate on each different 2-hybrid media. As with Sir4-9, Cdc68-1, and Cdc13-1, the San1 deletions had variable effects for each substrate interaction (Figure 6). Although many regions throughout San1's N- and C-terminal regions were involved in substrate interactions, the conserved segments in the C-terminal region – particularly those spanning residues 295-319, 340-368, 402-422, 454-473, and 505-521 – appear to be the most important and widely involved in San1's interactions with these substrates. For most substrates, the deletions had little effect on the interactions when cells were plated on the lowest stringency media (Figure 6, -his media), but showed greater effects as the stringency for the 2-hybrid interaction was increased (-ade and -his+3AT media). This result is consistent with San1 having multiple, variable affinity contacts with each substrate such that loss of one contact affects the affinity in a way that only becomes more apparent as the requirement for a stable 2-hybrid interaction increases.
Figure 6. San1 recognizes each substrate using elements throughout its N- and C-terminal regions.
Dendogram and heat maps of pair-wise interactions between deletion variants of GBD-San1C279S, listed on the left, and each GAD fusion, listed across the top.
Although each substrate had a distinct pattern of San1 interactions, we wanted to see if there might be commonalities that weren't obvious at first inspection. We applied the hierarchical clustering package provided by Mathematica to cluster the collection of substrates according to growth changes observed with the San1 deletions. From this analysis, we discerned that a number of substrates fell into well-separated clusters (Figure 6 dendogram). This suggests that San1 recognizes some substrates in similar ways perhaps because they share comparable features of aberrancy.
Substrate degradation is distinctly influenced by San1 deletions
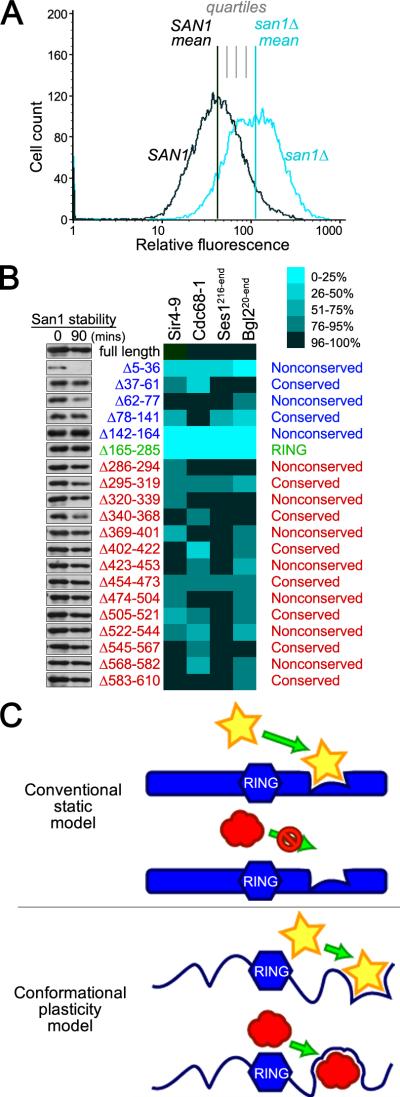
Lastly, we used substrate steady-state levels to measure the effect of a San1 deletion mutant on substrate degradation. A substrate's steady-state level reflects the balance between its synthesis and degradation and can be used to query subtle changes in the degradation rate that are difficult to observe by conventional decay-based degradation assays. Because we needed a facile way to quantify the steady-state levels of multiple substrates in the presence of all 20 San1 deletion variants, we replaced the GAD with GFP in select substrates (Sir4-9, Cdc68-1, Ses1216-end, and Bgl220-end). This allowed us to use flow cytometry to measure the mean substrate-GFP fluorescence in a population of cells expressing a particular San1 deletion, similar to the method used to query mean Hmg2-GFP fluorescence in cells expressing mutants of Hrd1 (Gardner et al., 2000).
To compute a quartile measure of San1's activity, we compared a substrate's mean steady-state level in cells expressing each San1 deletion to its mean level in cells that have San1 fully intact or completely deleted (Figure 7A). The mean for a substrate-San1 deletion pair was determined in triplicate. The stability of each San1 deletion was also assessed to insure that the differences measured weren't due to reduced San1 levels (Figure 7B). This analysis revealed that each substrate had a distinct steady-state profile with the San1 deletions, consistent with what we observed in the 2-hybrid interaction profiles. The majority of deletions had modest effects on steady-state levels (75-95% of full-length San1 activity). This result is consistent with the model that San1 engages each substrate using multiple recognition modules such that other modules can compensate to some extent for the loss of any single module.
Figure 7. Each substrate has a distinct degradation profile with the San1 deletions.
(A) Cartoon illustrating quartile scoring of steady-state effects. (B) Steady-state levels of the indicated substrate were examined by flow cytometry. The percent of normal San1 function was calculated by comparing each deletion steady-state level median with the median in SAN1 and san1Δ cells. Cycloheximide-chase assays of cells with each SAN1 deletion were performed to assess stability of San1. Western blots were probed with anti-HSV antibodies. Time after cycloheximide addition is indicated above each lane. (C) Model for how San1 uses the plasticity of disorder to interact with its substrates.
Note that the overall substrate steady-state profiles are similar but not identical to the 2-hybrid interaction profiles. We expect this to be the case for a few reasons. First, the different GFP and GAD fusions might impose different steric restrictions on how San1 “sees” its substrates. Second, the strength of interaction required to register a 2-hybrid readout will not necessarily be the same for ubiquitination and degradation; the 2-hybrid assay likely requires greater interaction strengths and, thus, will be more sensitive to subtle alterations in binding. Third, and most importantly, there will be regions in San1 that aren't required for substrate interactions but are required for substrate ubiquitination (the RING domain) or for controlling San1 function. For example, two deletion mutants, San1Δ5-36 and San1Δ142-164, were more pronounced in their effects than the other deletion mutants (0-50% the activity of full-length San1). San1Δ5-36 showed significantly decreased stability compared with full-length San1 (Figure 7B). Thus, this region controls San1 stability and the effects on substrate steady-state levels are likely due to reduced San1 steady-state levels. The 142-164 segment is immediately adjacent to the first Zn2+-coordinating Cys residue and actually constitutes part of the first ordered segment of San1's RING domain (Figure 2B and 3C). As such, the substrate stabilization observed with San1Δ142-164 is likely due to a compromised RING domain structure.
Discussion
The molecular mechanisms of enzyme-substrate interactions have been traditionally viewed through the lens of 3D structure whereby a structurally defined region within the enzyme fits a defined structural motif within the substrate protein. This view is difficult to reconcile with the mechanism of substrate recognition in PQC degradation because the proteins that become PQC substrates do so by having lost their native structures. Furthermore, PQC degradation systems must have a broadly inclusive binding mechanism that can recognize an immense diversity of unrelated and differently shaped misfolded proteins. How PQC degradation systems recognize substrates remains an open question.
San1's mode of substrate recognition
In exploring San1's function, we discovered that San1 uses highly disordered N- and C-terminal regions to bind its disparate substrates. Based on this, we propose the “disorder targets misorder” hypothesis whereby the intrinsic disorder of these regions endows San1 with the conformational plasticity to accommodate the binding of each distinctly shaped, misfolded substrate (Figure 7C, model 2). Although no single segment within the disordered regions was universally required for San1-substrate interactions, the conserved segments, particularly in the C-terminal region, were important for most interactions. From an evolutionary perspective, it makes sense that substrate-binding sites would be conserved because they encode essential substrate-recognition specificities. In contrast, flexible regions would not require sequence conservation, only that they remain flexible.
Our hypothesis for San1's substrate-binding mechanism has precedent in PQC. Intrinsic disorder has been documented in chaperones and small heat shock proteins (sHSPs) (Bhattacharyya and Das, 1999; Kim et al., 2002), and sHSPs use their disordered regions for substrate interactions (Haslbeck et al., 2004; Jaya et al., 2009; Stromer et al., 2004). The common use of disorder to bind substrates makes it tempting to speculate that San1 and sHSPs evolved from a common ancestral protein. The potential similarity in substrate recognition between San1 and sHSPs might also indicate that San1 recognizes exposed hydrophobicity analogous to sHSPs (van Montfort et al., 2001). Supporting this speculation, the gibberish sequences identified in the 2-hybrid selection are primarily hydrophobic (Figure 4A).
San1's plasticity in recognizing each substrate
While each San1-substrate interaction profile was distinct, a cluster analysis of the interaction data revealed that substrates could be grouped into well-separated clusters. This clustering suggests two possibilities concerning San1's plasticity in recognizing different substrates. First, San1 may possess a broad, but limited number of distinct substrate-binding modalities. Alternatively, the substrates in each cluster may share comparable abnormal features and shapes that are recognized by San1 in a similar way. To better understand San1's plasticity and range of San1-substrate interactions at the molecular level, we will need to develop methods to visualize exactly how San1 physically engages each substrate.
The spectrum of PQC substrates targeted by San1
The 2-hybrid selection allowed us to assemble the largest collection of substrates to date for any known PQC ubiquitin ligase. The lesions in these substrates consist of small truncations (<20 residues), large truncations (>20 residues), or gibberish sequences. These contrast with the temperature-sensitive missense mutations in Sir4-9, Cdc68-1, and Cdc13-1. While temperature-sensitive mutants have historically been a main class of substrate used to study PQC degradation systems, it is important to consider that truncated and gibberish proteins will also occupy the in vivo spectrum of substrates that PQC degradation systems will encounter in the cell. For example, truncated proteins can arise through a number of common mechanisms: frameshift mutations, nonsense mutations, failures in nonsense-mediated decay, premature transcription or translation termination, mRNA splicing defects, and the action of cellular proteases. In fact, proteolytic cleavage precedes aggregation and neurodegeneration in a number of tauopathies and polyQ-expansion disorders (Walsh et al., 2005; Wang et al., 2010). Gibberish proteins can also arise through a number of common mechanisms: termination codon read through, failure to suppress cryptic transcription, frameshift mutations, and mRNA splicing defects. Considering these important physiological failures, the substrates identified by the 2-hybrid selection represent additional classes of abnormal proteins targeted by San1 and thus provide a rich resource to probe San1's general mechanism of substrate recognition.
San1 targets abnormal proteins that are toxic to the cell
It is generally assumed PQC degradation systems target misfolded proteins that are toxic to the cell but there are few published examples of PQC degradation substrates that are toxic. It surprised us then that toxic proteins constituted such a large proportion of the San1 substrates identified by our 2-hybrid selection, particularly as this was performed without a selection for toxicity being imposed. Of the 25 2-hyrbid-derived substrates, 11 (44%) were toxic to some measurable degree. If we consider only the 14 substrates that undergo primarily San1-dependent degradation, the proportion of toxic proteins found increases to 78%. This means that either a significant proportion of all abnormal proteins produced are toxic or San1 has an exceptional proclivity for recognizing toxic abnormal proteins. Either way, we think the high proportion of toxic proteins identified in the 2-hybrid analysis reflects the fundamental raison d'être for PQC degradation – to protect the cell from harm caused by the accumulation of misfolded proteins. It will be interesting to learn if the feature of aberrancy San1 recognizes within its substrates is the same feature that confers toxicity to a protein. We speculate that PQC degradation systems likely evolved to recognize toxic aberrant features such as exposed hydrophobicity that can cause aggregation.
The San1 substrate-ubiquitination cascade can be reconstructed in bacteria
In this study, we developed a ubiquitination assay that reconstituted the San1 cascade in E. coli. Other groups have previously reconstituted a SUMOylation cascade in E. coli (Mencia and de Lorenzo, 2004; Okada et al., 2009; Uchimura et al., 2004), but a more complicated ubiquitination cascade had yet to be reconstituted in a similar manner. While the in coli ubiquitination assay provides a key tool to examine PQC ubiquitin ligase function, we think it could also have utility in understanding the interaction between other ubiquitin ligases and their substrates.
Experimental Procedures
Yeast strains
Parent yeast strains used in this study are PJ69-3D (James et al., 1996), BY4741 (Brachmann et al., 1998), RGY506 (BY4741 san1Δ), RGY187 and RGY649 (Gardner et al., 2005).
2-hybrid Assays
All interaction tests were performed in duplicate using two independent isolates. Growth of cells expressing a GAD fusion and a GBD-San1 deletion was scored relative to growth of cells expressing the identical GAD fusion and full-length GBD-San1 on each different stringency plate. Growth of cells expressing a San1 deletion that resulted in colony sizes larger than cells expressing full-length San1 was scored 5. Growth similar to cells expressing full-length was scored 4. Growth resulting in smaller colony sizes or reduced 10 fold in spotting was scored 3. Growth reduced 102-103 fold was scored 2. Growth reduced ≥104 fold was scored 1. For the dendogram in Figure 6, an agglomerative hierarchical clustering was computed for the interactions on -his+3AT media using Euclidean distance to quantify the dissimilarity of substrates and Ward linkage to quantify the dissimilarity of clusters. The hierarchical clustering package of Mathematica (Version 7.0, Wolfram Research Inc) was used to generate the dendrogram.
Mass spectrometry
Independent 3HSV-San1C279S or untagged San1C279S cultures were grown to ~1×107 cells/ml. Formaldehyde was added to 1% (v/v) and cells incubated for 2 minutes. Glycine was added to 125mM to quench crosslinking. Cells were lysed in SUME (8M urea, 1% SDS, 10mM MOPS, pH6.8, 10mM EDTA). Lysates were diluted 1:5 in IP buffer (15mM Na2HPO4, 150mM NaCl, 2% Triton X-100, 0.1% SDS, 0.5% deoxycholate, 10mM EDTA) and incubated for 16 hours at 4°C with 1:1000 mouse anti-HSV antibody (Novagen) bound to 1.25mg/ml Protein A Dynabeads (Invitrogen). Beads were washed 3x in IP buffer and 2x in IP wash buffer (50mM NaCl, 10mM Tris-Cl, pH7.5). Proteins were eluted by incubation at 65°C for 10 minutes in SUMEB (SUME + 0.01% bromophenol blue). Samples were run 1cm into an 8-16% SDS-PAGE gel and gel slices excised. Proteins in the slices were trypsin digested and peptides identified by tandem MS using an Orbitrap mass spectrometer (Fred Hutchinson Cancer Research Center Proteomics facility). We used Peptide Prophet (Keller et al., 2002) to evaluate the validity of peptide identifications eliminating peptides with a score <0.85 (<2.5% false discovery rate).
In vitro ubiquitination
0.27μM luciferase in 50mM Tris-HCl (pH7.5) was denatured by incubating at 23°C for 10 minutes in 4M urea. 15μl reactions containing 0.08μg ubiquitin activase (Uba1), 0.1μg ubiquitin conjugase (UbcH5a), 0.76μg San1, 2.5μg ubiquitin, 2mM Mg-ATP, 50mM Tris-HCl (pH7.5), 2.5mM MgCl2, and 0.9μg denatured or native luciferase were incubated at 30°C for 60 minutes. For heat denaturation, the reaction was incubated at 42°C. 15μl of SUMEB was added to each sample and incubated at 65°C for 10 minutes to stop the reaction.
In coli ubiquitination
Components of the San1 ubiquitination cascade were cloned behind T7lac promoters in Duet expression vectors (Novagen) that were maintained in T7 Express competent E. coli (New England Biolabs) with appropriate antibiotic selection. Cells were grown at 37°C to OD600 ~1.0 and induced with 800μM IPTG for 6 hours at 30°C. Cells were lysed in Bugbuster (Novagen) with 10mM PMSF. To immunoprecipitate the 2xHA-tagged substrates, soluble extract was incubated for 16 hours at 4°C with 1:2000 mouse anti-HA antibodies (Sigma) conjugated to 0.75mg/ml Protein A Dynabeads (Invitrogen) in IP buffer. Beads were washed 3x in IP buffer then incubated at 65°C for 10 minutes in SUMEB.
Expression and purification of San1
StrepII-San1-3HSV-8His was cloned into pACYCDuet and transformed into BL-21 cells. Cells were grown in LB + chloramphenicol to an OD600 ~1.0 and induced with 600μM IPTG overnight at 16°C. Cells were harvested and lysed in BugBuster with PMSF, aprotinin, leupeptin, and pepstatin A. Soluble extract was applied to TALON resin (Clontech) equilibrated in a 300mM NaCl, 50mM Na2HPO4 pH8.0 buffer and eluted with 1M imidazole added to the buffer. The elution fraction was applied to Strep-Tactin resin (Novagen) equilibrated with a 150mM NaCl, 100mM Tris-HCl, 1mM EDTA, pH8.0 buffer. San1 was eluted with 2.5mM desthiobiotin and concentrated using an Amicon Ultracel concentrator (cut-off 30 kDa). Purified San1 was centrifuged at 100,000×g for 45 minutes to eliminate aggregates prior to gel filtration.
Limited Proteolysis
Proteolysis of San1 and BSA was carried out by trypsin and thermolysin digestion at 25°C in digestion buffer (50mM NaCl and 15mM Na2HPO4, pH7.3). The enzyme to substrate ratio was 1:50. Proteolysis was stopped by addition of SUMEB and boiling for 10 minutes.
Circular dichroism spectroscopy
Far-UV CD spectra of purified San1 or BSA were obtained on an Aviv 62DS CD spectrometer. Spectra were recorded at 25°C with a 1mm optical cuvette from 260 to 190 nm. An average of three runs was obtained by sampling every 2nm with a 30 second averaging time. Each sample was in 50mM NaCl and 15mM Na2HPO4, pH7.3 and the protein concentrations were between 5 and 10μM. We subtracted the background buffer spectrum from each protein spectrum.
Degradation Assays
Cycloheximide-chase degradation assays were performed similar to previously described (Gardner et al., 2005). Cells were grown at 23°C or 30°C in media with 3% raffinose to ~1×107 cells/ml. Galactose was added to 3% and the cells incubated 3-5 hours at 23°C or 30°C. Cycloheximide was added to 50μg/ml and the cells further incubated at 30°C or 37°C for 0-3 hours. Cells were lysed at the appropriate time point in 200μl SUMEB.
Supplementary Material
Acknowledgements
We thank Laura Sheard and Ning Zheng for help with protein purification, and Fabio Parmeggiani and David Baker for help with CD analyses. R.G.G. thanks Larry G. for the years of devoted thumbs-up support. This work was supported by NIH/NIGMS grant R01GM043893 (D.E.G.), NIH/NIA grant R01AG031136 (R.G.G.), NIH/NCRR grant R21RR025787 (R.G.G), an Ellison Medical Foundation New Scholar Award in Aging (R.G.G), and a Marian E. Smith Junior Faculty Award (R.G.G).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bhattacharyya J, Das KP. Molecular chaperone-like properties of an unfolded protein, alpha(s)-casein. J Biol Chem. 1999;274:15505–15509. doi: 10.1074/jbc.274.22.15505. [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta A, Ramsey KL, Smith JS, Auble DT. Sir antagonist 1 (San1) is a ubiquitin ligase. J Biol Chem. 2004;279:26830–26838. doi: 10.1074/jbc.M400894200. [DOI] [PubMed] [Google Scholar]
- Denic V, Quan EM, Weissman JS. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 2006;126:349–359. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- Dosztanyi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- Dosztanyi Z, Meszaros B, Simon I. ANCHOR: web server for predicting protein binding regions in disordered proteins. Bioinformatics. 2009;25:2745–2746. doi: 10.1093/bioinformatics/btp518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker AK, Oldfield CJ, Meng J, Romero P, Yang JY, Chen JW, Vacic V, Obradovic Z, Uversky VN. The unfoldomics decade: an update on intrinsically disordered proteins. BMC Genomics. 2008;9(Suppl 2):S1. doi: 10.1186/1471-2164-9-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana A, de Laureto PP, Spolaore B, Frare E, Picotti P, Zambonin M. Probing protein structure by limited proteolysis. Acta Biochim Pol. 2004;51:299–321. [PubMed] [Google Scholar]
- Fu L, Gao YS, Tousson A, Shah A, Chen TL, Vertel BM, Sztul E. Nuclear aggresomes form by fusion of PML-associated aggregates. Mol Biol Cell. 2005;16:4905–4917. doi: 10.1091/mbc.E05-01-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RG, Nelson ZW, Gottschling DE. Degradation-mediated protein quality control in the nucleus. Cell. 2005;120:803–815. doi: 10.1016/j.cell.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Gardner RG, Swarbrick GM, Bays NW, Cronin SR, Wilhovsky S, Seelig L, Kim C, Hampton RY. Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control of Hrd1p by Hrd3p. J Cell Biol. 2000;151:69–82. doi: 10.1083/jcb.151.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauss R, Jarosch E, Sommer T, Hirsch C. A complex of Yos9p and the HRD ligase integrates endoplasmic reticulum quality control into the degradation machinery. Nat Cell Biol. 2006a;8:849–854. doi: 10.1038/ncb1445. [DOI] [PubMed] [Google Scholar]
- Gauss R, Sommer T, Jarosch E. The Hrd1p ligase complex forms a linchpin between ER-lumenal substrate selection and Cdc48p recruitment. Embo J. 2006b;25:1827–1835. doi: 10.1038/sj.emboj.7601088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield NJ. Using circular dichroism spectra to estimate protein secondary structure. Nat Protoc. 2006;1:2876–2890. doi: 10.1038/nprot.2006.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslbeck M, Ignatiou A, Saibil H, Helmich S, Frenzl E, Stromer T, Buchner J. A domain in the N-terminal part of Hsp26 is essential for chaperone function and oligomerization. J Mol Biol. 2004;343:445–455. doi: 10.1016/j.jmb.2004.08.048. [DOI] [PubMed] [Google Scholar]
- Heck JW, Cheung SK, Hampton RY. Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc Natl Acad Sci U S A. 2010;107:1106–1111. doi: 10.1073/pnas.0910591107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata A, Nagashima Y, Matsumoto L, Suzuki T, Yamanaka T, Date H, Deoka K, Nukina N, Tsuji S. Intra-nuclear degradation of polyglutamine aggregates by the ubiquitin proteasome system. J Biol Chem. 2009;284:9796–9803. doi: 10.1074/jbc.M809739200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janer A, Martin E, Muriel MP, Latouche M, Fujigasaki H, Ruberg M, Brice A, Trottier Y, Sittler A. PML clastosomes prevent nuclear accumulation of mutant ataxin-7 and other polyglutamine proteins. J Cell Biol. 2006;174:65–76. doi: 10.1083/jcb.200511045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaya N, Garcia V, Vierling E. Substrate binding site flexibility of the small heat shock protein molecular chaperones. Proc Natl Acad Sci U S A. 2009;106:15604–15609. doi: 10.1073/pnas.0902177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- Kim TD, Paik SR, Yang CH. Structural and functional implications of C-terminal regions of alpha-synuclein. Biochemistry. 2002;41:13782–13790. doi: 10.1021/bi026284c. [DOI] [PubMed] [Google Scholar]
- Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonough H, Patterson C. CHIP: a link between the chaperone and proteasome systems. Cell Stress Chaperones. 2003;8:303–308. doi: 10.1379/1466-1268(2003)008<0303:calbtc>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mencia M, de Lorenzo V. Functional transplantation of the sumoylation machinery into Escherichia coli. Protein Expr Purif. 2004;37:409–418. doi: 10.1016/j.pep.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Metzger MB, Maurer MJ, Dancy BM, Michaelis S. Degradation of a cytosolic protein requires endoplasmic reticulum-associated degradation machinery. J Biol Chem. 2008;283:32302–32316. doi: 10.1074/jbc.M806424200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nillegoda NB, Theodoraki MA, Mandal AK, Mayo KJ, Ren HY, Sultana R, Wu K, Johnson J, Cyr DM, Caplan AJ. Ubr1 and Ubr2 Function in a Quality Control Pathway for Degradation of Unfolded Cytosolic Proteins. Mol Biol Cell. 2010;21:2102–2016. doi: 10.1091/mbc.E10-02-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada S, Nagabuchi M, Takamura Y, Nakagawa T, Shinmyozu K, Nakayama J, Tanaka K. Reconstitution of Arabidopsis thaliana SUMO pathways in E. coli: functional evaluation of SUMO machinery proteins and mapping of SUMOylation sites by mass spectrometry. Plant Cell Physiol. 2009;50:1049–1061. doi: 10.1093/pcp/pcp056. [DOI] [PubMed] [Google Scholar]
- Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, Silman I, Sussman JL. FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics. 2005;21:3435–3438. doi: 10.1093/bioinformatics/bti537. [DOI] [PubMed] [Google Scholar]
- Receveur-Brechot V, Bourhis JM, Uversky VN, Canard B, Longhi S. Assessing protein disorder and induced folding. Proteins. 2006;62:24–45. doi: 10.1002/prot.20750. [DOI] [PubMed] [Google Scholar]
- Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Sato BK, Schulz D, Do PH, Hampton RY. Misfolded membrane proteins are specifically recognized by the transmembrane domain of the Hrd1p ubiquitin ligase. Mol Cell. 2009;34:212–222. doi: 10.1016/j.molcel.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skovronsky DM, Lee VM, Trojanowski JQ. Neurodegenerative diseases: new concepts of pathogenesis and their therapeutic implications. Annu Rev Pathol. 2006;1:151–170. doi: 10.1146/annurev.pathol.1.110304.100113. [DOI] [PubMed] [Google Scholar]
- Stromer T, Fischer E, Richter K, Haslbeck M, Buchner J. Analysis of the regulation of the molecular chaperone Hsp26 by temperature-induced dissociation: the N-terminal domail is important for oligomer assembly and the binding of unfolding proteins. J Biol Chem. 2004;279:11222–11228. doi: 10.1074/jbc.M310149200. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002;27:527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- Uchimura Y, Nakao M, Saitoh H. Generation of SUMO-1 modified proteins in E. coli: towards understanding the biochemistry/structural biology of the SUMO-1 pathway. FEBS Lett. 2004;564:85–90. doi: 10.1016/S0014-5793(04)00321-7. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- van Montfort RL, Basha E, Friedrich KL, Slingsby C, Vierling E. Crystal structure and assembly of a eukaryotic small heat shock protein. Nat Struct Biol. 2001;8:1025–1030. doi: 10.1038/nsb722. [DOI] [PubMed] [Google Scholar]
- Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh R, Storey E, Stefani D, Kelly L, Turnbull V. The roles of proteolysis and nuclear localisation in the toxicity of the polyglutamine diseases. A review. Neurotox Res. 2005;7:43–57. doi: 10.1007/BF03033775. [DOI] [PubMed] [Google Scholar]
- Wang Y, Garg S, Mandelkow EM, Mandelkow E. Proteolytic processing of tau. Biochem Soc Trans. 2010;38:955–961. doi: 10.1042/BST0380955. [DOI] [PubMed] [Google Scholar]
- Woulfe JM. Abnormalities of the nucleus and nuclear inclusions in neurodegenerative disease: a work in progress. Neuropathol Appl Neurobiol. 2007;33:2–42. doi: 10.1111/j.1365-2990.2006.00819.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.